

Chinese Bulletin of Botany ›› 2025, Vol. 60 ›› Issue (3): 342-353.DOI: 10.11983/CBB24047 cstr: 32102.14.CBB24047
• RESEARCH ARTICLES • Previous Articles Next Articles
Zhao Ling1, Guan Ju1, Liang Wenhua1, Zhang Yong2, Lu Kai1, Zhao Chunfang1, Li Yusheng1, Zhang Yadong1,*( )
)
Received:2024-03-28
Accepted:2024-05-27
Online:2025-05-10
Published:2024-05-30
Contact:
*E-mail: zhangyd@jaas.ac.cn
Zhao Ling, Guan Ju, Liang Wenhua, Zhang Yong, Lu Kai, Zhao Chunfang, Li Yusheng, Zhang Yadong. Mapping of QTLs for Heat Tolerance at the Seedling Stage in Rice Based on a High-density Bin Map[J]. Chinese Bulletin of Botany, 2025, 60(3): 342-353.

Figure 1 Heat tolerance of parents and some recombinant inbred lines (RILs) during the seedling stage (A) Heat tolerance of parents and RILs population during seedling stage (bars=2 cm); (B) Distribution of high-temperature seedling survival rates in the RILs. HTSR: High-temperature seedling survival rate
| QTL | Chr. | Interval (Mb) | Genes | Annotation | Reference |
|---|---|---|---|---|---|
| qHTSR1.1 | 1 | 1.90-1.95 | Os01g0134800* | Glycosyl hydrolase family | |
| Os01g0134900* | Glycosyl hydrolase family | ||||
| Os01g0135000 | Similar to 1,4-beta-D xylan xylanohydrolase | ||||
| Os01g0135700* | OsCML16, calmodulin, calcium signaling during abiotic stress | Yang et al., | |||
| Os01g0135800* | HSP20, heat shock protein | ||||
| Os01g0135900* | HSP20, heat shock protein | Sarkar et al., | |||
| Os01g0136000* | OsHSP16.9C, HSP17.5 heat shock protein | Sarkar et al., | |||
| Os01g0136050 | 16.9 kDa heat shock protein | ||||
| qHTSR4.1 | 4 | 15.72-15.92 | Os04g0336600 | Peptidase aspartic, catalytic domain containing protein | |
| Os04g0337000* | Aspartyl protease domain containing protein | ||||
| Os04g0337201* | SAD2, sensitive to ABA and drought2 | ||||
| Os04g0337300 | SAD2, sensitive to ABA and drought2 | ||||
| Os04g0337500 | Oxidoreductase | ||||
| Os04g0337800* | IN2-2 protein | ||||
| Os04g0338000* | Aldo/keto reductase family protein | Kim et al., | |||
| qHTSR5.1 | 5 | 20.41-20.48 | Os05g0417000* | DUF260 domain containing protein | |
| Os05g0417100* | Deg Protease Protein, chloroplast development and maintenance of PSII function under high temperatures | Zheng et al., | |||
| Os05g0417800* | ulp1 protease family | ||||
| qHTSR5.2 | 5 | 26.25-26.39 | Os05g0418000* | GDP-dissociation inhibitor | Shad et al., |
| Os05g0528500 | OsWRKY58, WRKY transcription factor | ||||
| Os05g0528600 | OsYUCCA2, flavin monooxygenase-like enzyme, auxin biosynthesis | Yamamoto et al., | |||
