

Chinese Bulletin of Botany ›› 2023, Vol. 58 ›› Issue (5): 743-749.DOI: 10.11983/CBB22183 cstr: 32102.14.CBB22183
• EXPERIMENTAL COMMUNICATIONS • Previous Articles Next Articles
Tian Chuanyu1,2, Fang Yanli1,2, Shen Qing3, Wang Hongjie1,2, Chen Xifeng1, Guo Wei1, Zhao Kaijun2, Wang Chunlian2,4( ), Ji Zhiyuan2(
), Ji Zhiyuan2( )
)
Received:2022-08-03
Accepted:2023-02-09
Online:2023-09-01
Published:2023-09-21
Contact:
*E-mail: wangchunlian@caas.cn;jizhiyuan@caas.cn
About author:First author contact:† These authors contributed equally to this paper.
Tian Chuanyu, Fang Yanli, Shen Qing, Wang Hongjie, Chen Xifeng, Guo Wei, Zhao Kaijun, Wang Chunlian, Ji Zhiyuan. Genotypic Diversity and Pathogenisity of Xanthomonas oryzae pv. oryzae Isolated from Southern China in 2019-2021[J]. Chinese Bulletin of Botany, 2023, 58(5): 743-749.
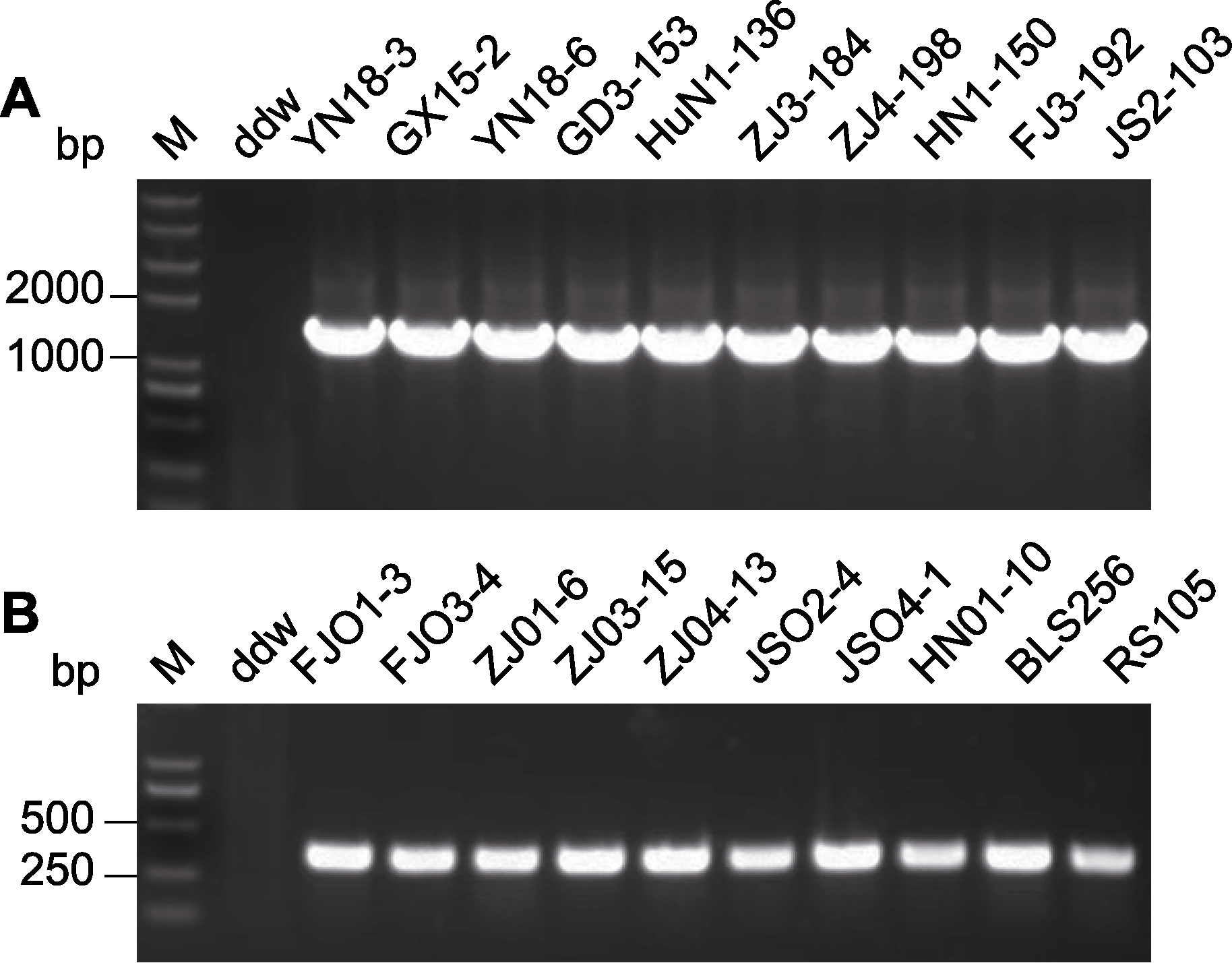
Figure 1 PCR-based detection of Xanthomonas oryzae strains isolated from diseased leaves of Southern China in 2019-2021 (A) Specific PCR detection from Xoo strains; (B) Specific PCR detection from Xoc strains. M: Marker; ddw: Control
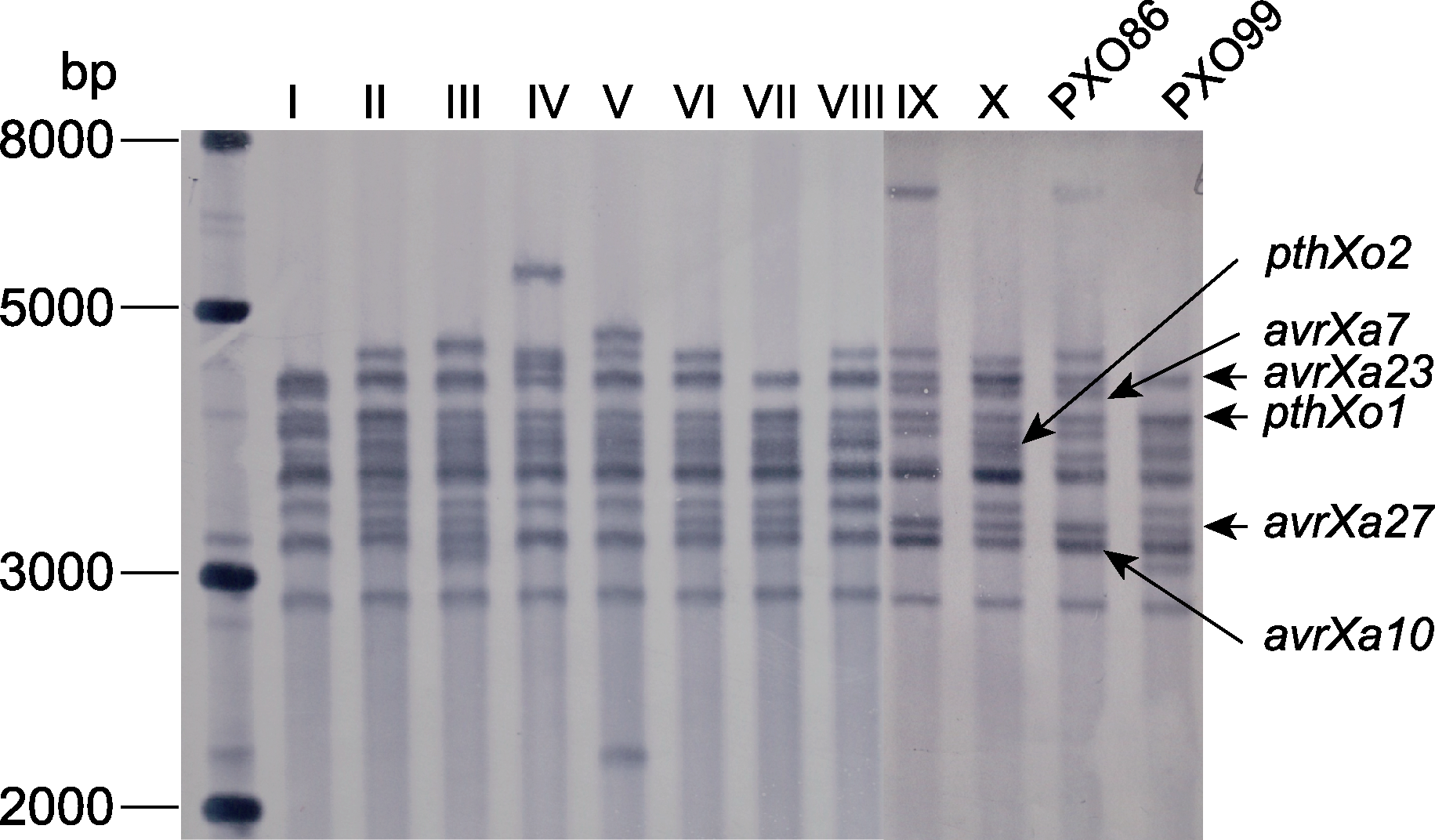
Figure 2 Southern blot analysis of tale genes of Xoo strains collected from Southern China in 2019-2021 I-X: Genotypes of isolated strains in this study; PXO86: Philippine race 2; PXO99: Philippine race 6
| tale genotype | Representative strains | Distribution | Number of strains | Frequency (%) | Putative tale gene |
|---|---|---|---|---|---|
| I | YN18-3 | Yunnan, Guangxi | 4 | 4.12 | pthXo1, avrXa7, avrXa23 |
| II | GX15-2, YN17-8 | Yunnan, Guangxi | 2 | 2.06 | pthXo1, pthXo2, avrXa23, avrXa27 |
