

Chinese Bulletin of Botany ›› 2023, Vol. 58 ›› Issue (3): 433-439.DOI: 10.11983/CBB22055 cstr: 32102.14.CBB22055
• TECHNIQUES AND METHODS • Previous Articles Next Articles
Shang Sun1, Yingying Hu1, Yangshuo Han2, Chao Xue2, Zhiyun Gong1,2( )
)
Received:2022-03-24
Accepted:2022-08-04
Online:2023-05-01
Published:2023-05-17
Contact:
*E-mail: zygong@yzu.edu.cn
Shang Sun, Yingying Hu, Yangshuo Han, Chao Xue, Zhiyun Gong. Double-stranded Labelled Oligo-FISH in Rice Chromosomes[J]. Chinese Bulletin of Botany, 2023, 58(3): 433-439.
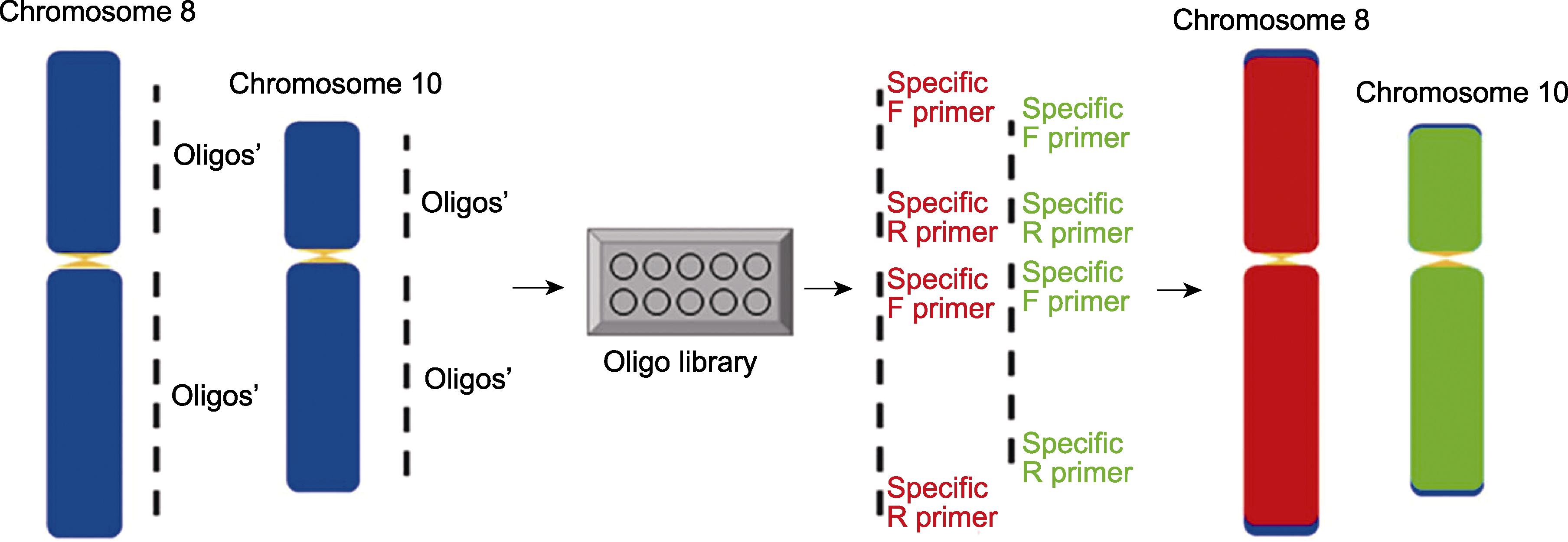
Figure 1 Establishment of double-stranded oligo-painting probes based on multiplex PCR in rice The blue rectangles are chromosomes; the small yellow triangles are centromeres; the red rectangles represent TAMRA-labelled chromosome 8 oligo signals; the green rectangles represent FAM-labelled chromosome 10 oligo signals. Specific forward and reverse primers for the amplification of different chromosomal segments represented by F and R, respectively. The dashed line represents the selected specific oligo set with connectors at both ends.
