

Chinese Bulletin of Botany ›› 2024, Vol. 59 ›› Issue (1): 1-9.DOI: 10.11983/CBB23071 cstr: 32102.14.CBB23071
• EXPERIMENTAL COMMUNICATIONS • Next Articles
Yanli Fang1,2,†, Chuanyu Tian1,2,†, Ruyi Su1, Yapei Liu2, Chunlian Wang2, Xifeng Chen1, Wei Guo1,*( ), Zhiyuan Ji2,*(
), Zhiyuan Ji2,*( )
)
Received:2023-05-31
Accepted:2023-12-19
Online:2024-01-10
Published:2024-01-10
Contact:
*E-mail: About author:†These authors contributed equally to this paper
Yanli Fang, Chuanyu Tian, Ruyi Su, Yapei Liu, Chunlian Wang, Xifeng Chen, Wei Guo, Zhiyuan Ji. Mining and Preliminary Mapping of Rice Resistance Genes Against Bacterial Leaf Streak[J]. Chinese Bulletin of Botany, 2024, 59(1): 1-9.

Figure 1 Diagram for the identification of rice germplasms that confer resistance to bacterial leaf streak (BLS) (A) Occurrence of bacterial leaf streak in paddy (bar=1 cm); (B) Syringe inoculation; (C) Rice materials for resistance identification
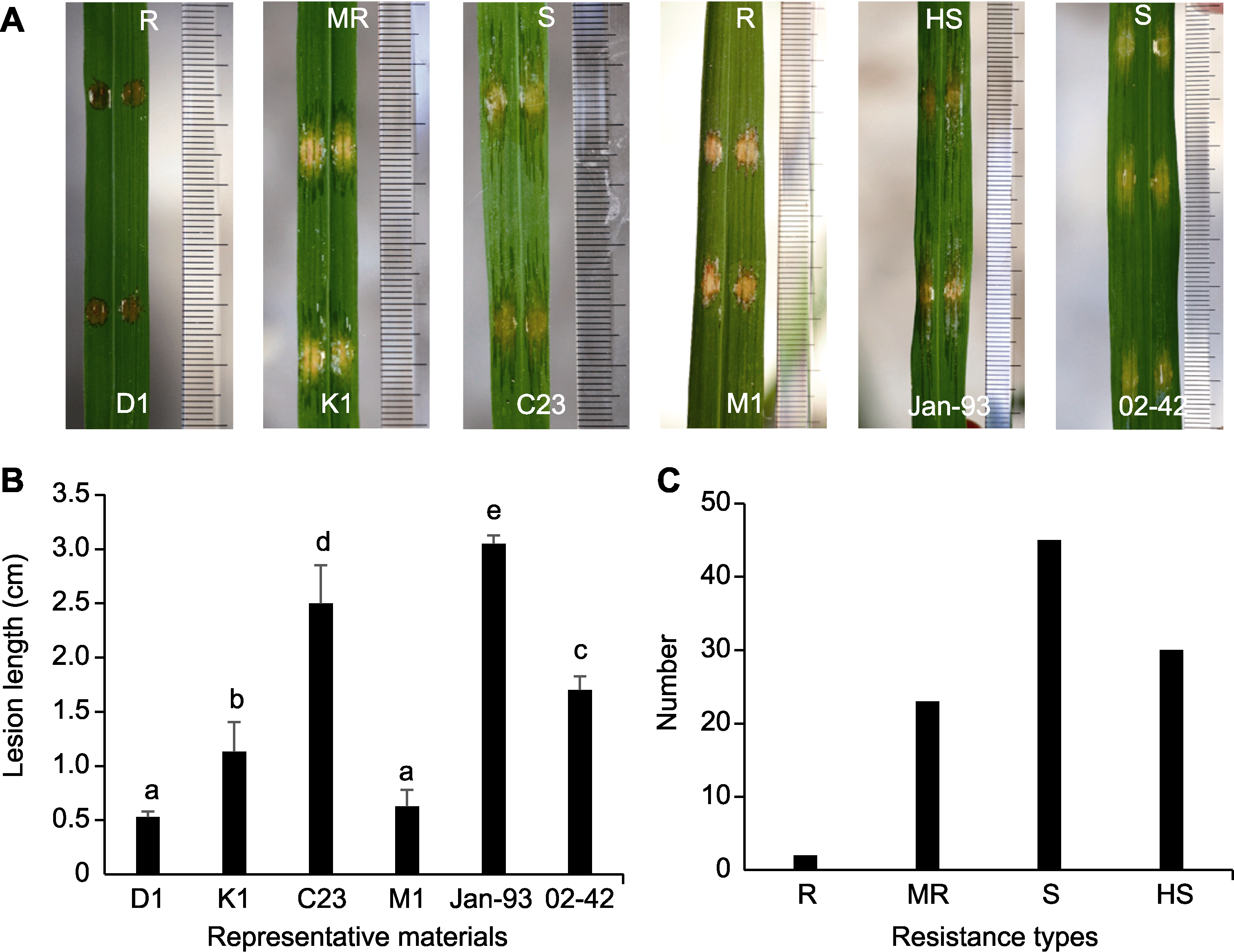
Figure 2 Identification of rice germplasms that confer resistance to bacterial leaf streak (BLS) with needleless syringes (A) Phenotype of representative rice materials; (B) Lesion length of representative rice materials (different lowercase letters above horizontal columns indicate significant statistical differences