INTRODUCTION: Nicotinamide mononucleotide (NMN) has important biological activities such as anti-cancer, anti-aging and improving crop stress resistance, and its importance as a nutritional health product has been established. However, the content of NMN in plants is low, and the metabolic pathway of NMN degradation is poorly understood. It has been reported that 5'-nucleotidases can catalyze the dephosphorylation of NMN in Saccharomyces cerevisiae. At present, 5'-nucleotidases have been isolated in plants, but whether they can catalyze the degradation of NMN remains unclear. Edible kale (Brassica oleracea var. acephala) has high nutritional value. It is important to analyze the metabolic pathway of NMN in kale and increase the content of NMN by blocking the degradation pathway.
RATIONALE: The degradation of NMN in plants is closely related to the NAD+ remediation synthesis (pyridine nucleotide cycle) pathway. Compared with bacteria and mammals, studies on the biosynthetic pathway of NAD+ remediation in plants mainly use isotope tracer method, lack specific gene and function analysis, and only a few related studies have been reported in plants. Eight 5'-nucleotidase genes were cloned from B. oleraceavar. acephala, heterologous expression of them was performed by Escherichia coli expression system, and the catalytic properties of 5'-nucleotidase were investigated by enzymological means in vitro.
RESULTS: In this study, ten 5'-nucleotidase genes were retrieved from the genome of B. oleracea. Based on these sequences, eight 5'-nucleotidase candidate genes were successfully cloned from B. oleracea var. acephala, which laid a foundation for further revealing the degradation pathway of NMN. Phylogenetic analysis revealed that 5'-nucleotidase is conserved in plants, suggesting that it may play an important role in plant nucleotide metabolism. The catalytic properties of 5'-nucleotides in kale were investigated by using the expression system of E. coli. In vitro enzymatic experiments showed that 5'-nucleotides can catalyze purine, pyrimidine and pyridine nucleotides, and have a wide range of substrate adaptability. Specifically, BolN2, BolN5-X1 and BolN6 can catalyze the dephosphorylation of NMN to the generation of NR, which proves that 5'-nucleotidase can catalyze the degradation of NMN in plants. In addition, BolN2, BolN5 and BolN6 can catalyze the hydrolysis of pyridine nucleotides NaMN, purine and pyrimidine nucleotides (including AMP, GMP, CMP and UMP). However, BolN7 and BolN8 had only weak catalytic activity against GMP.
CONCLUSION: The 5'-nucleotidase gene from the HAD and SurE families of B. oleracea var. acephalawas cloned and phylogenetic analysis showed that it was conserved in plants. The results of enzymatic reaction in vitro showed that BolN2, BolN5-X1 and BolN6 could catalyze the degradation of NMN to produce NR. In addition, BolN2, BolN5 and BolN6 have catalytic effects on NaNM, purine and pyrimidine nucleotides. This study further enhanced our understanding of the NMN metabolic pathway in kale, and provided a theoretical basis for creating edible kale new germplasm with high NMN content.
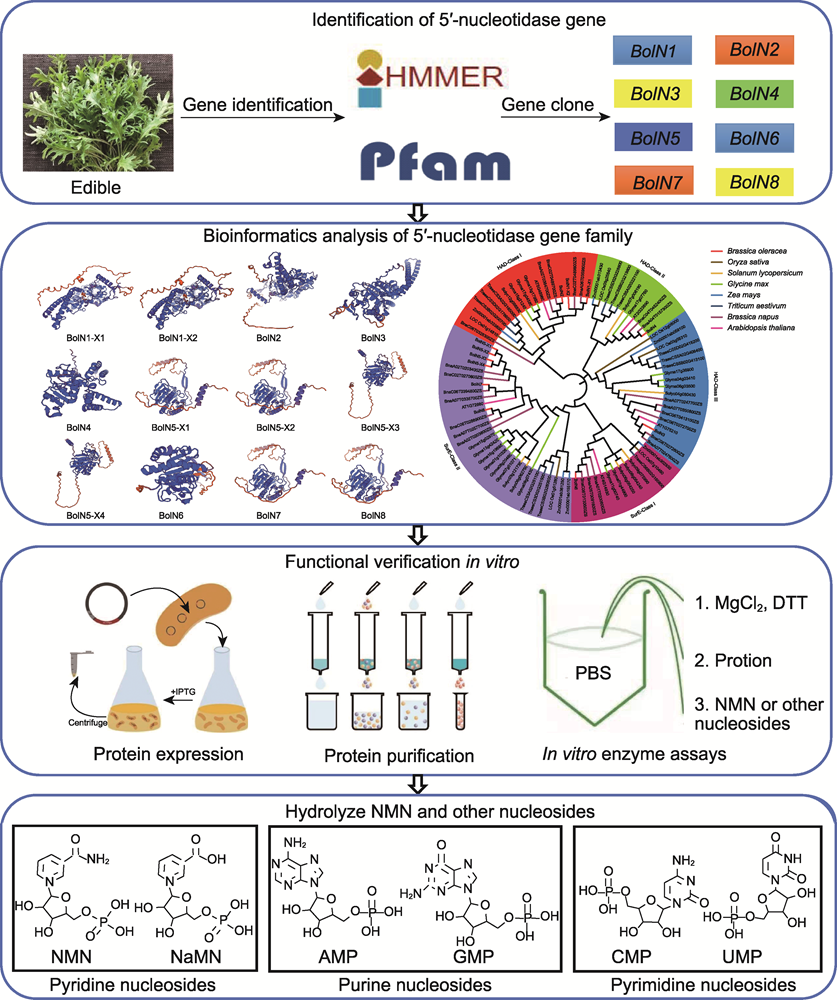
Cloning and functional analysis of 5'-nucleotidase gene catalyzing NMN degradation to NR inBrassica oleraceavar. acephala.The 5'-nucleotidase gene of B. oleraceavar. acephala was successfully cloned and phylogenetic analysis showed that it was conserved in plants. By constructing prokaryotic expression vector, 5'-nucleotidase was expressed and purified in Escherichia coli. In vitro enzymatic experiments showed that 5'-nucleotidase could catalyze the dephosphorylation of NMN to NR, and had catalytic effects on NaMN, purine nucleotides (AMP, GMP) and pyrimidine nucleotides (CMP, UMP).

 Table of Content
Table of Content
 Home
Home