

Chinese Bulletin of Botany ›› 2024, Vol. 59 ›› Issue (5): 738-751.DOI: 10.11983/CBB24065 cstr: 32102.14.CBB24065
Special Issue: 大食物观
• EXPERIMENTAL COMMUNICATIONS • Previous Articles Next Articles
Jinjin Lian1,†, Luyao Tang1,†, Yinuo Zhang1, Jiaxing Zheng1, Chaoyu Zhu1, Yuhan Ye1, Yuexing Wang2, Wennan Shang3, Zhenghao Fu1, Xinxuan Xu1, Richeng Wu1, Mei Lu1, Changchun Wang1,*( ), Yuchun Rao1,*(
), Yuchun Rao1,*( )
)
Received:2024-04-30
Accepted:2024-06-21
Online:2024-09-10
Published:2024-08-19
Contact:
Changchun Wang, Yuchun Rao
About author:First author contact: These authors contributed equally to this paper
Jinjin Lian, Luyao Tang, Yinuo Zhang, Jiaxing Zheng, Chaoyu Zhu, Yuhan Ye, Yuexing Wang, Wennan Shang, Zhenghao Fu, Xinxuan Xu, Richeng Wu, Mei Lu, Changchun Wang, Yuchun Rao. Genetic Locus Mining and Candidate Gene Analysis of Antioxidant Traits in Rice[J]. Chinese Bulletin of Botany, 2024, 59(5): 738-751.
| Physiological traits | Gene name | Forward primer (5'-3') | Reverse primer (5′-3′) |
|---|---|---|---|
| OsActin | TGGCATCTCAGCACATTCC | TGCACAATGGATGGGTCAGA | |
| Hydroxyl radical scavenging rate | LOC_Os06g01850 | ACATGGTCTTCAGCACCGAG | CTTTGCCTACTGGTCCGGTT |
| LOC_Os06g48000 | TTCAACCTCGACCAGCTCAC | GGAGGTTCTGGTAGAAGGCG | |
| LOC_Os06g48020 | GTTGTGACGCGTCGATCATG | CCGCTCTGGTAGACGGATTC | |
| Total phenolic content | LOC_Os04g45210 | AGCTGGTCAAGACGTTCCAG | GAAGTAGACCATCTCGCCGG |
| LOC_Os12g07820 | GTTCGGTTAGGGTGGCATGA | CATTGTTCAGGGGCAGCAAC | |
| LOC_Os12g07830 | CCTCTCCCCTCCCTTCTCTC | GTCATGCCACCCTAACCGAA | |
| Flavonoid content | LOC_Os05g12190 | GACCTCAAGCTGGCATCACT | CCCAAACAGTCCATGACCGA |
| LOC_Os05g12210 | ATGCCCTCCCTTGAAACTCG | TCACGGTTGTTCTCGGTGAG | |
| LOC_Os05g12240 | GGCCTCAACCCTTCTGTCTC | AGGACCATCTCAAACAGCGG | |
| LOC_Os05g45120 | CTCATGGTGGAGGACATGGG | CAACAGGAACTCGGCAAACG | |
| LOC_Os12g16260 | TGATACGCAACTTCTACACTGCT | AGATGGCTCAGGTTGGTTGG | |
| Anthocyanin content | LOC_Os03g60509 | TGTGTTCCGTGTCAGTGGAG | CAGGATCATCGTCACCCTCG |
| LOC_Os03g62480 | CCATCCCCGCCTACTTCTTC | ATCGAAGCTGTTCACGAGCA |
Table 1 Primer sequences for qRT-PCR
| Physiological traits | Gene name | Forward primer (5'-3') | Reverse primer (5′-3′) |
|---|---|---|---|
| OsActin | TGGCATCTCAGCACATTCC | TGCACAATGGATGGGTCAGA | |
| Hydroxyl radical scavenging rate | LOC_Os06g01850 | ACATGGTCTTCAGCACCGAG | CTTTGCCTACTGGTCCGGTT |
| LOC_Os06g48000 | TTCAACCTCGACCAGCTCAC | GGAGGTTCTGGTAGAAGGCG | |
| LOC_Os06g48020 | GTTGTGACGCGTCGATCATG | CCGCTCTGGTAGACGGATTC | |
| Total phenolic content | LOC_Os04g45210 | AGCTGGTCAAGACGTTCCAG | GAAGTAGACCATCTCGCCGG |
| LOC_Os12g07820 | GTTCGGTTAGGGTGGCATGA | CATTGTTCAGGGGCAGCAAC | |
| LOC_Os12g07830 | CCTCTCCCCTCCCTTCTCTC | GTCATGCCACCCTAACCGAA | |
| Flavonoid content | LOC_Os05g12190 | GACCTCAAGCTGGCATCACT | CCCAAACAGTCCATGACCGA |
| LOC_Os05g12210 | ATGCCCTCCCTTGAAACTCG | TCACGGTTGTTCTCGGTGAG | |
| LOC_Os05g12240 | GGCCTCAACCCTTCTGTCTC | AGGACCATCTCAAACAGCGG | |
| LOC_Os05g45120 | CTCATGGTGGAGGACATGGG | CAACAGGAACTCGGCAAACG | |
| LOC_Os12g16260 | TGATACGCAACTTCTACACTGCT | AGATGGCTCAGGTTGGTTGG | |
| Anthocyanin content | LOC_Os03g60509 | TGTGTTCCGTGTCAGTGGAG | CAGGATCATCGTCACCCTCG |
| LOC_Os03g62480 | CCATCCCCGCCTACTTCTTC | ATCGAAGCTGTTCACGAGCA |
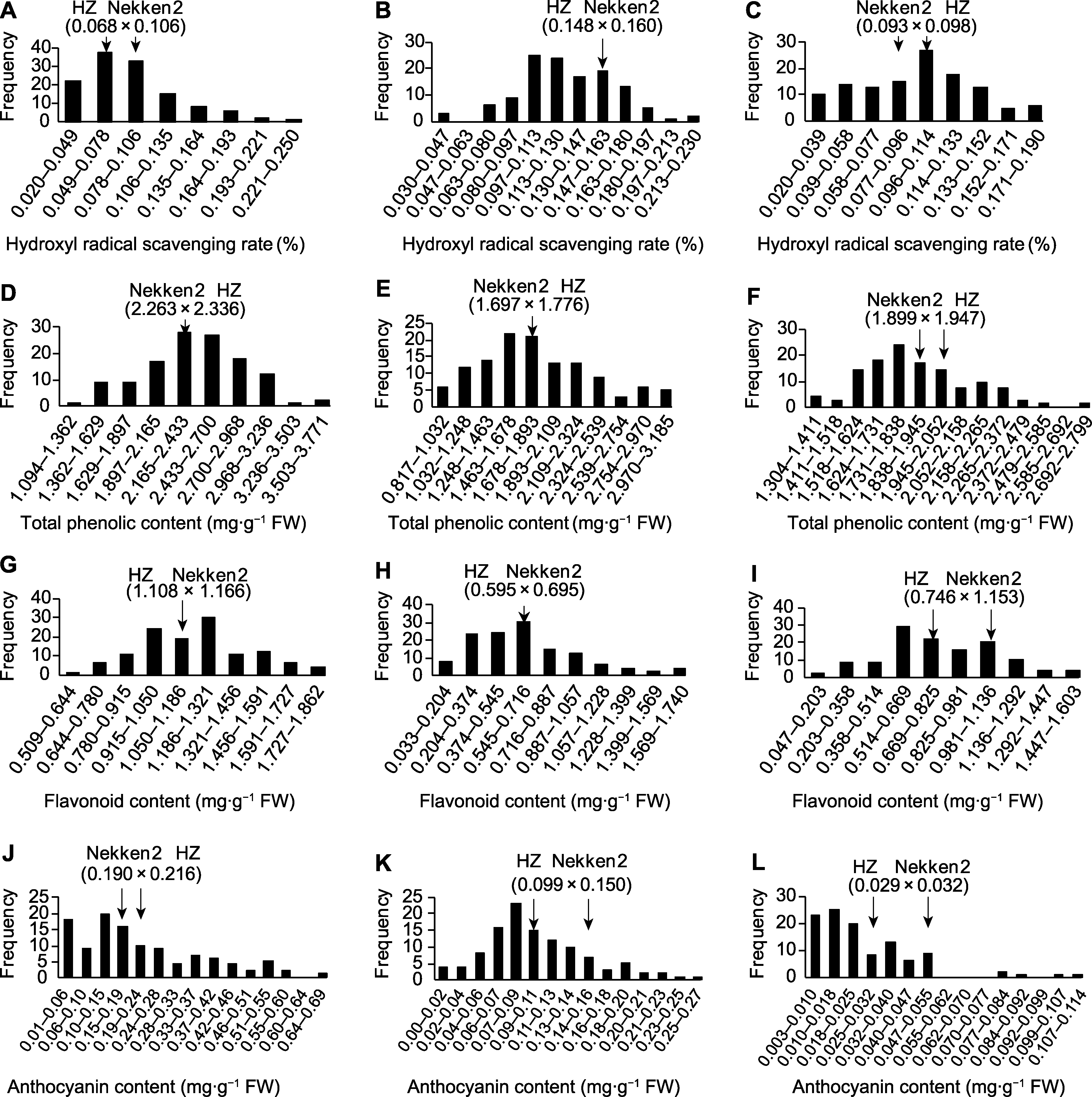