at P<0.01); (C) Distribution of rice materials in BLS disease reactions (R: High resistance; MR: Middle resistance; S: Susceptibility; HS: High susceptibility)
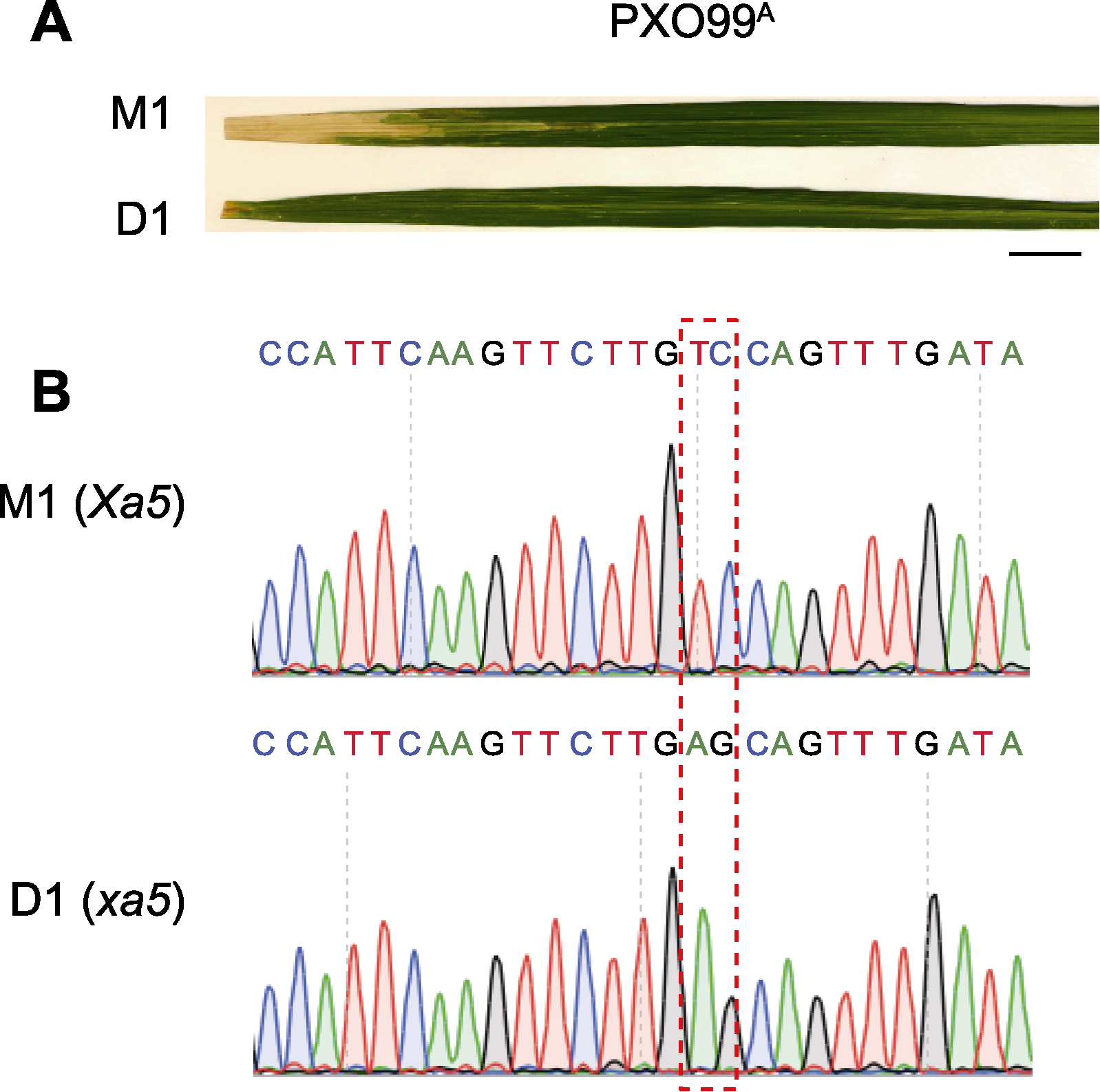
Figure 3 Identification of M1 and D1 that confer resistance to bacterial blight (BB) (A) Disease response of M1 and D1 inoculated with Xoo (PXO99A) (bar=1 cm); (B) Sequence of Xa5/xa5 locus in M1 and D1 (the red dashed box represents the mutation sites)
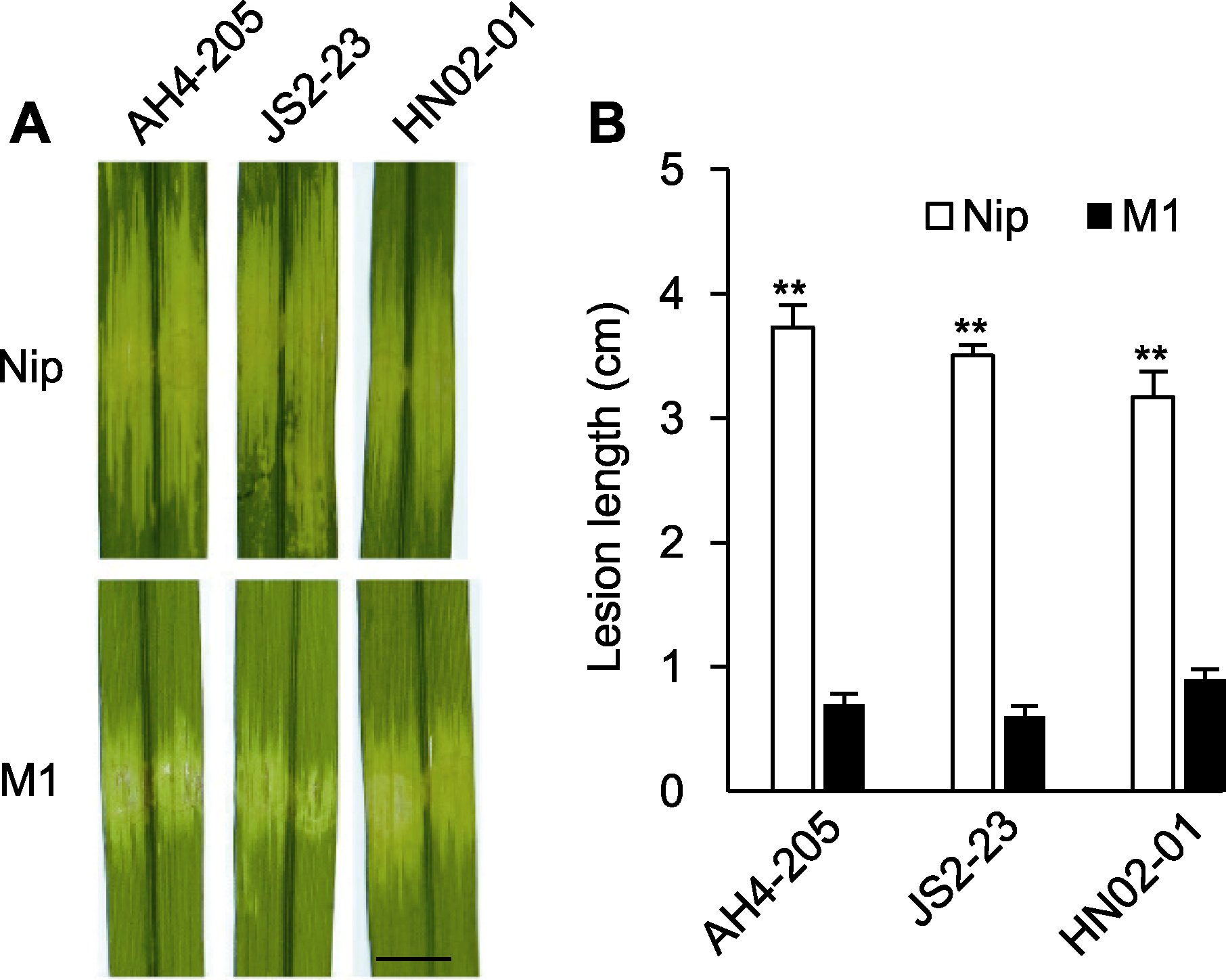
Figure 4 Bacterial leaf streak (BLS) resistance spectrum assay of M1 (A) Phenotype of hypervirulent Xoc strains in Nip and M1 (bar=0.5 cm); (B) Lesion