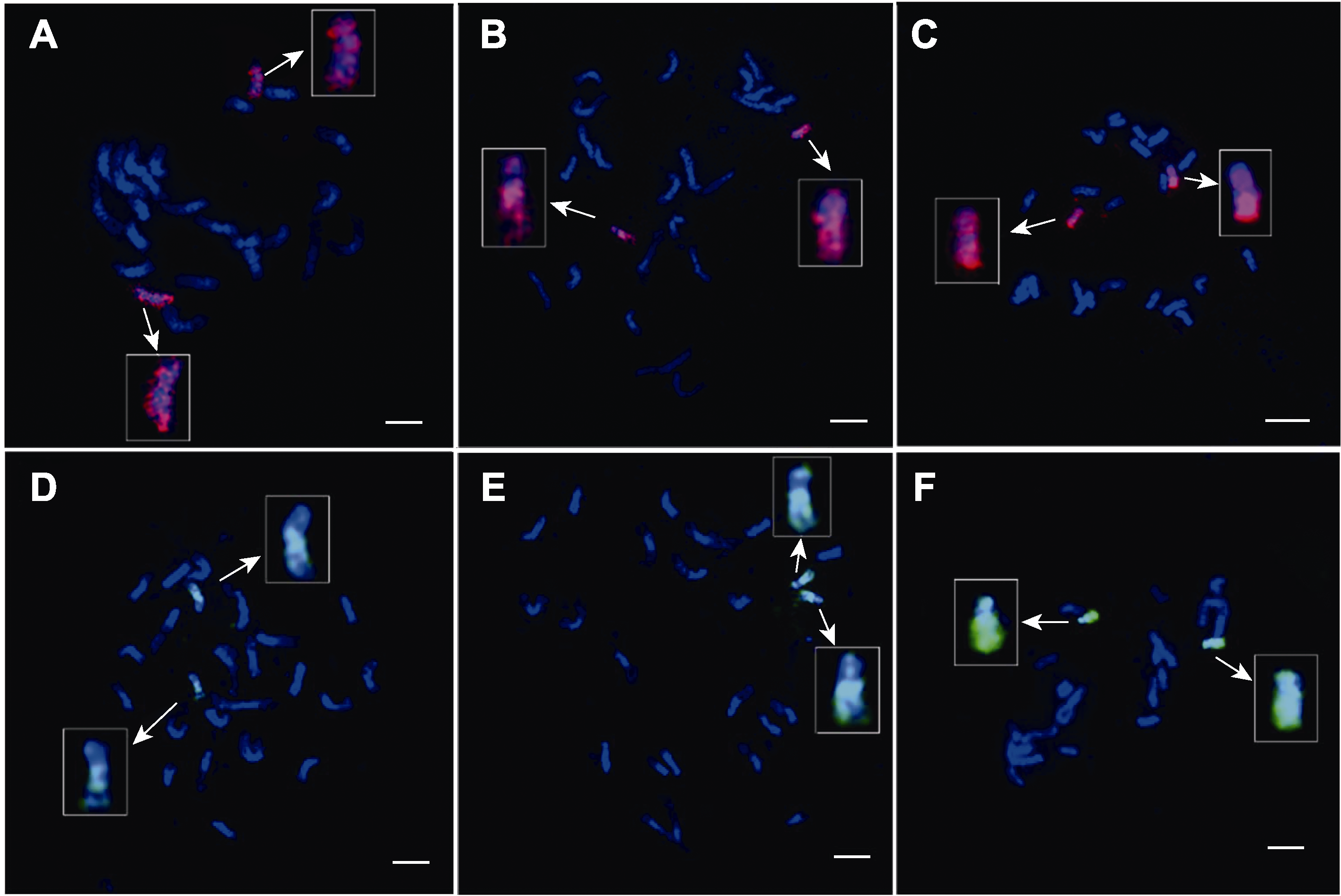
Figure 2 Exploring the denaturation time of rice chromosomes in double-stranded oligo-FISH (A)-(C) During the experiment, the effects of oligo signals labelled by TAMRA on chromosome 8 while the denaturation time at 1 min 30 s, 2 min 30 s and 3 min 30 s, respectively. (D)-(F) During the experiment, the effects of oligo signals labelled by FAM on chromosome 10 while the denaturation time at 1 min 30 s, 2 min 30 s and 3 min 30 s, respectively. The white arrow points to the painting-chromosomes after magnification. Chromosomes were counterstained with DAPI. Bars=5 μm
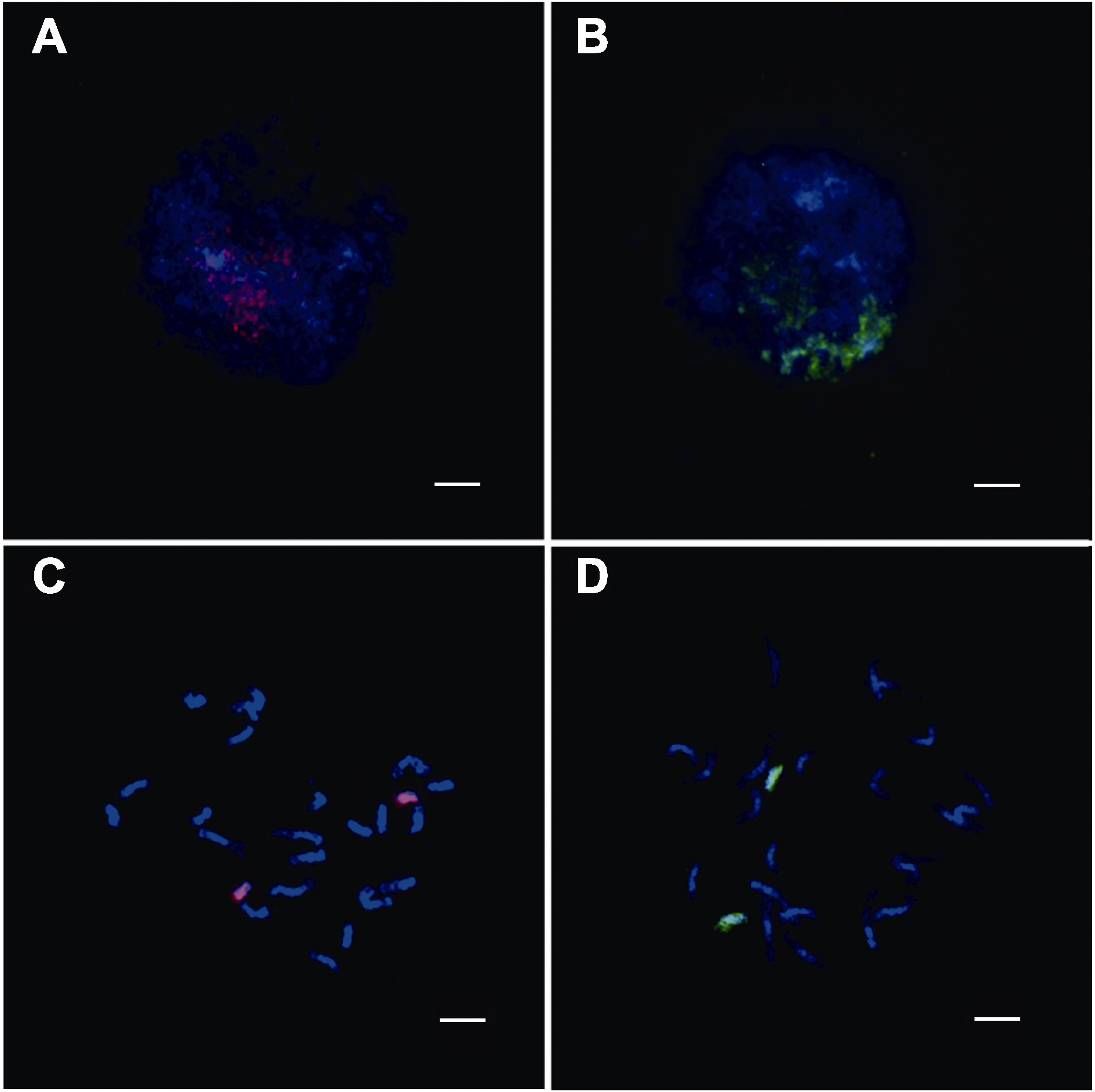
Figure 3 Double-stranded labelled oligo-FISH based on chromosome painting targets the mitosis of chromosome 8 and 10 analysis of Nipponbare (A) Oligo-painting analysis on mitotic interphase chromoso-me 8 of Nipponbare; (B) Oligo-painting analysis on mitotic interphase chromosome 10 of Nipponbare; (C) Oligo-painting analysis on mitotic prometaphase chromosome 8 of Nippon-bare; (D) Oligo-painting analysis on mitotic prometaphase chromosome 10 of Nipponbare. Red: TAMRA-labelled probe signals of whole chromosome 8; Green: FAM-labelled probe signals of whole chromosome 10. Chromosomes were coun-terstained with DAPI. Bars=5 μm
| [1] |
Anderson LK, Stack SM, Mitchell JB (1982). An investiga-tion of the basis of a current hypothesis for the lack of G- banding in plant chromosomes. Exp Cell Res 138, 433-436.