| Os05g0528900 | Ribosomal protein | ||||
| Os05g0529000* | OsCOLE1, regulation of intracellular auxin transport | Liu et al., | |||
| Os05g0529200 | Enoyl-CoA hydratase/isomerase family protein | ||||
| Os05g0529300 | ER lumen protein retaining receptor | ||||
| Os05g0529400* | Ubiquitin domain-containing protein | ||||
| Os05g0529600* | ATP binding protein | ||||
| Os05g0529700 | Electron transporter/heat shock protein binding protein | ||||
| Os05g0529900 | Cell cycle control protein, OsHsfA4d; Heat stress transcription factor | Mittal et al., | |||
| Os05g0530400* | Plant growth and balancing reactive oxygen species du- ring biotic and abiotic stress | ||||
| Os05g0530500 | Snf1 protein kinase; sucrose non-fermenting-1 related protein kinase 1 | Kanegae et al., | |||
| Os05g0530701 | Leucine-rich repeat | ||||
| Os05g0531000* | A member of S40 gene family, regulation of crosstalk among abiotic and biotic and developmental senescence | ||||
| Os05g0531100 | A member of S40 gene family, regulation of crosstalk among abiotic and biotic and developmental senescence | ||||
| Os05g0531200 | Pollen Ole e I allergen and extensin family protein precursor | ||||
| Os05g0531400 | Pollen Ole e I allergen and extensin family protein precursor | ||||
| Os05g0531500 | Hydrolase |
Table 1 Annotated genes within the interval of major QTLs
| QTL | Chr. | Interval (Mb) | Genes | Annotation | Reference |
|---|---|---|---|---|---|
| qHTSR1.1 | 1 | 1.90-1.95 | Os01g0134800* | Glycosyl hydrolase family | |
| Os01g0134900* | Glycosyl hydrolase family | ||||
| Os01g0135000 | Similar to 1,4-beta-D xylan xylanohydrolase | ||||
| Os01g0135700* | OsCML16, calmodulin, calcium signaling during abiotic stress | Yang et al., | |||
| Os01g0135800* | HSP20, heat shock protein | ||||
| Os01g0135900* | HSP20, heat shock protein | Sarkar et al., | |||
| Os01g0136000* | OsHSP16.9C, HSP17.5 heat shock protein | Sarkar et al., | |||
| Os01g0136050 | 16.9 kDa heat shock protein | ||||
| qHTSR4.1 | 4 | 15.72-15.92 | Os04g0336600 | Peptidase aspartic, catalytic domain containing protein | |
| Os04g0337000* | Aspartyl protease domain containing protein | ||||
| Os04g0337201* | SAD2, sensitive to ABA and drought2 | ||||
| Os04g0337300 | SAD2, sensitive to ABA and drought2 | ||||
| Os04g0337500 | Oxidoreductase | ||||
| Os04g0337800* | IN2-2 protein | ||||
| Os04g0338000* | Aldo/keto reductase family protein | Kim et al., | |||