length of hypervirulent Xoc strains in Nip and M1 (** indicate extremely significant differences (P<0.01))
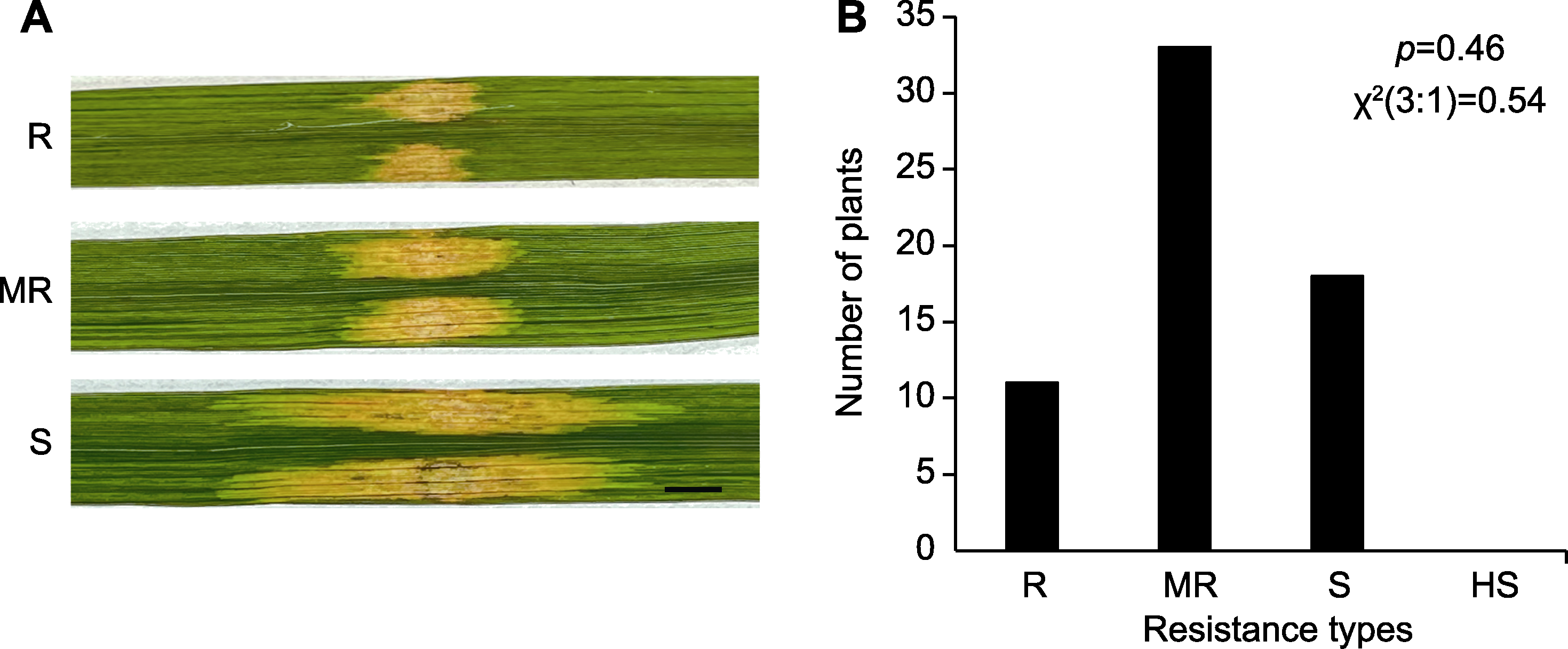
Figure 5 Genetic analysis of M1 that confers resistance to bacterial leaf streak (BLS) (A) Resistance identification of F2 plants derived from the cross between Nipponbare and M1 (bar=0.5 cm); (B) Statistical analysis of resistant and susceptible F2 plants. R, MR, S, and HS are the same as shown in Figure 2.
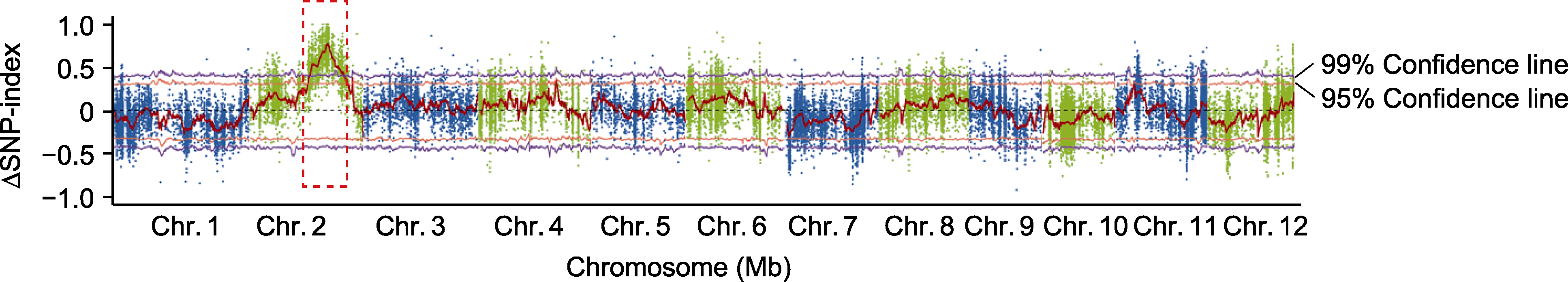
Figure 6 Mapping of bacterial leaf streak (BLS) resistance gene Xo-3 in M1 using BSA-Seq method The red dashed box represents the candidate gene region.