PMID |
| [2] | Ban C, Chung S, Park DS, Shim YB (2004). Detection of protein-DNA interaction with a DNA probe: distinction between single-strand and double-strand DNA-protein interaction. Nucleic Acids Res 32, e110. |
| [3] |
Bi YF, Zhao QZ, Yan WK, Li MX, Liu YX, Cheng CY, Zhang L, Yu XQ, Li J, Qian CT, Wu YF, Chen JF, Lou QF (2020). Flexible chromosome painting based on multiplex PCR of oligo-nucleotides and its application for comparative chro-mosome analyses in Cucumis. Plant J 102, 178-186.
DOI URL |
| [4] |
Braz GT, do Vale Martins L, Zhang T, Albert PS, Birchler JA, Jiang JM, (2020). A universal chromosome identifica-tion system for maize and wild Zea species. Chromosome Res 28, 183-194.
DOI |
| [5] |
Braz GT, He L, Zhao HN, Zhang T, Semrau K, Rouillard JM, Torres GA, Jiang JM (2018). Comparative oligo-FISH mapping: an efficient and powerful methodology to reveal Karyo-typic and chromosomal evolution. Genetics 208, 513-523.
DOI URL |
| [6] |
do Vale Martins L, Yu F, Zhao HN, Dennison T, Lauter N, Wang HY, Deng ZH, Thompson A, Semrau K, Rouillard JM, Birchler JA, Jiang JM (2019). Meiotic crossovers characterized by haplotype-specific chromosome painting in maize. Nat Commun 10, 4604.
DOI PMID |
| [7] |
Fuchs J, Houben A, Brandes A, Schubert I (1996). Chro-mosome ‘painting’ in plants—a feasible technique? Chromosoma 104, 315-320.
PMID |
| [8] |
Hou LL, Xu M, Zhang T, Xu ZH, Wang WY, Zhang JX, Yu MM, Ji W, Zhu CW, Gong ZY, Gu MH, Jiang JM, Yu HX (2018). Chromosome painting and its applications in culti-vated and wild rice. BMC Plant Biol 18, 110.
DOI |
| [9] |
Huang W, Du Y, Zhao X, Jin WW (2016). B chromosome contains active genes and impacts the transcription of A chromosomes in maize (Zea mays L.). BMC Plant Biol 16, 88.
DOI PMID |
| [10] |
Jiang J, Gill BS, Wang GL, Ronald PC, Ward DC (1995). Metaphase and interphase fluorescence in situ hybridiza-tion mapping of the rice genome with bacterial artificial chromosomes. Proc Natl Acad Sci USA 92, 4487-4491.
DOI PMID |
| [11] |
Jiang JM (2019). Fluorescence in situ hybridization in plants: recent developments and future applications. Chromosome Res 27, 153-165.
DOI |
| [12] |
Jiang JM, Gill BS (2006). Current status and the future of fluorescence in situ hybridization (FISH) in plant genome research. Genome 49, 1057-1068.
DOI URL |
| [13] |
Landegent JE, Jansen in de Wal N, Dirks RW, van der Ploeg M (1987). Use of whole cosmid cloned genomic sequences for chromosomal localization by non-radioactive in situ hybridization. Hum Genet 77, 366-370.
PMID |
| [14] |
Langer-Safer PR, Levine M, Ward DC (1982). Immunological method for mapping genes on drosophila polytene chromosomes. Proc Natl Acad Sci USA 79, 4381-4385.