| qHTSR5.1 | 5 | 20.41-20.48 | Os05g0417000* | DUF260 domain containing protein | |
| Os05g0417100* | Deg Protease Protein, chloroplast development and maintenance of PSII function under high temperatures | Zheng et al., | |||
| Os05g0417800* | ulp1 protease family | ||||
| qHTSR5.2 | 5 | 26.25-26.39 | Os05g0418000* | GDP-dissociation inhibitor | Shad et al., |
| Os05g0528500 | OsWRKY58, WRKY transcription factor | ||||
| Os05g0528600 | OsYUCCA2, flavin monooxygenase-like enzyme, auxin biosynthesis | Yamamoto et al., | |||
| Os05g0528900 | Ribosomal protein | ||||
| Os05g0529000* | OsCOLE1, regulation of intracellular auxin transport | Liu et al., | |||
| Os05g0529200 | Enoyl-CoA hydratase/isomerase family protein | ||||
| Os05g0529300 | ER lumen protein retaining receptor | ||||
| Os05g0529400* | Ubiquitin domain-containing protein | ||||
| Os05g0529600* | ATP binding protein | ||||
| Os05g0529700 | Electron transporter/heat shock protein binding protein | ||||
| Os05g0529900 | Cell cycle control protein, OsHsfA4d; Heat stress transcription factor | Mittal et al., | |||
| Os05g0530400* | Plant growth and balancing reactive oxygen species du- ring biotic and abiotic stress | ||||
| Os05g0530500 | Snf1 protein kinase; sucrose non-fermenting-1 related protein kinase 1 | Kanegae et al., | |||
| Os05g0530701 | Leucine-rich repeat | ||||
| Os05g0531000* | A member of S40 gene family, regulation of crosstalk among abiotic and biotic and developmental senescence | ||||
| Os05g0531100 | A member of S40 gene family, regulation of crosstalk among abiotic and biotic and developmental senescence | ||||
| Os05g0531200 | Pollen Ole e I allergen and extensin family protein precursor | ||||
| Os05g0531400 | Pollen Ole e I allergen and extensin family protein precursor | ||||
| Os05g0531500 | Hydrolase |
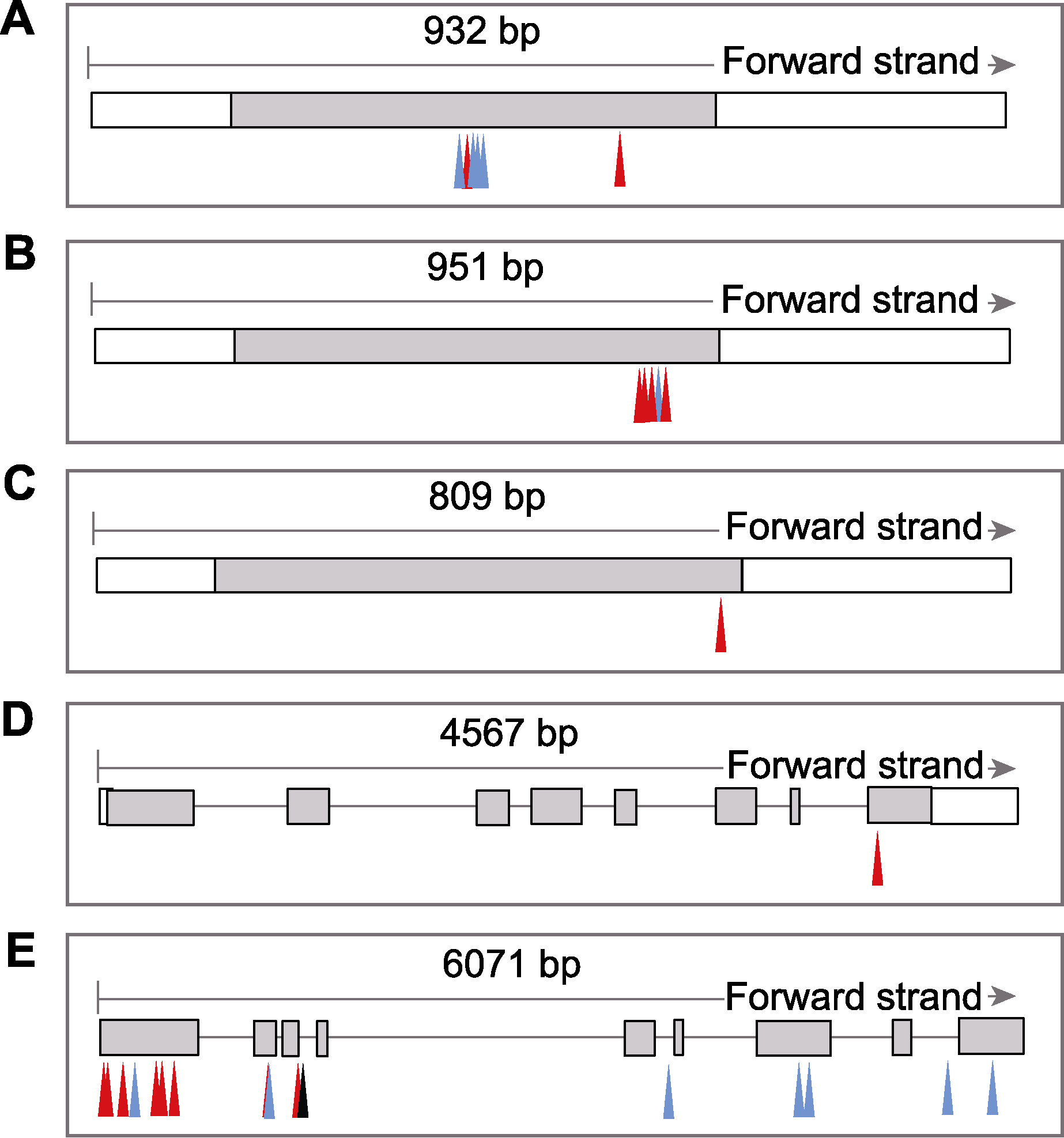
Figure 3 Gene structure and non-synonymous mutation of five candidate genes between TD70 and Kasalath (A) LOC_Os01g04340; (B) LOC_Os01g04350; (C) LOC_Os01g04360; (D) LOC_Os05g34460; (E) LOC_Os05g34520. Frames with black lines: Exon; Grey boxes: Protein coding sequence; Red arrow: Missense mutation; Blue arrow: Frameshift mutation; Black arrow: Synonymous mutation
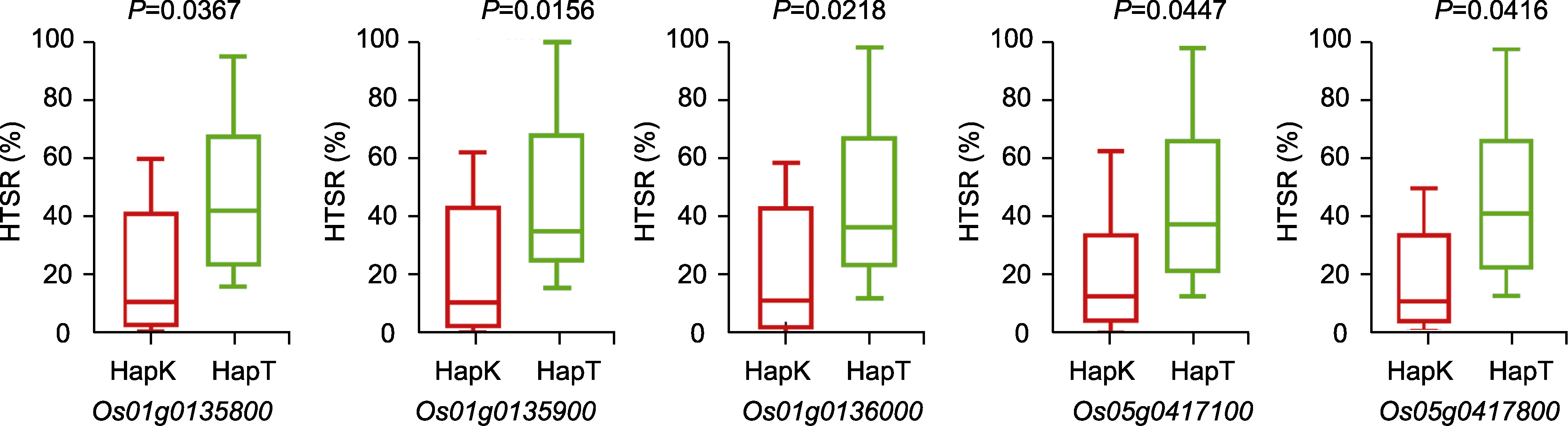
Figure 4 Effects of different Haps of candidate genes on the HTSR of recombinant inbred lines (RILs) HapT: The SNP of the candidate genes are the same as those in TD70; HapK: The SNP of the candidate genes are the same as those in Kasalath. HTSR is the same as shown in Figure 1.