Figure 2 Distribution of antioxidant traits in the recombinant inbred lines of rice (A) Hydroxyl radical scavenging rate at the tillering stage; (B) Hydroxyl radical scavenging rate at the grain filling stage; (C) Hydroxyl radical scavenging rate at the maturity stage; (D) Total phenolic content at the tillering stage; (E) Total phenolic content at the grain filling stage; (F) Total phenolic content at the maturity stage; (G) Flavonoid content at the tillering stage; (H) Flavonoid content at the grain filling stage; (I) Flavonoid content at the maturity stage; (J) Anthocyanin content at the tillering stage; (K) Anthocyanin content at the grain filling stage; (L) Anthocyanin content at the maturity stage
| Physiological traits | QTL | Chromosome | Physical distance (bp) | Position of support (cM) | Limit of detection |
|---|---|---|---|---|---|
| Hydroxyl radical scavenging rate | qHRSR1.1 | 1 | 2556386-2834929 | 10.96-12.15 | 2.14 |
| qHRSR2.1 | 2 | 3467582-4955784 | 14.87-21.24 | 4.36 | |
| qHRSR2.2 | 2 | 5098574-6084984 | 21.86-26.08 | 3.37 | |
| qHRSR2.3 | 2 | 27009300-27239796 | 115.78-116.77 | 2.75 | |
| qHRSR2.4 | 2 | 28427871-28664694 | 121.86-122.88 | 2.21 | |
| qHRSR3.1 | 3 | 12723031-12925845 | 54.54-55.41 | 2.22 | |
| qHRSR3.2 | 3 | 29949770-30321190 | 128.39-129.98 | 2.31 | |
| qHRSR4.1 | 4 | 23316226-23991142 | 99.95-102.84 | 3.35 | |
| qHRSR4.2 | 4 | 23680403-239113 17 | 101.51-102.50 | 2.00 | |
| qHRSR4.3 | 4 | 24725621-25022230 | 105.99-107.26 | 2.29 | |
| qHRSR5.1 | 5 | 26927610-27082669 | 115.43-116.10 | 2.30 | |
| qHRSR6.1 | 6 | 33441-781344 | 0.14-3.35 | 2.50 | |
| qHRSR6.2 | 6 | 26490081-27333693 | 113.56-117.17 | 2.24 | |
| qHRSR6.3 | 6 | 28674720-29130039 | 123.31-124.87 | 2.62 | |
| qHRSR8.1 | 8 | 1092742-2649021 | 4.68-11.36 | 2.97 | |
| qHRSR8.2 | 8 | 11086291-11153903 | 47.52-47.81 | 2.22 | |
| qHRSR8.3 | 8 | 14983464-15034055 | 64.23-64.45 | 2.36 | |
| qHRSR8.4 | 8 | 21388530-22202508 | 91.69-95.18 | 2.44 | |
| Total phenolic content | qTPC1.1 | 1 | 7667525-8235815 | 32.87-35.30 | 2.23 |
| qTPC3.1 | 3 | 15484683-16021103 | 66.38-68.68 | 3.07 | |
| qTPC3.2 | 3 | 16899768-17721978 | 72.45-75.97 | 3.58 | |
| qTPC3.3 | 3 | 25032115-25648379 | 107.31-109.95 | 2.50 | |
| qTPC4.1 | 4 | 26377931-27479589 | 113.07-117.80 | 2.44 | |
| qTPC12.1 | 12 | 2689107-3043767 | 11.53-13.05 | 2.52 | |
| qTPC12.2 | 12 | 3933801-4462301 | 16.86-19.13 | 2.08 | |
| qTPC12.3 | 12 | 11398960-11895586 | 48.86-50.99 | 2.00 | |
| qTPC12.4 | 12 | 13100229-14071703 | 56.16-60.32 | 2.00 | |
| qTPC12.5 | 12 | 14215593-15112427 | 60.94-64.78 | 2.11 | |
| qTPC12.6 | 12 | 16682032-1680484 | 71.51-72.19 | 2.47 | |
| Flavonoid content | qFC1.1 | 1 | 26148806-27178896 | 112.09-116.50 | 2.42 |
| qFC1.2 | 1 | 27178896-27631674 | 116.50-118.45 | 2.03 | |
| qFC1.3 | 1 | 37113012-37638053 | 159.09-161.34 | 2.12 | |
| qFC1.4 | 1 | 38157798-38395992 | 163.57-164.59 | 2.35 | |
| qFC2.1 | 2 | 21915215-22231364 | 93.94-95.30 | 2.30 | |
| qFC4.1 | 4 | 17472715-19028032 | 74.90-81.57 | 3.67 | |
| qFC4.2 | 4 | 19642144-19986484 | 84.20-85.68 | 2.70 | |
| qFC4.3 | 4 | 22019563-23235126 | 94.39-99.60 | 3.74 | |
| qFC4.4 | 4 | 26377931-26619819 | 113.07-114.11 | 2.87 | |
| qFC5.1 | 5 | 5206830-6439595 | 22.32-27.60 | 4.03 | |
| qFC5.2 | 5 | 6673100-7059585 | 28.61-30.26 | 4.12 | |
| qFC5.3 | 5 | 12422042-12794190 | 53.25-54.85 | 2.04 | |
| qFC5.4 | 5 | 26189817-26927610 | 112.27-115.43 | 2.59 | |
| qFC6.1 | 6 | 5187517-6015262 | 22.24-25.79 | 3.28 | |
| qFC6.2 | 6 | 6069227-6560451 | 26.02-28.12 | 2.68 | |
| qFC6.3 | 6 | 21068005-21507916 | 90.31-92.20 | 2.16 | |
| qFC6.4 | 6 | 27961504-28384769 | 119.86-121.68 | 2.51 | |
| qFC9.1 | 9 | 13119644-13620974 | 56.24-58.39 | 2.07 | |
| qFC10.1 | 10 | 21806620-22080253 | 93.48-94.65 | 2.02 | |