| [1] | 黄大辉, 岑贞陆, 刘驰, 贺文爱, 陈英之, 马增凤, 杨朗, 韦绍丽, 刘亚利, 黄思良, 杨新庆, 李容柏 (2008). 野生稻细菌性条斑病抗性资源筛选及遗传分析. 植物遗传资源学报 9, 11-14. |
| [2] | 林作晓, 龙梦玲, 唐洁瑜, 辛德育, 黄成宇 (2021). 广西2020年农作物主要病虫害发生实况. 广西植保 34, 25-32. |
| [3] |
马路, 方媛, 肖飒清, 周纯, 金哲伦, 叶雯澜, 饶玉春 (2018). 水稻条斑病抗性QTL的挖掘及相关基因的表达. 植物学报 53, 468-476.
DOI |
| [4] |
王田幸子, 朱峥, 陈悦, 刘玉晴, 燕高伟, 徐珊, 张彤, 马金姣, 窦世娟, 李莉云, 刘国振 (2021). 水稻OsWRKY42是Xa21介导的抗白叶枯病途径新元件. 植物学报 56, 687-698.
DOI |
| [5] |
Bogdanove AJ, Koebnik R, Lu H, Furutani A, Angiuoli SV, Patil PB, Van Sluys MA, Ryan RP, Meyer DF, Han SW, Aparna G, Rajaram M, Delcher AL, Phillippy AM, Puiu D, Schatz MC, Shumway M, Sommer DD, Trapnell C, Benahmed F, Dimitrov G, Madupu R, Radune D, Sullivan S, Jha G, Ishihara H, Lee SW, Pandey A, Sharma V, Sriariyanun M, Szurek B, Vera-Cruz CM, Dorman KS, Ronald PC, Verdier V, Dow JM, Sonti RV, Tsuge S, Brendel VP, Rabinowicz PD, Leach JE, White FF, Salzberg SL (2011). Two new complete genome sequences offer insight into host and tissue specificity of plant pathogenic Xanthomonas spp. J Bacteriol 193, 5450-5464.
DOI PMID |
| [6] |
Cai LL, Cao YY, Xu ZY, Ma WX, Zakria M, Zou LF, Cheng ZQ, Chen GY (2017). A transcription activator-like effector Tal7 of Xanthomonas oryzae pv. oryzicola activates rice gene Os09g29100 to suppress rice immunity. Sci Rep 7, 5089.
DOI |
| [7] |
Cernadas RA, Doyle EL, Niño-Liu DO, Wilkins KE, Bancroft T, Wang L, Schmidt CL, Caldo R, Yang B, White FF, Nettleton D, Wise RP, Bogdanove AJ (2014). Code-assisted discovery of TAL effector targets in bacterial leaf streak of rice reveals contrast with bacterial blight and a novel susceptibility gene. PLoS Pathog 10, e1003972.
DOI URL |
| [8] |
Ji CH, Ji ZY, Liu B, Cheng H, Liu H, Liu SZ, Yang B, Chen GY (2020). Xa1 allelic R genes activate rice blight resistance suppressed by interfering TAL effectors. Plant Commun 1, 100087.
DOI URL |
| [9] |
Ji ZY, Ji CH, Liu B, Zou LF, Chen GY, Yang B (2016). Interfering TAL effectors of Xanthomonas oryzae neutralize R-gene-mediated plant disease resistance. Nat Commun 7, 13435.
DOI |
| [10] |
Ji ZY, Wang CL, Zhao KJ (2018). Rice routes of countering Xanthomonas oryzae. Int J Mol Sci 19, 3008.
DOI URL |
| [11] |
Ji ZY, Zakria M, Zou LF, Xiong L, Li Z, Ji GH, Chen GY (2014). Genetic diversity of transcriptional activator-like effector genes in Chinese isolates of Xanthomonas oryzae pv. oryzicola. Phytopathology 104, 672-682.
DOI URL |
| [12] |
Jiang GH, Xia ZH, Zhou YL, Wan J, Li DY, Chen RS, Zhai WX, Zhu LH (2006). Testifying the rice bacterial blight resistance gene xa5 by genetic complementation and further analyzing xa5 (Xa5) in comparison with its homolog TFIIAγ1. Mol Genet Genomics 275, 354-366.
DOI URL |
| [13] |
Jiang N, Yan J, Liang Y, Shi YL, He ZZ, Wu YT, Zeng Q, Liu XL, Peng JH (2020). Resistance genes and their interactions with bacterial blight/leaf streak pathogens (Xanthomonas oryzae) in rice (Oryza sativa L.)—an updated review. Rice 13, 3.