| QTL | This study | Known QTLs or genes | ||||
|---|---|---|---|---|---|---|
| Chr. | Position (Mb) | Population | Traits | Position (Mb) | Reference | |
| qHTSR1.1 | 1 | 1.90-1.95 | Nipponbare/Kasalath backcross inbred lines (BILs) | HT during grain filling | 0.64-3.44 | 朱昌兰等, |
| Teqin/Yuanjiang Common Wild Rice BILs | HT at flowering stage | 0.38-2.18 | 奎丽梅等, | |||
| N22/IR64 recombinant inbred lines (RILs) | Shoot length under heat stress | 1.86-2.31 | Kilasi et al., | |||
| qHTSR1.2 | 1 | 39.82-39.86 | Longdao 5/Zhongyouzao 8RILs | HTSR | 37.6-39.08 | 刘进等, |
| Bala/Azucena RILs | HT at flowering stage | 38.35-40.66 | Jagadish et al., | |||
| qHTSR2.2 | 2 | 24.58-24.72 | 255 core materials | HTSR | 24.66 | 魏昭然, |
| qHTSR4.1 | 4 | 15.71-15.92 | 996/4628 RILs | HT at flowering stage | 15.74-18.82 | Xiao et al., |
| Bala/Azucena RILs | HT at flowering stage | 14.19-16.86 | Jagadish et al., | |||
| qHTSR 5.2 | 5 | 26.25-26.39 | Zhongyouzao 8/Fengjin RILs | HT at flowering stage | 3.08-26.85 | 张涛等, |
| Longdao 5/Zhongyouzao 8 RILs | HTSR | 25.78-26.28 | 刘进等, | |||
| R498/R3551 RILs | HT at flowering stage | 26.69-27.69 | 陶磊, | |||
| qHTSR9.1 | 9 | 9.42-9.46 | Nipponbare/Kasalath bils | Thermo-tolerance of amylose content | 5.27-21.85 | 朱昌兰等, |
| qHTSR10 | 10 | 22.57-22.63 | Bala/Azucena RILs | HT at flowering stage | 20.84-23.45 | Jagadish et al., |
| 233 core materials | HT during grain filling | 22.62 | 杨飞, | |||
| Sasanishiki/Habataki CSSLs | Pollen fertility under high temperature | 19.89-23.01 | Zhu et al., | |||
Table 2 Overlap of known QTLs contributing to heat resistance with the QTLs detected in this study
| QTL | This study | Known QTLs or genes | ||||
|---|---|---|---|---|---|---|
| Chr. | Position (Mb) | Population | Traits | Position (Mb) | Reference | |
| qHTSR1.1 | 1 | 1.90-1.95 | Nipponbare/Kasalath backcross inbred lines (BILs) | HT during grain filling | 0.64-3.44 | 朱昌兰等, |
| Teqin/Yuanjiang Common Wild Rice BILs | HT at flowering stage | 0.38-2.18 | 奎丽梅等, | |||
| N22/IR64 recombinant inbred lines (RILs) | Shoot length under heat stress | 1.86-2.31 | Kilasi et al., | |||
| qHTSR1.2 | 1 | 39.82-39.86 | Longdao 5/Zhongyouzao 8RILs | HTSR | 37.6-39.08 | 刘进等, |
| Bala/Azucena RILs | HT at flowering stage | 38.35-40.66 | Jagadish et al., | |||
| qHTSR2.2 | 2 | 24.58-24.72 | 255 core materials | HTSR | 24.66 | 魏昭然, |
| qHTSR4.1 | 4 | 15.71-15.92 | 996/4628 RILs | HT at flowering stage | 15.74-18.82 | Xiao et al., |
| Bala/Azucena RILs | HT at flowering stage | 14.19-16.86 | Jagadish et al., | |||
| qHTSR 5.2 | 5 | 26.25-26.39 | Zhongyouzao 8/Fengjin RILs | HT at flowering stage | 3.08-26.85 | 张涛等, |
| Longdao 5/Zhongyouzao 8 RILs | HTSR | 25.78-26.28 | 刘进等, | |||
| R498/R3551 RILs | HT at flowering stage | 26.69-27.69 | 陶磊, | |||
| qHTSR9.1 | 9 | 9.42-9.46 | Nipponbare/Kasalath bils | Thermo-tolerance of amylose content | 5.27-21.85 | 朱昌兰等, |
| qHTSR10 | 10 | 22.57-22.63 | Bala/Azucena RILs | HT at flowering stage | 20.84-23.45 | Jagadish et al., |
| 233 core materials | HT during grain filling | 22.62 | 杨飞, | |||
| Sasanishiki/Habataki CSSLs | Pollen fertility under high temperature | 19.89-23.01 | Zhu et al., | |||
| [1] |
曹志斌, 李瑶, 曾博虹, 毛凌华, 蔡耀辉, 吴晓峰, 袁林峰 (2020). 非洲栽培稻垩白粒率耐热性QTL的定位. 中国水稻科学 34, 135-142.