| qFC12.1 | 12 | 2351235-2689107 | 10.08-11.53 | 2.56 | |
| qFC12.2 | 12 | 4462301-6224960 | 19.13-26.68 | 2.95 | |
| qFC12.3 | 12 | 7508660-9990803 | 32.19-42.83 | 3.74 | |
| qFC12.4 | 12 | 11398960-14215593 | 48.86-60.94 | 3.04 | |
| qFC12.5 | 12 | 17056555-17574928 | 73.12-75.34 | 2.19 | |
| qFC12.6 | 12 | 23839134-24088353 | 102.19-103.26 | 2.39 | |
| Anthocyanin content | qAC2.1 | 2 | 35599368-35902152 | 152.60-153.90 | 2.42 |
| qAC3.1 | 3 | 31965505-32493480 | 137.03-138.32 | 2.25 | |
| qAC3.2 | 3 | 33386000-34509821 | 143.12-147.93 | 3.23 | |
| qAC3.3 | 3 | 35314343-35646346 | 151.38-152.81 | 2.30 | |
| qAC5.1 | 5 | 22147125-22524107 | 94.94-96.55 | 2.88 | |
| qAC11.1 | 11 | 9832804-10200412 | 42.15-43.73 | 2.28 | |
| qAC12.1 | 12 | 927518-2689107 | 3.98-11.53 | 2.31 | |
| qAC12.2 | 12 | 23013674-23469132 | 98.65-100.61 | 2.50 |
Table 2 QTL analysis of physiological indicators of antioxidant properties in the recombinant inbred line population of rice
| Physiological traits | QTL | Chromosome | Physical distance (bp) | Position of support (cM) | Limit of detection |
|---|---|---|---|---|---|
| Hydroxyl radical scavenging rate | qHRSR1.1 | 1 | 2556386-2834929 | 10.96-12.15 | 2.14 |
| qHRSR2.1 | 2 | 3467582-4955784 | 14.87-21.24 | 4.36 | |
| qHRSR2.2 | 2 | 5098574-6084984 | 21.86-26.08 | 3.37 | |
| qHRSR2.3 | 2 | 27009300-27239796 | 115.78-116.77 | 2.75 | |
| qHRSR2.4 | 2 | 28427871-28664694 | 121.86-122.88 | 2.21 | |
| qHRSR3.1 | 3 | 12723031-12925845 | 54.54-55.41 | 2.22 | |
| qHRSR3.2 | 3 | 29949770-30321190 | 128.39-129.98 | 2.31 | |
| qHRSR4.1 | 4 | 23316226-23991142 | 99.95-102.84 | 3.35 | |
| qHRSR4.2 | 4 | 23680403-239113 17 | 101.51-102.50 | 2.00 | |
| qHRSR4.3 | 4 | 24725621-25022230 | 105.99-107.26 | 2.29 | |
| qHRSR5.1 | 5 | 26927610-27082669 | 115.43-116.10 | 2.30 | |
| qHRSR6.1 | 6 | 33441-781344 | 0.14-3.35 | 2.50 | |
| qHRSR6.2 | 6 | 26490081-27333693 | 113.56-117.17 | 2.24 | |
| qHRSR6.3 | 6 | 28674720-29130039 | 123.31-124.87 | 2.62 | |
| qHRSR8.1 | 8 | 1092742-2649021 | 4.68-11.36 | 2.97 | |
| qHRSR8.2 | 8 | 11086291-11153903 | 47.52-47.81 | 2.22 | |
| qHRSR8.3 | 8 | 14983464-15034055 | 64.23-64.45 | 2.36 | |
| qHRSR8.4 | 8 | 21388530-22202508 | 91.69-95.18 | 2.44 | |
| Total phenolic content | qTPC1.1 | 1 | 7667525-8235815 | 32.87-35.30 | 2.23 |
| qTPC3.1 | 3 | 15484683-16021103 | 66.38-68.68 | 3.07 | |
| qTPC3.2 | 3 | 16899768-17721978 | 72.45-75.97 | 3.58 | |
| qTPC3.3 | 3 | 25032115-25648379 | 107.31-109.95 | 2.50 | |
| qTPC4.1 | 4 | 26377931-27479589 | 113.07-117.80 | 2.44 | |
| qTPC12.1 | 12 | 2689107-3043767 | 11.53-13.05 | 2.52 | |
| qTPC12.2 | 12 | 3933801-4462301 | 16.86-19.13 | 2.08 | |
| qTPC12.3 | 12 | 11398960-11895586 | 48.86-50.99 | 2.00 | |
| qTPC12.4 | 12 | 13100229-14071703 | 56.16-60.32 | 2.00 | |
| qTPC12.5 | 12 | 14215593-15112427 | 60.94-64.78 | 2.11 | |
| qTPC12.6 | 12 | 16682032-1680484 | 71.51-72.19 | 2.47 | |
| Flavonoid content | qFC1.1 | 1 | 26148806-27178896 | 112.09-116.50 | 2.42 |
| qFC1.2 | 1 | 27178896-27631674 | 116.50-118.45 | 2.03 | |
| qFC1.3 | 1 | 37113012-37638053 | 159.09-161.34 | 2.12 | |
| qFC1.4 | 1 | 38157798-38395992 | 163.57-164.59 | 2.35 | |
| qFC2.1 | 2 | 21915215-22231364 | 93.94-95.30 | 2.30 | |
| qFC4.1 | 4 | 17472715-19028032 | 74.90-81.57 | 3.67 | |
| qFC4.2 | 4 | 19642144-19986484 | 84.20-85.68 | 2.70 | |
| qFC4.3 | 4 | 22019563-23235126 | 94.39-99.60 | 3.74 | |
| qFC4.4 | 4 | 26377931-26619819 | 113.07-114.11 | 2.87 | |
| qFC5.1 | 5 | 5206830-6439595 | 22.32-27.60 | 4.03 | |
| qFC5.2 | 5 | 6673100-7059585 | 28.61-30.26 | 4.12 | |