DOI PMID |
| [14] |
Jones JDG, Dangl JL (2006). The plant immune system. Nature 444, 323-329.
DOI |
| [15] |
Li CY, Zhou L, Wu B, Li SH, Zha WJ, Li W, Zhou ZH, Yang LF, Shi L, Lin YJ, You AQ (2022). Improvement of bacterial blight resistance in two conventionally cultivated rice varieties by editing the noncoding region. Cells 11, 2535.
DOI URL |
| [16] |
Liu HF, Chang QL, Feng WJ, Zhang BG, Wu T, Li N, Yao FY, Ding XH, Chu ZH (2014). Domain dissection of AvrRxo1 for suppressor, avirulence and cytotoxicity functions. PLoS One 9, e113875.
DOI URL |
| [17] |
Ma ZF, Qin G, Zhang YX, Liu C, Wei MY, Cen ZL, Yan Y, Luo TP, Li ZJ, Liang HF, Huang DH, Deng GF (2021). Bacterial leaf streak 1 encoding a mitogen-activated protein kinase confers resistance to bacterial leaf streak in rice. Plant J 107, 1084-1101.
DOI URL |
| [18] |
Makino S, Sugio A, White F, Bogdanove AJ (2006). Inhibition of resistance gene-mediated defense in rice by Xanthomonas oryzae pv. oryzicola. Mol Plant Microbe Interact 19, 240-249.
DOI URL |
| [19] |
Meyer M, Cox JA, Hitchings MDT, Burgin L, Hort MC, Hodson DP, Gilligan CA (2017). Quantifying airborne dispersal routes of pathogens over continents to safeguard global wheat supply. Nat Plants 3, 780-786.
DOI PMID |
| [20] |
Ni Z, Cao YQ, Jin X, Fu ZM, Li JY, Mo XY, He YQ, Tang JL, Huang S (2021). Engineering resistance to bacterial blight and bacterial leaf streak in rice. Rice 14, 38.
DOI PMID |
| [21] |
Niño-Liu DO, Ronald PC, Bogdanove AJ (2006). Xanthomonas oryzae pathovars: model pathogens of a model crop. Mol Plant Pathol 7, 303-324.
DOI PMID |
| [22] |
Pruitt RN, Schwessinger B, Joe A, Thomas N, Liu FR, Albert M, Robinson MR, Chan LJG, Luu DD, Chen HM, Bahar O, Daudi A, De Vleesschauwer D, Caddell D, Zhang WG, Zhao XX, Li X, Heazlewood JL, Ruan DL, Majumder D, Chern M, Kalbacher H, Midha S, Patil PB, Sonti RV, Petzold CJ, Liu CC, Brodbelt JS, Felix G, Ronald PC (2015). The rice immune receptor XA21 recognizes a tyrosine-sulfated protein from a Gram-negative bacterium. Sci Adv 1, e1500245.
DOI URL |
| [23] |
Shidore T, Broeckling CD, Kirkwood JS, Long JJ, Miao JM, Zhao BY, Leach JE, Triplett LR (2017). The effector AvrRxo1 phosphorylates NAD in planta. PLoS Pathog 13, e1006442.
DOI URL |
| [24] |
Streubel J, Pesce C, Hutin M, Koebnik R, Boch J, Szurek B (2013). Five phylogenetically close rice SWEET genes confer TAL effector-mediated susceptibility to Xanthomonas oryzae pv. oryzae. New Phytol 200, 808-819.
DOI PMID |
| [25] | Tall H, Lachaux M, Diallo A, Wonni I, Tékété C, Verdier V, Szurek B, Hutin M (2022). Confirmation report of bacterial leaf streak disease of rice caused by Xanthomonas oryzae pv. oryzicola in senegal. Plant Dis 106, 2253. |
| [26] |
Tian JJ, Hui SG, Shi YR, Yuan M (2019). The key residues of OsTFIIAγ5/Xa5 protein captured by the arginine-rich TFB domain of TALEs compromising rice susceptibility and bacterial pathogenicity. J Integr Agric 18, 1178-1188.
DOI URL |
| [27] |
Timilsina S, Potnis N, Newberry EA, Liyanapathiranage P, Iruegas-Bocardo F, White FF, Goss EM, Jones JB (2020). Xanthomonas diversity, virulence and plant-pathogen interactions. Nat Rev Microbiol 18, 415-427.
DOI PMID |
| [28] |
Triplett LR, Hamilton JP, Buell CR, Tisserat NA, Verdier V, Zink F, Leach JE (2011). Genomic analysis of Xanthomonas oryzae isolates from rice grown in the United States reveals substantial divergence from known X. oryzae pathovars. Appl Environ Microb 77, 3930-3937.
DOI PMID |
| [29] |
Triplett LR, Shidore T, Long J, Miao JM, Wu SC, Han Q, Zhou CH, Ishihara H, Li JY, Zhao BY, Leach JE (2016). AvrRxo1 is a bifunctional type III secreted effector and toxin-antitoxin system component with homologs in diverse environmental contexts. PLoS One 11, e0158856.