DOI |
| [2] | 郭虹霞, 王创云, 赵丽, 王陆军, 张丽光, 邓妍 (2019). 水稻中2个小分子热激蛋白基因启动子的序列分析及功能鉴定. 西北农业学报 28, 1079-1086. |
| [3] |
郝立生, 马宁, 何丽烨 (2022). 2022年长江中下游夏季异常干旱高温事件之环流异常特征. 干旱气象 40, 721-732.
DOI |
| [4] |
胡时开, 钱前 (2016). RNA解旋酶调控rRNA内稳态: 水稻耐热新机制、分子育种新资源. 植物学报 51, 283-286.
DOI |
| [5] | 阚义, 林鸿宣 (2022). 水稻高温感知及响应机制的研究进展. 自然杂志 44, 411-421. |
| [6] | 奎丽梅, 谭禄宾, 涂建, 卢义宣, 孙传清 (2008). 云南元江野生稻抽穗开花期耐热QTL定位. 农业生物技术学报 16, 461-464. |
| [7] | 李林浩 (2023). 基于Meta-QTL和RNA-seq的整合分析挖掘水稻持续耐高温候选基因. 硕士论文. 荆州: 长江大学. pp. 26-30. |
| [8] |
栗振义, 龙瑞才, 张铁军, 杨青川, 康俊梅 (2016). 植物热激蛋白研究进展. 生物技术通报 32(2), 7-13.
DOI |
| [9] | 刘进, 崔迪, 余丽琴, 张立娜, 周慧颖, 马小定, 胡佳晓, 韩冰, 韩龙植, 黎毛毛 (2022a). 水稻苗期耐热种质资源筛选及QTL定位. 中国水稻科学 36, 259-268. |
| [10] | 刘进, 胡佳晓, 马小定, 陈武, 勒思, Sumin J, 崔迪, 周慧颖, 张立娜, Dongjin S, 黎毛毛, 韩龙植, 余丽琴 (2022b). 水稻RIL群体高密度遗传图谱的构建及苗期耐热性QTL定位. 中国农业科学 55, 4327-4341. |
| [11] | 盘毅, 陈立云, 肖应辉 (2008). 水稻耐热遗传育种及热激蛋白的研究综述. 作物研究 22(S1), 363-367. |
| [12] |
盘毅, 罗丽华, 邓化冰, 张桂莲, 唐文邦, 陈立云, 肖应辉 (2011). 水稻开花期高温胁迫下的花粉育性QTL定位. 中国水稻科学 25, 99-102.
DOI |
| [13] |
沈泓, 姚栋萍, 吴俊, 罗秋红, 吴志鹏, 雷东阳, 邓启云, 柏斌 (2022). 灌浆期不同时段高温对稻米淀粉理化特性的影响. 中国水稻科学 36, 377-387.
DOI |
| [14] |
宋有金, 吴超, 李子煜, 唐设, 李刚华, 王绍华, 丁艳锋 (2021). 水稻产量对生殖生长阶段不同时期高温的响应差异. 中国水稻科学 35, 177-186.
DOI |
| [15] | 陶磊 (2020). 水稻开花期耐高温QTL分析及精细定位. 硕士论文. 成都: 四川农业大学. pp. 25-27. |
| [16] |
王建康 (2009). 数量性状基因的完备区间作图方法. 作物学报 35, 239-245.
DOI |
| [17] | 魏昭然 (2020). 水稻苗期高温关键候选位点鉴定. 硕士论文. 泰安: 山东农业大学. pp. 21-23. |
| [18] | 杨飞 (2020). 水稻灌浆期耐热性及主要农艺性状的全基因组关联分析. 硕士论文. 武汉: 华中农业大学. pp. 27-29. |
| [19] |
杨军, 章毅之, 贺浩华, 李迎春, 陈小荣, 边建民, 金国花, 李翔翔, 黄淑娥 (2020). 水稻高温热害的研究现状与进展. 应用生态学报 31, 2817-2830.
DOI |
| [20] | 杨梯丰, 刘斌 (2009). 水稻耐热性QTL鉴定的研究进展. 广东农业科学 (6), 16-20. |
| [21] | 俞佳虹, 冯坤, 程远, 叶青静, 阮美颖, 王荣青, 李志邈, 周国治, 姚祝平, 魏家香, 杨悦俭, 万红建 (2017). 植物小热激蛋白的研究进展. 分子植物育种 15, 3016-3023. |
| [22] | 张涛, 杨莉, 蒋开锋, 黄敏, 孙群, 陈温福, 郑家奎 (2008). 水稻抽穗扬花期耐热性的QTL分析. 分子植物育种 6, 867-873. |
| [23] |
张亚东, 梁文化, 赫磊, 赵春芳, 朱镇, 陈涛, 赵庆勇, 赵凌, 姚姝, 周丽慧, 路凯, 王才林 (2021). 水稻RIL群体高密度遗传图谱构建及粒型QTL定位. 中国农业科学 54, 5163-5176.
DOI |
| [24] |
赵凌, 张勇, 魏晓东, 梁文化, 赵春芳, 周丽慧, 姚姝, 王才林, 张亚东 (2022). 利用高密度Bin图谱定位水稻抽穗期剑叶叶绿素含量QTL. 中国农业科学 55, 825-836.