| qFC5.3 | 5 | 12422042-12794190 | 53.25-54.85 | 2.04 | |
| qFC5.4 | 5 | 26189817-26927610 | 112.27-115.43 | 2.59 | |
| qFC6.1 | 6 | 5187517-6015262 | 22.24-25.79 | 3.28 | |
| qFC6.2 | 6 | 6069227-6560451 | 26.02-28.12 | 2.68 | |
| qFC6.3 | 6 | 21068005-21507916 | 90.31-92.20 | 2.16 | |
| qFC6.4 | 6 | 27961504-28384769 | 119.86-121.68 | 2.51 | |
| qFC9.1 | 9 | 13119644-13620974 | 56.24-58.39 | 2.07 | |
| qFC10.1 | 10 | 21806620-22080253 | 93.48-94.65 | 2.02 | |
| qFC12.1 | 12 | 2351235-2689107 | 10.08-11.53 | 2.56 | |
| qFC12.2 | 12 | 4462301-6224960 | 19.13-26.68 | 2.95 | |
| qFC12.3 | 12 | 7508660-9990803 | 32.19-42.83 | 3.74 | |
| qFC12.4 | 12 | 11398960-14215593 | 48.86-60.94 | 3.04 | |
| qFC12.5 | 12 | 17056555-17574928 | 73.12-75.34 | 2.19 | |
| qFC12.6 | 12 | 23839134-24088353 | 102.19-103.26 | 2.39 | |
| Anthocyanin content | qAC2.1 | 2 | 35599368-35902152 | 152.60-153.90 | 2.42 |
| qAC3.1 | 3 | 31965505-32493480 | 137.03-138.32 | 2.25 | |
| qAC3.2 | 3 | 33386000-34509821 | 143.12-147.93 | 3.23 | |
| qAC3.3 | 3 | 35314343-35646346 | 151.38-152.81 | 2.30 | |
| qAC5.1 | 5 | 22147125-22524107 | 94.94-96.55 | 2.88 | |
| qAC11.1 | 11 | 9832804-10200412 | 42.15-43.73 | 2.28 | |
| qAC12.1 | 12 | 927518-2689107 | 3.98-11.53 | 2.31 | |
| qAC12.2 | 12 | 23013674-23469132 | 98.65-100.61 | 2.50 |
| Antioxidant physiological indicators | Gene ID | QTL locus | Functional annotation | Gene name |
|---|---|---|---|---|
| Hydroxyl radical scavenging rate | LOC_Os06g01850 | qHRSR6.1 | Leaf-type ferredoxin-NADP+ oxidoreductase | OsLFNR2 (Da et al., |
| LOC_Os06g48000 | qHRSR6.3 | Peroxidase precursor | ||
| LOC_Os06g48020 | qHRSR6.3 | Peroxidase precursor | ||
| Total phenolic content | LOC_Os04g45210 | qTPC4.1 | Peroxisomal biogenesis factor 11 | |
| LOC_Os12g07820 | qTPC12.2 | Ascorbate peroxidase gene | OsAPx6 (Chou et al., | |
| LOC_Os12g07830 | qTPC12.2 | Ascorbate peroxidase gene | OsAPx5 (Hong et al., | |
| Flavonoid content | LOC_Os05g12190 | qFC5.2 | Chalcone synthase | |
| LOC_Os05g12210 | qFC5.2 | Chalcone synthase | ||
| LOC_Os05g12240 | qFC5.2 | Chalcone synthase | ||
| LOC_Os05g45120 | qFC5.4 | Anthocyanidin 5,3-O-glucosyltransferase | ||
| LOC_Os12g16260 | qFC12.3 | Isoflavone reductase | ||
| Anthocyanin content | LOC_Os03g60509 | qAC3.2 | Chalcone isomerase gene | OsCHI (Lam et al., |
| LOC_Os03g62480 | qAC3.3 | UV-B-responsive glycosyltransferase; flavonoid 7-O-glycosyltransferase |
Table 3 Functions of candidate genes related to physiological indicators of antioxidant properties in rice
| Antioxidant physiological indicators | Gene ID | QTL locus | Functional annotation | Gene name |
|---|---|---|---|---|
| Hydroxyl radical scavenging rate | LOC_Os06g01850 | qHRSR6.1 | Leaf-type ferredoxin-NADP+ oxidoreductase | OsLFNR2 (Da et al., |
| LOC_Os06g48000 | qHRSR6.3 | Peroxidase precursor | ||
| LOC_Os06g48020 | qHRSR6.3 | Peroxidase precursor | ||
| Total phenolic content | LOC_Os04g45210 | qTPC4.1 | Peroxisomal biogenesis factor 11 | |
| LOC_Os12g07820 | qTPC12.2 | Ascorbate peroxidase gene | OsAPx6 (Chou et al., | |
| LOC_Os12g07830 | qTPC12.2 | Ascorbate peroxidase gene | OsAPx5 (Hong et al., | |
| Flavonoid content | LOC_Os05g12190 | qFC5.2 | Chalcone synthase | |
| LOC_Os05g12210 | qFC5.2 | Chalcone synthase | ||
| LOC_Os05g12240 | qFC5.2 | Chalcone synthase | ||
| LOC_Os05g45120 | qFC5.4 | Anthocyanidin 5,3-O-glucosyltransferase | ||
| LOC_Os12g16260 | qFC12.3 | Isoflavone reductase | ||
| Anthocyanin content | LOC_Os03g60509 | qAC3.2 | Chalcone isomerase gene | OsCHI (Lam et al., |
