INTRODUCTION Trehalose-6-phosphate synthase (TPS) is a key enzyme involved in the synthesis of trehalose and has been reported to participate in regulating photosynthesis, carbohydrate metabolism, growth and development, and stress responses in various species. Currently, reports on TPS genes in soybean are scarce.
RATIONALE TPS is a stable non-reducing disaccharide, whose synthesis, decomposition and regulation not only provide energy for plant, but also play an important role in plant growth and development and stress tolerance. The in-depth study of soybean TPS genes and its relationships with salt stress is of great significance in elucidating the molecular mechanism of soybean salt tolerance and improving soybean yield.
RESULTS This study identified 20 soybean TPS genes and their associated 10 conserved protein motifs in the soybean genome. Molecular analysis of the promoter elements revealed that the TPS gene promoters are rich in stress-responsive elements. After salt stress treatment, the expression of 17 TPS genes changed, with 12 genes up-regulated and 5 genes down-regulated. Haplotype and selection analyses revealed two major allelic variations in TPS8, TPS13, TPS15, TPS17, and TPS18. Notably, variants carrying TPS15H2, TPS13H2, TPS17H2, and TPS18H2 were significantly enriched in improved cultivars that underwent strong artificial selection.
CONCLUSION This study reveals the molecular characteristics of the soybean TPS gene family, their expression patterns under salt stress, and their evolutionary history, providing a theoretical basis and genetic material for further elucidating the functions of soybean TPS genes and breeding salt-tolerant soybean varieties.
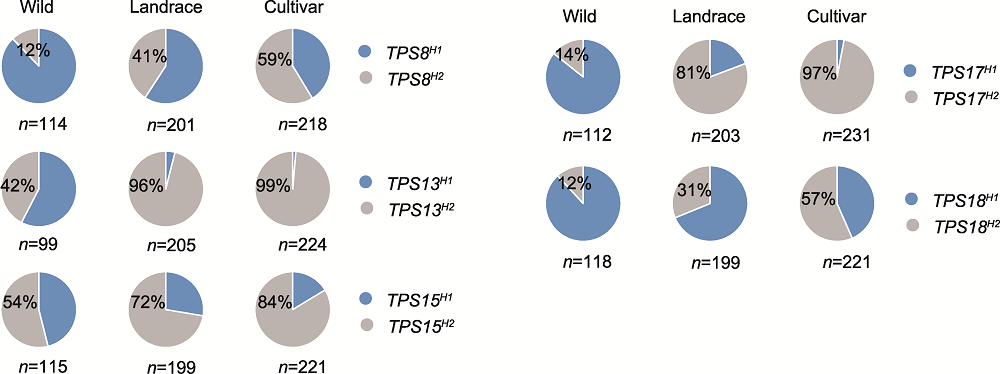
TPS genes were subjected to intense artificial selection. The natural variations of TPS8, TPS13, TPS15, TPS17, and TPS18 have been subjected to strong artificial selection during soybean domestication and improvement, with the variants carrying TPS15H2, TPS13H2, TPS17H2, and TPS18H2 being heavily enriched in improved cultivars.