DOI URL |
| [30] |
Wang CC, Yu H, Huang J, Wang WS, Faruquee M, Zhang F, Zhao XQ, Fu BY, Chen K, Zhang HL, Tai SS, Wei CC, McNally KL, Alexandrov N, Gao XY, Li JY, Li ZK, Xu JL, Zheng TQ (2020). Towards a deeper haplotype mining of complex traits in rice with RFGB v2.0. Plant Biotechnol J 18, 14-16.
DOI URL |
| [31] |
Wang CL, Qin TF, Yu HM, Zhang XP, Che JY, Gao Y, Zheng CK, Yang B, Zhao KJ (2014). The broad bacterial blight resistance of rice line CBB23 is triggered by a novel transcription activator-like (TAL) effector of Xanthomonas oryzae pv. oryzae. Mol Plant Pathol 15, 333-341.
DOI URL |
| [32] | Wang WS, Mauleon R, Hu ZQ, Chebotarov D, Tai SS, Wu ZC, Li M, Zheng TQ, Fuentes RR, Zhang F, Mansueto L, Copetti D, Sanciangco M, Palis KC, Xu JL, Sun C, Fu BY, Zhang HL, Gao YM, Zhao XQ, Shen F, Cui X, Yu H, Li ZC, Chen ML, Detras J, Zhou YL, Zhang XY, Zhao Y, Kudrna D, Wang CC, Li R, Jia B, Lu JY, He XC, Dong ZT, Xu JB, Li YH, Wang M, Shi JX, Li J, Zhang DB, Lee S, Hu WS, Poliakov A, Dubchak I, Ulat VJ, Borja FN, Mendoza JR, Ali J, Li J, Gao Q, Niu YC, Yue Z, Naredo MEB, Talag J, Wang XQ, Li JJ, Fang XD, Yin Y, Glaszmann JC, Zhang JW, Li JY, Hamilton RS, Wing RA, Ruan J, Zhang GY, Wei CC, Alexandrov N, McNally KL, Li ZK, Leung H (2018). Genomic variation in 3,010 diverse accessions of Asian cultivated rice. Nature 557, 43-49. |
| [33] |
White FF, Yang B (2009). Host and pathogen factors controlling the rice-Xanthomonas oryzae interaction. Plant Physiol 150, 1677-1686.
DOI URL |
| [34] |
Wu T, Zhang HM, Yuan B, Liu HF, Kong LG, Chu ZH, Ding XH (2022). Tal2b targets and activates the expression of OsF3H03g to hijack OsUGT74H4 and synergistically interfere with rice immunity. New Phytol 233, 1864-1880.
DOI URL |
| [35] |
Xu XM, Xu ZY, Li ZY, Zakria M, Zou LF, Chen GY (2021). Increasing resistance to bacterial leaf streak in rice by editing the promoter of susceptibility gene OsSULRT3;6. Plant Biotechnol J 19, 1101-1103.
DOI URL |
| [36] |
Xu ZY, Zou LF, Ma WX, Cai LL, Yang YY, Chen GY (2017). Action modes of transcription activator-like effectors (TALEs) of Xanthomonas in plants. J Integr Agric 16, 2736-2745.
DOI URL |
| [37] |
Yuan M, Ke YG, Huang RY, Ma L, Yang ZY, Chu ZH, Xiao JH, Li XH, Wang SP (2016). A host basal transcription factor is a key component for infection of rice by TALE- carrying bacteria. eLife 5, e19605.
DOI URL |
| [38] |
Yugander A, Ershad M, Muthuraman PP, Prakasam V, Ladhalakshmi D, Sheshu Madhav M, Srinivas Prasad M, Sundaram RM, Laha GS (2022). Understanding the variability of rice bacterial blight pathogen, Xanthomonas oryzae pv. oryzae in Andhra Pradesh, India. J Basic Microbiol 62, 185-196.
DOI URL |
| [39] |
Zhang SH, He XY, Zhao JL, Cheng YS, Xie ZM, Chen YH, Yang TF, Dong JF, Wang XF, Liu Q, Liu W, Mao XX, Fu H, Chen ZM, Liao YP, Liu B (2017). Identification and validation of a novel major QTL for harvest index in rice (Oryza sativa L.). Rice 10, 44.
DOI URL |
| [40] |
Zhao BY, Lin XH, Poland J, Trick H, Leach J, Hulbert S (2005). A maize resistance gene functions against bacterial streak disease in rice. Proc Natl Acad Sci USA 102, 15383-15388.
DOI PMID |
| [1] |
Juan Cui, Xiaoyu Yu, Yuejiao Yu, Chengwei Liang, Jian Sun, Wenfu Chen.