DOI |
| [25] | 朱昌兰, 江玲, 张文伟, 王春明, 翟虎渠, 万建民 (2006). 稻米直链淀粉含量和胶稠度对高温耐性的QTL分析. 中国水稻科学 20, 248-252. |
| [26] | 朱昌兰, 肖应辉, 王春明, 江玲, 翟虎渠, 万建民 (2005). 水稻灌浆期耐热害的数量性状基因位点分析. 中国水稻科学 19, 117-121. |
| [27] |
Bauer D, Meinhold S, Jakob RP, Stigler J, Merkel U, Maier T, Rief M, Žoldák G (2018). A folding nucleus and minimal ATP binding domain of HSP70 identified by single-molecule force spectroscopy. Proc Natl Acad Sci USA 115, 4666-4671.
DOI PMID |
| [28] | Cao ZB, Li Y, Tang HW, Zeng BH, Tang XY, Long QZ, Wu XF, Cai YH, Yuan LF, Wan JL (2020). Fine mapping of the qHTB1-1 QTL, which confers heat tolerance at the booting stage, using an Oryza rufipogon Griff. introgression line. Theor Appl Genet 133, 1161-1175. |
| [29] | Chen L, Wang Q, Tang MY, Zhang XL, Pan YH, Yang XH, Gao GQ, Lv RH, Tao W, Jiang LG, Liang TF (2021). QTL mapping and identification of candidate genes for heat tolerance at the flowering stage in rice. Front Genet 11, 621871. |
| [30] | Chen XG, Chen S (2018). China feels the heat: negative impacts of high temperatures on China's rice sector. Aust J Agric Resour Econ 62, 576-588. |
| [31] | Haq ul S, Khan A, Ali M, Khattak AM, Gai WX, Zhang HX, Wei AM, Gong ZH (2019). Heat shock proteins: dynamic biomolecules to counter plant biotic and abiotic stresses. Int J Mol Sci 20, 5321. |
| [32] | IPCC (2021). Climate Change 2021:The Physical Science Basis. Contribution of Working Group I to the Sixth Assessment Report of the Intergovernmental Panel on Climate Change. Cambridge: Cambridge University Press. pp. 199-200. |
| [33] |
Jacob P, Hirt H, Bendahmane A (2017). The heat-shock protein/chaperone network and multiple stress resistance. Plant Biotechnol J 15, 405-414.
DOI PMID |
| [34] | Jagadish SVK, Cairns J, Lafitte R, Wheeler TR, Price AH, Craufurd PQ (2010). Genetic analysis of heat tolerance at anthesis in rice. Crop Sci 50, 1633-1641. |
| [35] |
Kan Y, Mu XR, Zhang H, Gao J, Han JX, Ye WW, Lin HX (2022). TT2 controls rice thermotolerance through SCT1- dependent alteration of wax biosynthesis. Nat Plants 8, 53-67.
DOI PMID |
| [36] | Kanegae H, Miyoshi K, Hirose T, Tsuchimoto S, Mori M, Nagato Y, Takano M (2005). Expressions of rice sucrose non-fermenting-1 related protein kinase 1 genes are differently regulated during the caryopsis development. Plant Physiol Biochem 43, 669-679. |
| [37] | Khan S, Anwar S, Ashraf MY, Khaliq B, Sun M, Hussain S, Gao ZQ, Noor H, Alam S (2019). Mechanisms and adaptation strategies to improve heat tolerance in rice. Plants 8, 508. |
| [38] | Kilasi NL, Singh J, Vallejos CE, Ye CR, Jagadish SVK, Kusolwa P, Rathinasabapathi B (2018). Heat stress tolerance in rice (Oryza sativa L.): identification of quantitative trait loci and candidate genes for seedling growth under heat stress. Front Plant Sci 9, 1578. |
| [39] | Kim JH, Lim SD, Jang CS (2019). Oryza sativa heat-induced RING finger protein 1 (OsHIRP1) positively regulates plant response to heat stress. Plant Mol Biol 99, 545-559. |
| [40] |
Lee BH, Won SH, Lee HS, Miyao M, Chung WI, Kim IJ, Jo J (2000). Expression of the chloroplast-localized small heat shock protein by oxidative stress in rice. Gene 245, 283-290.
PMID |
| [41] | Li XM, Chao DY, Wu Y, Huang XH, Chen K, Cui LG, Su L, Ye WW, Chen H, Chen HC, Dong NQ, Guo T, Shi M, Feng Q, Zhang P, Han B, Shan JX, Gao JP, Lin HX (2015). Natural alleles of a proteasome α2 subunit gene contribute to thermotolerance and adaptation of African rice. Nat Genet 47, 827-833. |
| [42] | Liu F, Zhang L, Luo YZ, Xu MY, Fan YL, Wang L (2016a). Interactions of Oryza sativa OsCONTINUOUS VASCULAR RING-LIKE 1 (OsCOLE1) and OsCOLE1-INTERACTING PROTEIN reveal a novel intracellular auxin transport mechanism. New Phytol 212, 96-107. |
| [43] | Liu JP, Zhang CC, Wei CC, Liu X, Wang MG, Yu FF, Xie Q, Tu JM (2016b). The RING finger ubiquitin E3 ligase OsHTAS enhances heat tolerance by promoting H2O2-induced stomatal closure in rice. Plant Physiol 170, 429-443. |
| [44] | McCouch SR, Cho YG, Yano M, Paul E, Blinstrub M, Morishima H, Kinoshita T (1997). Report on QTL nomenclature. Rice Genet Newsl 14, 11-13. |
| [45] |
Meng L, Li HH, Zhang LY, Wang JK (2015). QTL ICIMAPPING: integrated software for genetic linkage map construction and quantitative trait locus mapping in biparental populations. Crop J 3, 269-283.