| III | YN17-2, YN18-6 | Yunnan, Guangdong | 8 | 8.25 | pthXo1, pthXo2, avrXa10, avrXa23, avrXa27 |
| IV | GX15-4, GD3-153 | Guangxi, Guangdong, Yunnan | 18 | 18.56 | pthXo1, pthXo2, avrXa23 |
| V | ZJ1-121, HuN1-136 | Zhejiang, Hunan, Jiangsu | 27 | 27.84 | pthXo1, pthXo2, avrXa23 |
| VI | ZJ3-184 | Zhejiang, Guangdong | 5 | 5.15 | pthXo1, pthXo2, avrXa23, avrXa27 |
| VII | ZJ4-198 | Zhejiang, Jiangsu | 4 | 4.12 | pthXo1, pthXo2, avrXa23, avrXa27 |
| VIII | GD1-105, HN1-150 | Guangdong, Hainan, Zhejiang, Guangxi | 22 | 22.68 | pthXo1, pthXo2, avrXa23 |
| IX | FJ3-192 | Fujian, Guangxi | 4 | 4.12 | pthXo1, avrXa7, avrXa10, avrXa23, avrXa27 |
| X | JS2-103 | Jiangsu, Zhejiang | 3 | 3.09 | pthXo1, pthXo2, avrXa7, avrXa10, avrXa23, avrXa27 |
Table 1 tale genotypes of Xoo strains collected from Southern China in 2019-2021
| tale genotype | Representative strains | Distribution | Number of strains | Frequency (%) | Putative tale gene |
|---|---|---|---|---|---|
| I | YN18-3 | Yunnan, Guangxi | 4 | 4.12 | pthXo1, avrXa7, avrXa23 |
| II | GX15-2, YN17-8 | Yunnan, Guangxi | 2 | 2.06 | pthXo1, pthXo2, avrXa23, avrXa27 |
| III | YN17-2, YN18-6 | Yunnan, Guangdong | 8 | 8.25 | pthXo1, pthXo2, avrXa10, avrXa23, avrXa27 |
| IV | GX15-4, GD3-153 | Guangxi, Guangdong, Yunnan | 18 | 18.56 | pthXo1, pthXo2, avrXa23 |
| V | ZJ1-121, HuN1-136 | Zhejiang, Hunan, Jiangsu | 27 | 27.84 | pthXo1, pthXo2, avrXa23 |
| VI | ZJ3-184 | Zhejiang, Guangdong | 5 | 5.15 | pthXo1, pthXo2, avrXa23, avrXa27 |
| VII | ZJ4-198 | Zhejiang, Jiangsu | 4 | 4.12 | pthXo1, pthXo2, avrXa23, avrXa27 |
| VIII | GD1-105, HN1-150 | Guangdong, Hainan, Zhejiang, Guangxi | 22 | 22.68 | pthXo1, pthXo2, avrXa23 |
| IX | FJ3-192 | Fujian, Guangxi | 4 | 4.12 | pthXo1, avrXa7, avrXa10, avrXa23, avrXa27 |