DOI PMID |
| [15] |
Li ZA, Bi YF, Wang X, Wang YZ, Yang SQ, Zhang ZT, Chen JF, Lou QF (2018). Chromosome identification in Cucumis anguria revealed by cross-species single-copy gene FISH. Genome 61, 397-404.
DOI URL |
| [16] |
Liu XY, Sun S, Wu Y, Zhou Y, Gu SW, Yu HX, Yi CD, Gu MH, Jiang JM, Liu B, Zhang T, Gong ZY (2020). Dual- color oligo-FISH can reveal chromosomal variations and evolution in Oryza species. Plant J 101, 112-121.
DOI URL |
| [17] |
Lou QF, Zhang YX, He YH, Li J, Jia L, Cheng CY, Guan W, Yang SQ, Chen JF (2014). Single-copy gene-based chromosome painting in cucumber and its application for chromosome rearrangement analysis in Cucumis. Plant J 78, 169-179.
DOI URL |
| [18] |
Mukai Y, Nakahara Y, Yamamoto M (1993). Simultaneous discrimination of the three genomes in hexaploid wheat by multicolor fluorescence in situ hybridization using total ge-nomic and highly repeated DNA probes. Genome 36, 489-494.
DOI PMID |
| [19] | Ouyang S, Zhu W, Hamilton J, Lin HN, Campbell M, Childs K, Thibaud-Nissen F, Malek RL, Lee Y, Zheng L, Orvis J, Haas B, Wortman J, Buell CR (2007). The TIGR rice genome annotation resource: improvements and new features. Nucleic Acids Res 35, D883-D887. |
| [20] |
Šimonikova D, Němečková A, Čížková J, Brown A, Swennen R, Doležel J, Hřibová E (2020). Chromosome painting in cultivated bananas and their wild relatives (Musa spp.) reveals differences in chromosome structure. Int J Mol Sci 21, 7915.
DOI URL |
| [21] |
Tang XM, Bao WD, Zhang WL, Cheng ZK (2007). Identifica-tion of chromosomes from multiple rice genomes using a universal molecular cytogenetic marker system. J Integr Plant Biol 49, 953-960.
DOI URL |
| [22] | Trent JM, Thompson FH (1987). Methods for chromosome banding of human and experimental tumors in vitro. Methods Enzymol 151, 267-279. |
| [23] |
Xin HY, Zhang T, Wu YF, Zhang WL, Zhang PD, Xi ML, Jiang JM (2020). An extraordinarily stable karyotype of the woody Populus species revealed by chromosome painting. Plant J 101, 253-264.
DOI URL |
| [24] |
Zhang T, Liu GQ, Zhao HN, Braz GT, Jiang JM (2021). Chorus2: design of genome-scale oligonucleotide-based probes for fluorescence in situ hybridization. Plant Biotechnol J 19, 1967-1978.
DOI PMID |
| [25] |
Zhao QZ, Meng Y, Wang PQ, Qin XD, Cheng CY, Zhou JG, Yu XQ, Li J, Lou QF, Jahn M, Chen JF (2021). Reconst-ruction of ancestral karyotype illuminates chromosome evolution in the genus Cucumis. Plant J 107, 1243-1259.
DOI URL |
| [1] |
Juan Cui, Xiaoyu Yu, Yuejiao Yu, Chengwei Liang, Jian Sun, Wenfu Chen.