| LOC_Os03g62480 | qAC3.3 | UV-B-responsive glycosyltransferase; flavonoid 7-O-glycosyltransferase |

Figure 4 Differences in the expression of candidate genes for physiological indicators of antioxidant properties in both parents (A) Tillering stage; (B) Grain filling stage; (C) Maturity stage. *P<0.05; ** P<0.01
| [1] | Abdel-Aal ESM, Hucl P (1999). A rapid method for quantifying total anthocyanins in blue aleurone and purple pericarp wheats. Cereal Chem 76, 350-354. |
| [2] | Cai YZ, Luo Q, Sun M, Corke H (2004). Antioxidant activity and phenolic compounds of 112 traditional Chinese medicinal plants associated with anticancer. Life Sci 74, 2157-2184. |
| [3] | Chen MH, Pinson SRM, Jackson AK, Edwards JD (2023). Genetic loci regulating the concentrations of anthocyanins and proanthocyanidins in the pericarps of purple and red rice. Plant Genome 16, e20338. |
| [4] | Chou TS, Chao YY, Kao CH (2012). Involvement of hydrogen peroxide in heat shock- and cadmium-induced expression of ascorbate peroxidase and glutathione reductase in leaves of rice seedlings. J Plant Physiol 169, 478-486. |
| [5] | Da XW, Guo JF, Yan P, Yang C, Zhao HF, Li W, Kong YZ, Jiang RR, He Y, Xu JM, Xu OY, Mao CZ, Mo XR (2023). Characterizing membrane anchoring of leaf-form ferredoxin-NADP+oxidoreductase in rice. Plant Cell Environ 46, 1195-1206. |
| [6] | Dai WD, Tie NY, Ma LY, Zhang C, Tang R, Wang SM, Yang B, Wang LL, Rao Y, Huang ZS (2021). QTL mapping of leaf anthocyanin and chlorophyll content in Brassica napus. Southwest China J Agric Sci 34, 2547-2556. (in Chinese) |
| 代文东, 铁拿优, 玛丽亚, 张超, 唐容, 王少铭, 杨斌, 王璐璐, 饶勇, 黄泽素 (2021). 甘蓝型油菜叶片花青素及叶绿素含量的QTL定位. 西南农业学报 34, 2547-2556. | |
| [7] | Fang CX, Li LL, Zhang PL, Wang DH, Yang LK, Reza BM, Lin WX (2019). Lsi1 modulates the antioxidant capacity of rice and protects against ultraviolet-B radiation. Plant Sci 278, 96-106. |
| [8] | Higuchi-Takeuchi M, Ichikawa T, Kondou Y, Matsui K, Hasegawa Y, Kawashima M, Sonoike K, Mori M, Hirochika, Matsui M (2011). Functional analysis of two isoforms of leaf-type ferredoxin-NADP+-oxidoreductase in rice using the heterologous expression system of Arabidopsis. Plant Physiol 157, 96-108. |
| [9] | Hong CY, Hsu YT, Tsai YC, Kao CH (2007). Expression of ASCORBATE PEROXIDASE 8 in roots of rice (Oryza sativa L.) seedlings in response to NaCl. J Exp Bot 58, 3273-3283. |
| [10] | Hong LL, Qian Q, Tang D, Wang KJ, Li M, Cheng ZK (2012). A mutation in the rice chalcone isomerase gene causes the golden hull and internode 1phenotype. Planta 236, 141-151. |
| [11] | Huang J, Zhu L, Xue PB, Fu Q (2022). Research on mechanism and QTL mapping associated with cadmium accumulation in rice leaves and grains. Biotechnol Bull 38(8), 118-126. (in Chinese) |
| 黄婧, 朱亮, 薛蓬勃, 付强 (2022). 水稻叶和籽粒镉积累机制及QTL定位研究. 生物技术通报 38( 8), 118-126. | |
| [12] | Huang YJ, Yang ZY, Rao ZM, Deng JL, Liu YB (2000). Active oxygen damage effect and regulation of chlorophyll degradation in rice leaves at filling stage under high temperature stress. Acta Agric Univ Jiangxiensis 22(5), 1-6. (in Chinese) |
| 黄英金, 杨芝燕, 饶志明, 邓接楼, 刘宜柏 (2000). 灌浆期高温胁迫下水稻叶片叶绿素降解的活性氧损伤及调控研究. 江西农业大学学报 22(5), 1-6. | |
| [13] | Islam F, Khan MSS, Ahmed S, Abdullah M, Hannan F, Chen J (2023). OsLPXC negatively regulates tolerance to cold stress via modulating oxidative stress, antioxidant defense and JA accumulation in rice. Free Radical Biol Med 199, 2-16. |
| [14] | Jaksomsak P, Rerkasem B, Prom-U-Thai C (2021). Variation in nutritional quality of pigmented rice varieties under different water regimes. Plant Prod Sci 24, 244-255. |