| X | JS2-103 | Jiangsu, Zhejiang | 3 | 3.09 | pthXo1, pthXo2, avrXa7, avrXa10, avrXa23, avrXa27 |
| Strains | tale genotype | IR24 | JG30 | IRBB3 | IRBB4 | IRBB5 | IRBB7 | IRBB21 | IRBB27 | CBB23 | KΔSWEET | 5024 | 4059 | 4064 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| YN18-3 | I | 2.17± 1.21 b | 11.00± 1.25 a | 0.50± 0.32 b | 0.50± 0.69 b | 0.50± 0.04 b | 0.10± 0.00 b | 0.50± 0.31 b | 0.50± 0.01 b | 0.30± 0.01 b | 1.90± 0.40 b | 0.10± 0.00 b | 0.35± 0.01 b | 0.35± 0.05 b |
| GX15-2 | II | 9.00± 0.69 b | 14.00± 0.82 a | 8.67± 2.75 b | 7.17± 0.05 c | 1.67± 0.02 c | 2.50± 0.01 d | 4.44± 0.45 c | 0.10± 0.00 d | 1.88± 0.03 d | 2.13± 0.45 d | 0.10± 0.00 d | 0.11± 0.02 d | 14.33± 1.25 a |
| YN18-6 | III | 9.83± 0.02 b | 18.83± 0.96 a | 15.67± 0.01 a | 3.50± 0.03 c | 0.40± 0.01 d | 0.48± 0.17 d | 0.50± 0.00 d | 0.32± 0.01 d | 0.10± 0.02 d | 1.40± 0.01 d | 0.20± 0.01 d | 0.60± 0.01 d | 2.42± 0.01 d |
| GD3-153 | IV | 8.17± 0.90 b | 17.00± 2.58 a | 8.67± 2.98 b | 0.50± 0.01 d | 0.10± 0.00 d | 0.10± 0.02 d | 3.38± 0.02 c | 0.50± 0.02 d | 0.10± 0.01 d | 1.08± 1.92 d | 0.10± 0.00 d | 0.10± 0.01 d | 7.00± 1.63 b |
| HuN1-136 | V | 9.50± 0.58 b | 11.33± 2.08 a | 5.17± 0.94 c | 6.75± 1.34 c | 0.10± 0.02 d | 0.10± 0.01 d | 14.83± 1.25 a | 0.20± 0.01 d | 0.10± 0.10 d | 0.75± 0.22 d | 3.50± 0.01 c | 1.17± 0.02 d | 9.92± 1.21 b |
| ZJ3-184 | VI | 6.33± 0.37 b | 14.50± 1.83 a | 7.75± 0.02 b | 0.20± 0.03 c | 0.10± 0.00 c | 2.75± 0.02 c | 0.10± 0.00 c | 0.10± 0.00 c | 0.88± 0.32 c | 0.35± 0.15 c | 0.10± 0.00 c | 1.33± 0.01 c | 2.50± 0.14 c |