Analysis of Texture Factors and Genetic Basis Influencing the Differences in Eating Quality between Northeast China and Japanese Japonica Rice [J]. Chinese Bulletin of Botany, 2025, 60(4): 1-0. |
| [2] | Zhao Ling, Guan Ju, Liang Wenhua, Zhang Yong, Lu Kai, Zhao Chunfang, Li Yusheng, Zhang Yadong. Mapping of QTLs for Heat Tolerance at the Seedling Stage in Rice Based on a High-density Bin Map [J]. Chinese Bulletin of Botany, 2025, 60(3): 342-353. |
| [3] | Xinyu Li, Yue Gu, Feifei Xu, Jinsong Bao. Research Progress on Post-translational Modifications of Starch Biosynthesis-related Proteins in Rice Endosperm [J]. Chinese Bulletin of Botany, 2025, 60(2): 256-270. |
| [4] | Jianguo Li, Yi Zhang, Wenjun Zhang. Iron Plaque Formation and Its Effects on Phosphorus Absorption in Rice Roots [J]. Chinese Bulletin of Botany, 2025, 60(1): 132-143. |
| [5] | Ruifeng Yao, Daoxin Xie. Activation and Termination of Strigolactone Signal Perception in Rice [J]. Chinese Bulletin of Botany, 2024, 59(6): 873-877. |
| [6] | Jinjin Lian, Luyao Tang, Yinuo Zhang, Jiaxing Zheng, Chaoyu Zhu, Yuhan Ye, Yuexing Wang, Wennan Shang, Zhenghao Fu, Xinxuan Xu, Richeng Wu, Mei Lu, Changchun Wang, Yuchun Rao. Genetic Locus Mining and Candidate Gene Analysis of Antioxidant Traits in Rice [J]. Chinese Bulletin of Botany, 2024, 59(5): 738-751. |
| [7] | Jiahui Huang, Huimin Yang, Xinyu Chen, Chaoyu Zhu, Yanan Jiang, Chengxiang Hu, Jinjin Lian, Tao Lu, Mei Lu, Weilin Zhang, Yuchun Rao. Response Mechanism of Rice Mutant pe-1 to Low Light Stress [J]. Chinese Bulletin of Botany, 2024, 59(4): 574-584. |
| [8] | Jianmin Zhou. A Combat Vehicle with a Smart Brake [J]. Chinese Bulletin of Botany, 2024, 59(3): 343-346. |
| [9] | Chaoyu Zhu, Chengxiang Hu, Zhenan Zhu, Zhining Zhang, Lihai Wang, Jun Chen, Sanfeng Li, Jinjin Lian, Luyao Tang, Qianqian Zhong, Wenjing Yin, Yuexing Wang, Yuchun Rao. Mapping of QTLs Associated with Rice Panicle Traits and Candidate Gene Analysis [J]. Chinese Bulletin of Botany, 2024, 59(2): 217-230. |
| [10] | Yanli Fang, Chuanyu Tian, Ruyi Su, Yapei Liu, Chunlian Wang, Xifeng Chen, Wei Guo, Zhiyuan Ji. Mining and Preliminary Mapping of Rice Resistance Genes Against Bacterial Leaf Streak [J]. Chinese Bulletin of Botany, 2024, 59(1): 1-9. |
| [11] | Bao Zhu, Jiangzhe Zhao, Kewei Zhang, Peng Huang. OsCKX9 is Involved in Regulating the Rice Lamina Joint Development and Leaf Angle [J]. Chinese Bulletin of Botany, 2024, 59(1): 10-21. |
| [12] | Tian Chuanyu, Fang Yanli, Shen Qing, Wang Hongjie, Chen Xifeng, Guo Wei, Zhao Kaijun, Wang Chunlian, Ji Zhiyuan. Genotypic Diversity and Pathogenisity of Xanthomonas oryzae pv. oryzae Isolated from Southern China in 2019-2021 [J]. Chinese Bulletin of Botany, 2023, 58(5): 743-749. |
| [13] | Dai Ruohui, Qian Xinyu, Sun Jinglei, Lu Tao, Jia Qiwei, Lu Tianqi, Lu Mei, Rao Yuchun. Research Progress on the Mechanisms of Leaf Color Regulation and Related Genes in Rice [J]. Chinese Bulletin of Botany, 2023, 58(5): 799-812. |
| [14] | Yuqiang Liu, Jianmin Wan. The Host Controls the Protein Level of Insect Effectors to Balance Immunity and Growth [J]. Chinese Bulletin of Botany, 2023, 58(3): 353-355. |
| [15] | Jiayi Jin, Yiting Luo, Huimin Yang, Tao Lu, Hanfei Ye, Jiyi Xie, Kexin Wang, Qianyu Chen, Yuan Fang, Yuexing Wang, Yuchun Rao. QTL Mapping and Expression Analysis on Candidate Genes Related to Chlorophyll Content in Rice [J]. Chinese Bulletin of Botany, 2023, 58(3): 394-403. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||