| [15] | Jia QW, Zhong QQ, Gu YJ, Lu TQ, Li W, Yang S, Zhu CY, Hu CX, Li SF, Wang YX, Rao YC (2023). Mapping of QTL for cell wall related components in rice stem and analysis of candidate genes. Chin Bull Bot 58, 882-892. (in Chinese) |
| 贾绮玮, 钟芊芊, 顾育嘉, 陆天麒, 李玮, 杨帅, 朱超宇, 胡程翔, 李三峰, 王跃星, 饶玉春 (2023). 水稻茎秆细胞壁相关组分含量QTL定位及候选基因分析. 植物学报 58, 882- 892. | |
| [16] | Jiang HH, Wang T, Chen N, Yu SL, Chi XY, Wang M, Qi PS (2019). Research progress in PGPR improving plant’s resistance to salt and alkali. Biotechnol Bull 35(10), 189- 197. (in Chinese) |
| 姜焕焕, 王通, 陈娜, 禹山林, 迟晓元, 王冕, 祁佩时 (2019). 根际促生菌提高植物抗盐碱性的研究进展. 生物技术通报 35( 10), 189-197. | |
| [17] | Jin JY, Luo YT, Yang HM, Lu T, Ye HF, Xie JY, Wang KX, Chen QY, Fang Y, Wang YX, Rao YC (2023). Mapping of QTL for cell wall related components in rice stem and analysis of candidate genes. Chin Bull Bot 58, 394-403. (in Chinese) |
| 金佳怡, 罗怿婷, 杨惠敏, 芦涛, 叶涵斐, 谢继毅, 王珂欣, 陈芊羽, 方媛, 王跃星, 饶玉春 (2023). 水稻叶绿素含量QTL定位与候选基因表达分析. 植物学报 58, 394-403. | |
| [18] | Jin L, Xiao P, Lu Y, Shao YF, Shen Y, Bao JS (2009). Quantitative trait loci for brown rice color, phenolics, flavonoid contents, and antioxidant capacity in rice grain. Cereal Chem 86, 609-615. |
| [19] | Kim B, Shim S, Zhang HJ, Lee C, Jang S, Jin Z, Seo J, Kwon SW, Koh HJ (2023). Dynamic transcriptome changes driven by the mutation of OsCOP1 underlie flavonoid biosynthesis and embryogenesis in the developing rice seed. J Plant Growth Regul 42, 4436-4452. |
| [20] | Lam PY, Wang LX, Lui ACW, Liu HJ, Takeda-Kimura Y, Chen MX, Zhu FY, Zhang JH, Umezawa T, Tobimatsu Y, Lo C (2022). Deficiency in flavonoid biosynthesis genes CHS, CHI, and CHIL alters rice flavonoid and lignin profiles. Plant Physiol 188, 1993-2011. |
| [21] | Li SC (2023). Novel insight into functions of ascorbate peroxidase in higher plants: more than a simple antioxidant enzyme. Redox Biol 64, 102789. |
| [22] | Li XY, Yin H, Zhan SY, Qiao Y (2017). Effects of enhanced UV-B radiation on photosynthetic characteristics and anti- oxidation of rice flag leaves in filling stage. J Shenyang Agric Univ 48, 271-276. (in Chinese) |
| 李雪莹, 殷红, 战莘晔, 乔媛 (2017). 添加UV-B辐射对灌浆期水稻剑叶光合特性及抗氧化能力的影响. 沈阳农业大学学报 48, 271-276. | |
| [23] | Liu M, Chu MJ, Ding YF, Wang SH, Liu ZH, Tang S, Ding CQ, Li GH (2015). Exogenous spermidine alleviates oxidative damage and reduce yield loss in rice submerged at tillering stage. Front Plant Sci 6, 919. |
| [24] | Liu Y, Xue XX, Zhao CL, Zhang J, Liu M, Li XY, Li YQ, Gao X (2021). Cloning and functional characterization of chalcone isomerase genes involved in anthocyanin biosynthesis in Clivia miniata. Ornam Plant Res 1, 2. |
| [25] | Livak KJ, Schmittgen TD (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-∆∆CT method. Methods 25, 402-408. |
| [26] | Lu T, Ye HF, Chu XJ, Lin H, Wang S, Pan CY, Li SF, Wang YX, Rao YC (2022). Identification of QTL brown rice rate to submergence in rice. J Zhejiang Normal Univ (Nat Sci) 45, 323-328. (in Chinese) |
| 芦涛, 叶涵斐, 褚晓洁, 林晗, 王盛, 潘晨阳, 李三峰, 王跃星, 饶玉春 (2022). 水稻糙米率QTL检测及候选基因分析. 浙江师范大学学报(自然科学版) 45, 323-328. | |
| [27] | McCouch SR, Cho YG, Yano M, Paul E, Blinstrub M, Morishima H, Kinoshita T (1997). Report on QTL nomenclature. Rice Genet News 14, 11-13. |
| [28] | Mi YZ (2021). Mechanism of Anthocyanin Alleviates Cd Oxidative Damage of Rice Leaves and Inhibits Cd Accumulation in Rice Grains. PhD dissertation. Hefei: Anhui Agricultural University. pp. 42. (in Chinese) |