| ZJ4-198 | VII | 9.50± 1.21 b | 12.00± 1.07 a | 6.67± 1.70 c | 7.33± 1.61 c | 1.75± 0.32 d | 0.10± 0.01 d | 2.42± 0.03 d | 0.10± 0.00 d | 0.10± 0.01 d | 7.83± 0.21 c | 0.10± 0.02 d | 0.50± 0.01 d | 5.83± 0.25 c |
| HN1-150 | VIII | 10.33± 0.69 b | 17.17± 1.29 a | 14.00± 0.94 a | 6.17± 1.12 c | 0.33± 0.01 d | 0.53± 0.25 d | 3.86± 0.01 d | 8.67± 3.34 c | 1.21± 0.24 d | 1.10± 0.24 d | 4.00± 0.95 d | 4.83± 0.05 c | 8.17± 0.14 c |
| FJ3-192 | IX | 6.67± 1.97 b | 8.17± 1.34 ab | 4.75± 1.15 bc | 4.92± 0.69 bc | 0.10± 0.00 c | 5.08± 0.37 b | 1.00± 0.01 c | 0.10± 0.02 c | 1.42± 1.28 c | 1.50± 0.37 c | 0.83± 0.01 c | 2.67± 0.01 c | 10.33± 0.17 a |
| JS2-103 | X | 4.83± 0.96 c | 12.83± 0.75 a | 7.17± 0.69 b | 1.67± 1.18 d | 0.50± 0.00 d | 1.93± 0.07 d | 2.17± 0.00 d | 0.10± 0.00 d | 0.25± 0.07 d | 1.58± 0.43 d | 0.50± 0.02 d | 0.10± 0.00 d | 4.83± 2.89 c |
Table 2 Virulence evaluation of 10 tale genotypes of Xoo strains in rice cultivars containing different R genes
| Strains | tale genotype | IR24 | JG30 | IRBB3 | IRBB4 | IRBB5 | IRBB7 | IRBB21 | IRBB27 | CBB23 | KΔSWEET | 5024 | 4059 | 4064 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| YN18-3 | I | 2.17± 1.21 b | 11.00± 1.25 a | 0.50± 0.32 b | 0.50± 0.69 b | 0.50± 0.04 b | 0.10± 0.00 b | 0.50± 0.31 b | 0.50± 0.01 b | 0.30± 0.01 b | 1.90± 0.40 b | 0.10± 0.00 b | 0.35± 0.01 b | 0.35± 0.05 b |
| GX15-2 | II | 9.00± 0.69 b | 14.00± 0.82 a | 8.67± 2.75 b | 7.17± 0.05 c | 1.67± 0.02 c | 2.50± 0.01 d | 4.44± 0.45 c | 0.10± 0.00 d | 1.88± 0.03 d | 2.13± 0.45 d | 0.10± 0.00 d | 0.11± 0.02 d | 14.33± 1.25 a |
| YN18-6 | III | 9.83± 0.02 b | 18.83± 0.96 a | 15.67± 0.01 a | 3.50± 0.03 c | 0.40± 0.01 d | 0.48± 0.17 d | 0.50± 0.00 d | 0.32± 0.01 d | 0.10± 0.02 d | 1.40± 0.01 d | 0.20± 0.01 d | 0.60± 0.01 d | 2.42± 0.01 d |