Analysis of Texture Factors and Genetic Basis Influencing the Differences in Eating Quality between Northeast China and Japanese Japonica Rice [J]. Chinese Bulletin of Botany, 2025, 60(4): 1-0. |
| [2] | Tong Miao, Wang Huan, Zhang Wenshuang, Wang Chao, Song Jianxiao. Distribution characteristics of antibiotic resistance genes in soil bacterial communities exposed to heavy metal pollution [J]. Biodiv Sci, 2025, 33(3): 24101-. |
| [3] | Zhao Ling, Guan Ju, Liang Wenhua, Zhang Yong, Lu Kai, Zhao Chunfang, Li Yusheng, Zhang Yadong. Mapping of QTLs for Heat Tolerance at the Seedling Stage in Rice Based on a High-density Bin Map [J]. Chinese Bulletin of Botany, 2025, 60(3): 342-353. |
| [4] | Xinyu Li, Yue Gu, Feifei Xu, Jinsong Bao. Research Progress on Post-translational Modifications of Starch Biosynthesis-related Proteins in Rice Endosperm [J]. Chinese Bulletin of Botany, 2025, 60(2): 256-270. |
| [5] | Jianguo Li, Yi Zhang, Wenjun Zhang. Iron Plaque Formation and Its Effects on Phosphorus Absorption in Rice Roots [J]. Chinese Bulletin of Botany, 2025, 60(1): 132-143. |
| [6] | Ruifeng Yao, Daoxin Xie. Activation and Termination of Strigolactone Signal Perception in Rice [J]. Chinese Bulletin of Botany, 2024, 59(6): 873-877. |
| [7] | Jinjin Lian, Luyao Tang, Yinuo Zhang, Jiaxing Zheng, Chaoyu Zhu, Yuhan Ye, Yuexing Wang, Wennan Shang, Zhenghao Fu, Xinxuan Xu, Richeng Wu, Mei Lu, Changchun Wang, Yuchun Rao. Genetic Locus Mining and Candidate Gene Analysis of Antioxidant Traits in Rice [J]. Chinese Bulletin of Botany, 2024, 59(5): 738-751. |
| [8] | Jiahui Huang, Huimin Yang, Xinyu Chen, Chaoyu Zhu, Yanan Jiang, Chengxiang Hu, Jinjin Lian, Tao Lu, Mei Lu, Weilin Zhang, Yuchun Rao. Response Mechanism of Rice Mutant pe-1 to Low Light Stress [J]. Chinese Bulletin of Botany, 2024, 59(4): 574-584. |
| [9] | Jianmin Zhou. A Combat Vehicle with a Smart Brake [J]. Chinese Bulletin of Botany, 2024, 59(3): 343-346. |
| [10] | Chaoyu Zhu, Chengxiang Hu, Zhenan Zhu, Zhining Zhang, Lihai Wang, Jun Chen, Sanfeng Li, Jinjin Lian, Luyao Tang, Qianqian Zhong, Wenjing Yin, Yuexing Wang, Yuchun Rao. Mapping of QTLs Associated with Rice Panicle Traits and Candidate Gene Analysis [J]. Chinese Bulletin of Botany, 2024, 59(2): 217-230. |
| [11] | Bao Zhu, Jiangzhe Zhao, Kewei Zhang, Peng Huang. OsCKX9 is Involved in Regulating the Rice Lamina Joint Development and Leaf Angle [J]. Chinese Bulletin of Botany, 2024, 59(1): 10-21. |
| [12] | Xiaoning Zeng, Penghang Wang, Mengfan Zhang, Jing Su, Zhiyuan Shi, Fuling Gao, Jiamei Li. List of the wild woody plants in Henan Province [J]. Biodiv Sci, 2023, 31(6): 22306-. |
| [13] | Dai Ruohui, Qian Xinyu, Sun Jinglei, Lu Tao, Jia Qiwei, Lu Tianqi, Lu Mei, Rao Yuchun. Research Progress on the Mechanisms of Leaf Color Regulation and Related Genes in Rice [J]. Chinese Bulletin of Botany, 2023, 58(5): 799-812. |
| [14] | Tian Chuanyu, Fang Yanli, Shen Qing, Wang Hongjie, Chen Xifeng, Guo Wei, Zhao Kaijun, Wang Chunlian, Ji Zhiyuan. Genotypic Diversity and Pathogenisity of Xanthomonas oryzae pv. oryzae Isolated from Southern China in 2019-2021 [J]. Chinese Bulletin of Botany, 2023, 58(5): 743-749. |
| [15] | Yuping Yan, Xiaoqi Yu, Deyong Ren, Qian Qian. Genetic Mechanisms and Breeding Utilization of Grain Number Per Panicle in Rice [J]. Chinese Bulletin of Botany, 2023, 58(3): 359-372. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||