| 米雅竹 (2021). 花青素缓解Cd胁迫下水稻叶片氧化损伤及抑制籽粒Cd累积机理研究. 博士论文. 合肥: 安徽农业大学. pp. 42. | |
| [29] | Ran YD, Moursy M, Hider RC, Cilibrizzi A (2023). The colorimetric detection of the hydroxyl radical. Int J Mol Sci 24, 4162. |
| [30] | Rao YC, Dai ZJ, Zhu YT, Jiang JJ, Ma RY, Wang YY, Wang YX (2020). Advances in research of drought resistance in rice. J Zhejiang Normal Univ (Nat Sci) 43, 417- 429. (in Chinese) |
| 饶玉春, 戴志俊, 朱怡彤, 姜嘉骥, 马若盈, 王予烨, 王跃星 (2020). 水稻抗干旱胁迫的研究进展. 浙江师范大学学报(自然科学版) 43, 417-429. | |
| [31] | Shi H, Wang RC, Wu XY, Liu Q (2018). Protective effect of Actinidia arguta flavonoid on hydrogen peroxide-induced injury in HaCaT cells. Food Sci 39(13), 229-234. (in Chinese) |
| 石浩, 王仁才, 吴小燕, 刘琼 (2018). 软枣猕猴桃黄酮对过氧化氢诱导HaCaT细胞损伤的保护作用. 食品科学 39(13), 229-234. | |
| [32] | Wang CC (2015). Study of Novel Functions of Arabidopsis AtSPXI Through Multi-dimension Omics Data-mining, and the Role of OsSPX1 Involved in Oxidative Stresses in Rice Seedlings. PhD dissertation. Beijing: China Agricultural University. pp. 77-79. (in Chinese) |
| 王春超 (2015). 基于多维组学数据挖掘研究拟南芥AtSPX1基因的新功能以及OsSPX1对苗期水稻抗氧化性的影响. 博士论文. 北京: 中国农业大学. pp. 77-79. | |
| [33] | Wang L, Huang LC, Dai LP, Yang YL, Xu J, Leng YJ, Zhang GH, Hu J, Zhu L, Gao ZY, Dong GJ, Guo LB, Qian Q, Zeng DL (2014). QTL analysis for rice leaf morphology at maturity stage using a recombinant inbred line population derived from a cross between Nipponbare and 9311. Chin J Rice Sci 28, 589-597. (in Chinese) |
| 王兰, 黄李超, 代丽萍, 杨窑龙, 徐杰, 冷语佳, 张光恒, 胡江, 朱丽, 高振宇, 董国军, 郭龙彪, 钱前, 曾大力 (2014). 利用日本晴/9311重组自交系群体定位水稻成熟期叶形相关性状QTL. 中国水稻科学 28, 589-597. | |
| [34] | Wang LP, Zhou SM, Dai DL, Cao JS (2010). Progress in plant phenolic compounds. Acta Agric Zhejiangensis 22, 696-701. (in Chinese) |
| 王玲平, 周生茂, 戴丹丽, 曹家树 (2010). 植物酚类物质研究进展. 浙江农业学报 22, 696-701. | |
| [35] | Wang WS (2020). Identification and Function Analysis of Glycosyltransferase Genes Related to Flavonoids and Anthocyanins Metabolism in Rice. Master’s thesis. Jinan: Shandong University. pp. 11-13. (in Chinese) |
| 王文帅 (2020). 水稻黄酮和花青素代谢相关糖基转移酶基因的鉴定与作用分析. 硕士论文. 济南: 山东大学. pp. 11-13. | |
| [36] | Xie WB, Feng Q, Yu HH, Huang XH, Zhao Q, Xing YZ, Yu SB, Han B, Zhang QF (2010). Parent-independent genotyping for constructing an ultrahigh-density linkage map based on population sequencing. Proc Natl Acad Sci USA 107, 10578-10583. |
| [37] | Yang HX, Chen JL, Liu W (2019). Research progress on cadmium toxicity and detoxification mechanism in plants. Jiangsu Agric Sci 47(2), 1-8. (in Chinese) |
| 杨红霞, 陈俊良, 刘崴 (2019). 镉对植物的毒害及植物解毒机制研究进展. 江苏农业科学 47(2), 1-8. | |
| [38] | Zhao MY, Kang QF, Zhao Y, Li XY (2023). Identification and bioinformatic analyses of genes related to anthocyanin biosynthesis in Setaria italica. Mol Plant Breed 1-17. (in Chinese) |
| 赵孟瑶, 康庆芳, 赵影, 李雪垠 (2023). 谷子花青苷合成相关基因的鉴定与生物信息学分析. 分子植物育种 1-17. | |
| [39] | Zhong YH, Guo ZJ, Wei MY, Wang JC, Song SW, Chi BJ, Zhang YC, Liu JW, Li J, Zhu XY, Tang HC, Song LY, Xu CQ, Zheng HL (2023). Hydrogen sulfide upregulates the alternative respiratory pathway in mangrove plant Avicennia marina to attenuate waterlogging-induced oxidative stress and mitochondrial damage in a calcium-dependent manner. Plant Cell Environ 46, 1521-1539. |
| [40] | Zhou H, Finkemeier I, Guan WX, Tossounian MA, Wei B, Young D, Huang JJ, Messens J, Yang XB, Zhu J, Wilson MH, Shen WB, Xie YJ, Foyer CH (2018). Oxidative stress-triggered interactions between the succinyl- and acetyl-proteomes of rice leaves. Plant Cell Environ 41, 1139-1153. |
| [1] |
Juan Cui, Xiaoyu Yu, Yuejiao Yu, Chengwei Liang, Jian Sun, Wenfu Chen.