| GD3-153 | IV | 8.17± 0.90 b | 17.00± 2.58 a | 8.67± 2.98 b | 0.50± 0.01 d | 0.10± 0.00 d | 0.10± 0.02 d | 3.38± 0.02 c | 0.50± 0.02 d | 0.10± 0.01 d | 1.08± 1.92 d | 0.10± 0.00 d | 0.10± 0.01 d | 7.00± 1.63 b |
| HuN1-136 | V | 9.50± 0.58 b | 11.33± 2.08 a | 5.17± 0.94 c | 6.75± 1.34 c | 0.10± 0.02 d | 0.10± 0.01 d | 14.83± 1.25 a | 0.20± 0.01 d | 0.10± 0.10 d | 0.75± 0.22 d | 3.50± 0.01 c | 1.17± 0.02 d | 9.92± 1.21 b |
| ZJ3-184 | VI | 6.33± 0.37 b | 14.50± 1.83 a | 7.75± 0.02 b | 0.20± 0.03 c | 0.10± 0.00 c | 2.75± 0.02 c | 0.10± 0.00 c | 0.10± 0.00 c | 0.88± 0.32 c | 0.35± 0.15 c | 0.10± 0.00 c | 1.33± 0.01 c | 2.50± 0.14 c |
| ZJ4-198 | VII | 9.50± 1.21 b | 12.00± 1.07 a | 6.67± 1.70 c | 7.33± 1.61 c | 1.75± 0.32 d | 0.10± 0.01 d | 2.42± 0.03 d | 0.10± 0.00 d | 0.10± 0.01 d | 7.83± 0.21 c | 0.10± 0.02 d | 0.50± 0.01 d | 5.83± 0.25 c |
| HN1-150 | VIII | 10.33± 0.69 b | 17.17± 1.29 a | 14.00± 0.94 a | 6.17± 1.12 c | 0.33± 0.01 d | 0.53± 0.25 d | 3.86± 0.01 d | 8.67± 3.34 c | 1.21± 0.24 d | 1.10± 0.24 d | 4.00± 0.95 d | 4.83± 0.05 c | 8.17± 0.14 c |
| FJ3-192 | IX | 6.67± 1.97 b | 8.17± 1.34 ab | 4.75± 1.15 bc | 4.92± 0.69 bc | 0.10± 0.00 c | 5.08± 0.37 b | 1.00± 0.01 c | 0.10± 0.02 c | 1.42± 1.28 c | 1.50± 0.37 c | 0.83± 0.01 c | 2.67± 0.01 c | 10.33± 0.17 a |
| JS2-103 | X | 4.83± 0.96 c | 12.83± 0.75 a | 7.17± 0.69 b | 1.67± 1.18 d | 0.50± 0.00 d | 1.93± 0.07 d | 2.17± 0.00 d | 0.10± 0.00 d | 0.25± 0.07 d | 1.58± 0.43 d | 0.50± 0.02 d | 0.10± 0.00 d | 4.83± 2.89 c |
| [1] | 张华, 姜英华, 胡白石, 刘凤权, 许志刚 (2008). 利用PCR技术专化性检测水稻细菌性条斑病菌. 植物病理学报 38, 1-5. |
| [2] |