DOI |
| [46] | Mittal D, Chakrabarti S, Sarkar A, Singh A, Grover A (2009). Heat shock factor gene family in rice: genomic organization and transcript expression profiling in response to high temperature, low temperature and oxidative stresses. Plant Physiol Biochem 47, 785-795. |
| [47] | Murakami T, Matsuba S, Funatsuki H, Kawaguchi K, Saruyama H, Tanida M, Sato Y (2004). Over-expression of a small heat shock protein, sHSP17.7, confers both heat tolerance and UV-B resistance to rice plants. Mol Breed 13, 165-175. |
| [48] | Murthy VS, Ravishankar KV (2016). Molecular mechanisms of heat shock proteins and thermotolerance in plants. In: SrinivasaRao NK, ShivashankaraKS, LaxmanRH, Abiotic Stress Physiology of Horticultural Crops.eds. New Delhi: Springer. pp. 71-83. |
| [49] |
Sarkar NK, Kim YK, Grover A (2009). Rice sHsp genes: genomic organization and expression profiling under stress and development. BMC Genomics 10, 393.
DOI PMID |
| [50] | Sarkar NK, Kotak S, Agarwal M, Kim YK, Grover A (2020). Silencing of class I small heat shock proteins affects seed-related attributes and thermotolerance in rice seedlings. Planta 251, 26. |
| [51] | Shad MA, Wang YX, Zhang H, Zhai SS, Shalmani A, Li YB (2023). Genetic analysis of GEFs and GDIs in rice reveals the roles of OsGEF5, OsGDI1, and OsGEF3 in the regulation of grain size and plant height. Crop J 11, 345-360. |
| [52] | Siddique M, Gernhard S, von Koskull-Döring P, Vierling E, Scharf KD (2008). The plant sHSP superfamily: five new members in Arabidopsis thaliana with unexpected properties. Cell Stress Chaperones 13, 183-197. |
| [53] | Wang D, Qin BX, Li X, Tang D, Zhang YE, Cheng ZK, Xue YB (2016). Nucleolar DEAD-Box RNA helicase TOGR1 regulates thermos tolerant growth as a Pre-rRNA chaperone in rice. PLoS Genet 12, e1005844. |
| [54] | Xiao YH, Pan Y, Luo LH, Zhang GL, Deng HB, Dai LY, Liu XL, Tang WB, Chen LY, Wang GL (2011). Quantitative trait loci associated with seed set under high temperature stress at the flowering stage in rice (Oryza sativa L.). Euphytica 178, 331-338. |
| [55] |
Yamamoto Y, Kamiya N, Morinaka Y, Matsuoka M, Sazuka T (2007). Auxin biosynthesis by the YUCCA genes in rice. Plant Physiol 143, 1362-1371.
DOI PMID |
| [56] | Yang J, Ji LX, Zhu BH, Yuan XJ, Jin DM, Xie GS (2018). OsCML16 interacts with a novel CC-NBS-LRR protein OsPi304 in the Ca2+/Mg2+dependent and independent manner in rice. Biochem Biophys Res Commun 504, 346-351. |
| [57] |
Zhang KM, Ezemaduka AN, Wang Z, Hu HL, Shi XD, Liu C, Lu XP, Fu XM, Chang ZY, Yin CC (2015). A novel mechanism for small heat shock proteins to function as molecular chaperones. Sci Rep 5, 8811.
DOI PMID |
| [58] | Zhang Y, Zou BH, Lu S, Ding Y, Liu H, Hua J (2016). Expression and promoter analysis of the OsHSP16.9C gene in rice. Biochem Biophys Res Commun 479, 260-265. |
| [59] | Zheng KL, Zhao J, Lin DZ, Chen JY, Xu JL, Zhou H, Teng S, Dong YJ (2016). The rice TCM5 gene encoding a novel Deg protease protein is essential for chloroplast development under high temperatures. Rice 9, 13. |
| [60] | Zhu S, Huang RL, Wai HP, Xiong HL, Shen XH, He HH, Yan S (2017). Mapping quantitative trait loci for heat tolerance at the booting stage using chromosomal segment substitution lines in rice. Physiol Mol Biol Plants 23, 817-825. |
| [1] | Yanan Jiang, Yuqing Xu, Yiting Wei, Jun Chen, Rongwan Zhang, Beibei Zhao, Yuxiang Lin, Yuchun Rao. Research Progress on the Regulatory Mechanism of Rice Disease Resistance [J]. Chinese Bulletin of Botany, 2025, 60(5): 1-0. |
| [2] | Jun Chen, Jiangmin Xu, Yinan Zhou, Yanan Jiang, Chengxiang Hu, Qianyun Jin, Beibei Zhao, Zhenan Zhu, Yuqing Xu, Luyi Zhang, Xiaoyan Liu, Jun Liu, Sanfeng Li, Yuexing Wang, Yuchun Rao. Mapping of QTLs Associated with Rice Resistance to Bacterial Blight and Candidate Gene Analysis [J]. Chinese Bulletin of Botany, 2025, 60(5): 1-0. |
| [3] |
Juan Cui, Xiaoyu Yu, Yuejiao Yu, Chengwei Liang, Jian Sun, Wenfu Chen.