Analysis of Texture Factors and Genetic Basis Influencing the Differences in Eating Quality between Northeast China and Japanese Japonica Rice [J]. Chinese Bulletin of Botany, 2025, 60(4): 1-0. |
| [2] | Zhao Ling, Guan Ju, Liang Wenhua, Zhang Yong, Lu Kai, Zhao Chunfang, Li Yusheng, Zhang Yadong. Mapping of QTLs for Heat Tolerance at the Seedling Stage in Rice Based on a High-density Bin Map [J]. Chinese Bulletin of Botany, 2025, 60(3): 342-353. |
| [3] | Xinyu Li, Yue Gu, Feifei Xu, Jinsong Bao. Research Progress on Post-translational Modifications of Starch Biosynthesis-related Proteins in Rice Endosperm [J]. Chinese Bulletin of Botany, 2025, 60(2): 256-270. |
| [4] | Jianguo Li, Yi Zhang, Wenjun Zhang. Iron Plaque Formation and Its Effects on Phosphorus Absorption in Rice Roots [J]. Chinese Bulletin of Botany, 2025, 60(1): 132-143. |
| [5] | Ruifeng Yao, Daoxin Xie. Activation and Termination of Strigolactone Signal Perception in Rice [J]. Chinese Bulletin of Botany, 2024, 59(6): 873-877. |
| [6] | Jiahui Huang, Huimin Yang, Xinyu Chen, Chaoyu Zhu, Yanan Jiang, Chengxiang Hu, Jinjin Lian, Tao Lu, Mei Lu, Weilin Zhang, Yuchun Rao. Response Mechanism of Rice Mutant pe-1 to Low Light Stress [J]. Chinese Bulletin of Botany, 2024, 59(4): 574-584. |
| [7] | Jianmin Zhou. A Combat Vehicle with a Smart Brake [J]. Chinese Bulletin of Botany, 2024, 59(3): 343-346. |
| [8] | Chaoyu Zhu, Chengxiang Hu, Zhenan Zhu, Zhining Zhang, Lihai Wang, Jun Chen, Sanfeng Li, Jinjin Lian, Luyao Tang, Qianqian Zhong, Wenjing Yin, Yuexing Wang, Yuchun Rao. Mapping of QTLs Associated with Rice Panicle Traits and Candidate Gene Analysis [J]. Chinese Bulletin of Botany, 2024, 59(2): 217-230. |
| [9] | Bao Zhu, Jiangzhe Zhao, Kewei Zhang, Peng Huang. OsCKX9 is Involved in Regulating the Rice Lamina Joint Development and Leaf Angle [J]. Chinese Bulletin of Botany, 2024, 59(1): 10-21. |
| [10] | Yanli Fang, Chuanyu Tian, Ruyi Su, Yapei Liu, Chunlian Wang, Xifeng Chen, Wei Guo, Zhiyuan Ji. Mining and Preliminary Mapping of Rice Resistance Genes Against Bacterial Leaf Streak [J]. Chinese Bulletin of Botany, 2024, 59(1): 1-9. |
| [11] | Qiwei Jia, Qianqian Zhong, Yujia Gu, Tianqi Lu, Wei Li, Shuai Yang, Chaoyu Zhu, Chengxiang Hu, Sanfeng Li, Yuexing Wang, Yuchun Rao. Mapping of QTL for Cell Wall Related Components in Rice Stem and Analysis of Candidate Genes [J]. Chinese Bulletin of Botany, 2023, 58(6): 882-892. |
| [12] | Tian Chuanyu, Fang Yanli, Shen Qing, Wang Hongjie, Chen Xifeng, Guo Wei, Zhao Kaijun, Wang Chunlian, Ji Zhiyuan. Genotypic Diversity and Pathogenisity of Xanthomonas oryzae pv. oryzae Isolated from Southern China in 2019-2021 [J]. Chinese Bulletin of Botany, 2023, 58(5): 743-749. |
| [13] | Dai Ruohui, Qian Xinyu, Sun Jinglei, Lu Tao, Jia Qiwei, Lu Tianqi, Lu Mei, Rao Yuchun. Research Progress on the Mechanisms of Leaf Color Regulation and Related Genes in Rice [J]. Chinese Bulletin of Botany, 2023, 58(5): 799-812. |
| [14] | Jingjing Zhao, Haibin Jia, Tien Ming Lee. Market status and the sustainable utilization strategy of wild earthworm (earth dragon) for medicinal use [J]. Biodiv Sci, 2023, 31(3): 22478-. |
| [15] | Shang Sun, Yingying Hu, Yangshuo Han, Chao Xue, Zhiyun Gong. Double-stranded Labelled Oligo-FISH in Rice Chromosomes [J]. Chinese Bulletin of Botany, 2023, 58(3): 433-439. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||