Antony G, Zhou JH, Huang S, Li T, Liu B, White F, Yang B (2010). Rice xa13 recessive resistance to bacterial blight is defeated by induction of the disease susceptibility gene Os-11N3. Plant Cell 22, 3864-3876.
DOI URL |
| [3] | Boch J, Bonas U, Lahaye T (2014). TAL effectors-pathogen strategies and plant resistance engineering. New Phy- tol 204, 823-832. |
| [4] |
Breia R, Conde A, Badim H, Fortes AM, Gerós H, Granell A (2021). Plant SWEETs: from sugar transport to plant-pathogen interaction and more unexpected physiological roles. Plant Physiol 186, 836-852.
DOI PMID |
| [5] |
Doyle EL, Stoddard BL, Voytas DF, Bogdanove AJ (2013). TAL effectors: highly adaptable phytobacterial virulence factors and readily engineered DNA-targeting proteins. Trends Cell Biol 23, 390-398.
DOI PMID |
| [6] |
Ji CH, Ji ZY, Liu B, Cheng H, Liu H, Liu SZ, Yang B, Chen GY (2020). Xa1 Allelic R genes activate rice blight resistance suppressed by interfering TAL effectors. Plant Commun 1, 100087.
DOI URL |
| [7] |
Ji ZY, Guo W, Chen XF, Wang CL, Zhao KJ (2022). Plant executor genes. Int J Mol Sci 23, 1524.
DOI URL |
| [8] |
Ji ZY, Ji CH, Liu B, Zou LF, Chen GY, Yang B (2016). Interfering TAL effectors of Xanthomonas oryzae neutralize R-gene-mediated plant disease resistance. Nat Commun 7, 13435.
DOI |
| [9] |
Ji ZY, Wang CL, Zhao KJ (2018). Rice routes of countering Xanthomonas oryzae. Int J Mol Sci 19, 3008.
DOI URL |
| [10] |
Ji ZY, Zakria M, Zou LF, Xiong L, Li Z, Ji GH, Chen GY (2014). Genetic diversity of transcriptional activator-like effector genes in Chinese isolates of Xanthomonas oryzae pv. oryzicola. Phytopathology 104, 672-682.
DOI URL |
| [11] |
Niño-Liu DO, Ronald PC, Bogdanove AJ (2006). Xanthomonas oryzae pathovars: model pathogens of a model crop. Mol Plant Pathol 7, 303-324.