Analysis of Texture Factors and Genetic Basis Influencing the Differences in Eating Quality between Northeast China and Japanese Japonica Rice [J]. Chinese Bulletin of Botany, 2025, 60(4): 1-0. |
| [4] | Xinyu Li, Yue Gu, Feifei Xu, Jinsong Bao. Research Progress on Post-translational Modifications of Starch Biosynthesis-related Proteins in Rice Endosperm [J]. Chinese Bulletin of Botany, 2025, 60(2): 256-270. |
| [5] | Jianguo Li, Yi Zhang, Wenjun Zhang. Iron Plaque Formation and Its Effects on Phosphorus Absorption in Rice Roots [J]. Chinese Bulletin of Botany, 2025, 60(1): 132-143. |
| [6] | Hengyu Yan, Zhaoxia Li, Yubin Li. Research Progress on Heat Stress Impact on Maize Growth and Heat-Tolerant Maize Screening in China [J]. Chinese Bulletin of Botany, 2024, 59(6): 1007-1023. |
| [7] | Ruifeng Yao, Daoxin Xie. Activation and Termination of Strigolactone Signal Perception in Rice [J]. Chinese Bulletin of Botany, 2024, 59(6): 873-877. |
| [8] | Jinjin Lian, Luyao Tang, Yinuo Zhang, Jiaxing Zheng, Chaoyu Zhu, Yuhan Ye, Yuexing Wang, Wennan Shang, Zhenghao Fu, Xinxuan Xu, Richeng Wu, Mei Lu, Changchun Wang, Yuchun Rao. Genetic Locus Mining and Candidate Gene Analysis of Antioxidant Traits in Rice [J]. Chinese Bulletin of Botany, 2024, 59(5): 738-751. |
| [9] | Jiahui Huang, Huimin Yang, Xinyu Chen, Chaoyu Zhu, Yanan Jiang, Chengxiang Hu, Jinjin Lian, Tao Lu, Mei Lu, Weilin Zhang, Yuchun Rao. Response Mechanism of Rice Mutant pe-1 to Low Light Stress [J]. Chinese Bulletin of Botany, 2024, 59(4): 574-584. |
| [10] | Jianmin Zhou. A Combat Vehicle with a Smart Brake [J]. Chinese Bulletin of Botany, 2024, 59(3): 343-346. |
| [11] | Chaoyu Zhu, Chengxiang Hu, Zhenan Zhu, Zhining Zhang, Lihai Wang, Jun Chen, Sanfeng Li, Jinjin Lian, Luyao Tang, Qianqian Zhong, Wenjing Yin, Yuexing Wang, Yuchun Rao. Mapping of QTLs Associated with Rice Panicle Traits and Candidate Gene Analysis [J]. Chinese Bulletin of Botany, 2024, 59(2): 217-230. |
| [12] | Bao Zhu, Jiangzhe Zhao, Kewei Zhang, Peng Huang. OsCKX9 is Involved in Regulating the Rice Lamina Joint Development and Leaf Angle [J]. Chinese Bulletin of Botany, 2024, 59(1): 10-21. |
| [13] | Yanli Fang, Chuanyu Tian, Ruyi Su, Yapei Liu, Chunlian Wang, Xifeng Chen, Wei Guo, Zhiyuan Ji. Mining and Preliminary Mapping of Rice Resistance Genes Against Bacterial Leaf Streak [J]. Chinese Bulletin of Botany, 2024, 59(1): 1-9. |
| [14] | Qiwei Jia, Qianqian Zhong, Yujia Gu, Tianqi Lu, Wei Li, Shuai Yang, Chaoyu Zhu, Chengxiang Hu, Sanfeng Li, Yuexing Wang, Yuchun Rao. Mapping of QTL for Cell Wall Related Components in Rice Stem and Analysis of Candidate Genes [J]. Chinese Bulletin of Botany, 2023, 58(6): 882-892. |
| [15] | Tian Chuanyu, Fang Yanli, Shen Qing, Wang Hongjie, Chen Xifeng, Guo Wei, Zhao Kaijun, Wang Chunlian, Ji Zhiyuan. Genotypic Diversity and Pathogenisity of Xanthomonas oryzae pv. oryzae Isolated from Southern China in 2019-2021 [J]. Chinese Bulletin of Botany, 2023, 58(5): 743-749. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||