DOI PMID |
| [12] |
Nowack MK, Holmes DR, Lahaye T (2022). TALE-induced cell death executors: an origin outside immunity? Trends Plant Sci 27, 536-548.
DOI URL |
| [13] |
Oliva R, Ji CH, Atienza-Grande G, Huguet-Tapia JC, Perez-Quintero A, Li T, Eom JS, Li CH, Nguyen H, Liu B, Auguy F, Sciallano C, Luu VT, Dossa GS, Cunnac S, Schmidt SM, Slamet-Loedin IH, Vera Cruz C, Szurek B, Frommer WB, White FF, Yang B (2019). Broad- spectrum resistance to bacterial blight in rice using genome editing. Nat Biotechnol 37, 1344-1350.
DOI |
| [14] |
Perez-Quintero AL, Szurek B (2019). A decade decoded: spies and hackers in the history of TAL effectors research. Annu Rev Phytopathol 57, 459-481.
DOI PMID |
| [15] | Sambrook J, Fritsch EF, Maniatis T (1989). Molecular Cloning: A Laboratory Manual, 2nd edn. Cold Spring Harbor: Cold Spring Harbor Laboratory. |
| [16] | Wang CL, Qin TF, Yu HM, Zhang XP, Che JY, Gao Y, Zheng CK, Yang B, Zhao KJ (2014). The broad bacterial blight resistance of rice line CBB23 is triggered by a novel transcription activator-like (TAL) effector of Xanthomonas oryzae pv. oryzae. Mol Plant Pathol 15, 333-341. |
| [17] |
White FF, Yang B (2009). Host and pathogen factors controlling the rice-Xanthomonas oryzae interaction. Plant Physiol 150, 1677-1686.
DOI URL |
| [18] |
Xu ZY, Xu XM, Gong Q, Li ZY, Li Y, Wang S, Yang YY, Ma WX, Liu LY, Zhu B, Zou LF, Chen GY (2019). Engineering broad-spectrum bacterial blight resistance by simultaneously disrupting variable TALE-binding elements of multiple susceptibility genes in rice. Mol Plant 12, 1434-1446.
DOI PMID |
| [19] |
Xu ZY, Zou LF, Ma WX, Cai LL, Yang YY, Chen GY (2017). Action modes of transcription activator-like effectors (TALEs) of Xanthomonas in plants. J Integr Agric 16, 2736-2745.
DOI URL |
| [20] |
Yang B, Sugio A, White FF (2006). Os8N3 is a host disease-susceptibility gene for bacterial blight of rice. Proc Natl Acad Sci USA 103, 10503-10508.
DOI URL |
| [21] |
Zhang BM, Zhang HT, Li F, Ouyang YD, Yuan M, Li XH, Xiao JH, Wang SP (2020). Multiple alleles encoding atypical NLRs with unique central tandem repeats in rice confer resistance to Xanthomonas oryzae pv. oryzae. Plant Commun 1, 100088.
DOI URL |
| [22] |
Zhou JH, Peng Z, Long JY, Sosso D, Liu B, Eom JS, Huang S, Liu SZ, Vera Cruz C, Frommer WB, White FF, Yang B (2015). Gene targeting by the TAL effector PthXo2 reveals cryptic resistance gene for bacterial blight of rice. Plant J 82, 632-643.
DOI URL |
| [23] |
Zhou JM, Zhang YL (2020). Plant immunity: danger perception and signaling. Cell 181, 978-989.
DOI URL |
| [1] |
Juan Cui, Xiaoyu Yu, Yuejiao Yu, Chengwei Liang, Jian Sun, Wenfu Chen.
Analysis of Texture Factors and Genetic Basis Influencing the Differences in Eating Quality between Northeast China and Japanese Japonica Rice [J]. Chinese Bulletin of Botany, 2025, 60(4): 1-0. |
| [2] | Tong Miao, Wang Huan, Zhang Wenshuang, Wang Chao, Song Jianxiao. Distribution characteristics of antibiotic resistance genes in soil bacterial communities exposed to heavy metal pollution [J]. Biodiv Sci, 2025, 33(3): 24101-. |
| [3] | Zhao Ling, Guan Ju, Liang Wenhua, Zhang Yong, Lu Kai, Zhao Chunfang, Li Yusheng, Zhang Yadong. Mapping of QTLs for Heat Tolerance at the Seedling Stage in Rice Based on a High-density Bin Map [J]. Chinese Bulletin of Botany, 2025, 60(3): 342-353. |
| [4] | Xinyu Li, Yue Gu, Feifei Xu, Jinsong Bao. Research Progress on Post-translational Modifications of Starch Biosynthesis-related Proteins in Rice Endosperm [J]. Chinese Bulletin of Botany, 2025, 60(2): 256-270. |
| [5] | Jianguo Li, Yi Zhang, Wenjun Zhang. Iron Plaque Formation and Its Effects on Phosphorus Absorption in Rice Roots [J]. Chinese Bulletin of Botany, 2025, 60(1): 132-143. |
| [6] | Ruifeng Yao, Daoxin Xie. Activation and Termination of Strigolactone Signal Perception in Rice [J]. Chinese Bulletin of Botany, 2024, 59(6): 873-877. |
| [7] | Jinjin Lian, Luyao Tang, Yinuo Zhang, Jiaxing Zheng, Chaoyu Zhu, Yuhan Ye, Yuexing Wang, Wennan Shang, Zhenghao Fu, Xinxuan Xu, Richeng Wu, Mei Lu, Changchun Wang, Yuchun Rao. Genetic Locus Mining and Candidate Gene Analysis of Antioxidant Traits in Rice [J]. Chinese Bulletin of Botany, 2024, 59(5): 738-751. |
| [8] | Jiahui Huang, Huimin Yang, Xinyu Chen, Chaoyu Zhu, Yanan Jiang, Chengxiang Hu, Jinjin Lian, Tao Lu, Mei Lu, Weilin Zhang, Yuchun Rao. Response Mechanism of Rice Mutant pe-1 to Low Light Stress [J]. Chinese Bulletin of Botany, 2024, 59(4): 574-584. |
| [9] | Jianmin Zhou. A Combat Vehicle with a Smart Brake [J]. Chinese Bulletin of Botany, 2024, 59(3): 343-346. |
| [10] | Chaoyu Zhu, Chengxiang Hu, Zhenan Zhu, Zhining Zhang, Lihai Wang, Jun Chen, Sanfeng Li, Jinjin Lian, Luyao Tang, Qianqian Zhong, Wenjing Yin, Yuexing Wang, Yuchun Rao. Mapping of QTLs Associated with Rice Panicle Traits and Candidate Gene Analysis [J]. Chinese Bulletin of Botany, 2024, 59(2): 217-230. |
| [11] | Bao Zhu, Jiangzhe Zhao, Kewei Zhang, Peng Huang. OsCKX9 is Involved in Regulating the Rice Lamina Joint Development and Leaf Angle [J]. Chinese Bulletin of Botany, 2024, 59(1): 10-21. |
| [12] | Yanli Fang, Chuanyu Tian, Ruyi Su, Yapei Liu, Chunlian Wang, Xifeng Chen, Wei Guo, Zhiyuan Ji. Mining and Preliminary Mapping of Rice Resistance Genes Against Bacterial Leaf Streak [J]. Chinese Bulletin of Botany, 2024, 59(1): 1-9. |
| [13] | Dai Ruohui, Qian Xinyu, Sun Jinglei, Lu Tao, Jia Qiwei, Lu Tianqi, Lu Mei, Rao Yuchun. Research Progress on the Mechanisms of Leaf Color Regulation and Related Genes in Rice [J]. Chinese Bulletin of Botany, 2023, 58(5): 799-812. |
| [14] | Yuping Yan, Xiaoqi Yu, Deyong Ren, Qian Qian. Genetic Mechanisms and Breeding Utilization of Grain Number Per Panicle in Rice [J]. Chinese Bulletin of Botany, 2023, 58(3): 359-372. |
| [15] | Shang Sun, Yingying Hu, Yangshuo Han, Chao Xue, Zhiyun Gong. Double-stranded Labelled Oligo-FISH in Rice Chromosomes [J]. Chinese Bulletin of Botany, 2023, 58(3): 433-439. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||