

Chinese Bulletin of Botany ›› 2024, Vol. 59 ›› Issue (6): 883-902.DOI: 10.11983/CBB24089 cstr: 32102.14.CBB24089
Special Issue: 大食物观; 粮食安全; 玉米生物学与分子设计(2024年59卷6期)
• INVITED REVIEWS • Previous Articles Next Articles
Ziyang Wang, Shengxue Liu, Zhirui Yang, Feng Qin*( )
)
Received:2024-06-07
Accepted:2024-07-23
Online:2024-11-10
Published:2024-08-01
Contact:
*E-mail: qinfeng@cau.edu.cn
Ziyang Wang, Shengxue Liu, Zhirui Yang, Feng Qin. Genetic Dissection of Drought Resistance in Maize[J]. Chinese Bulletin of Botany, 2024, 59(6): 883-902.
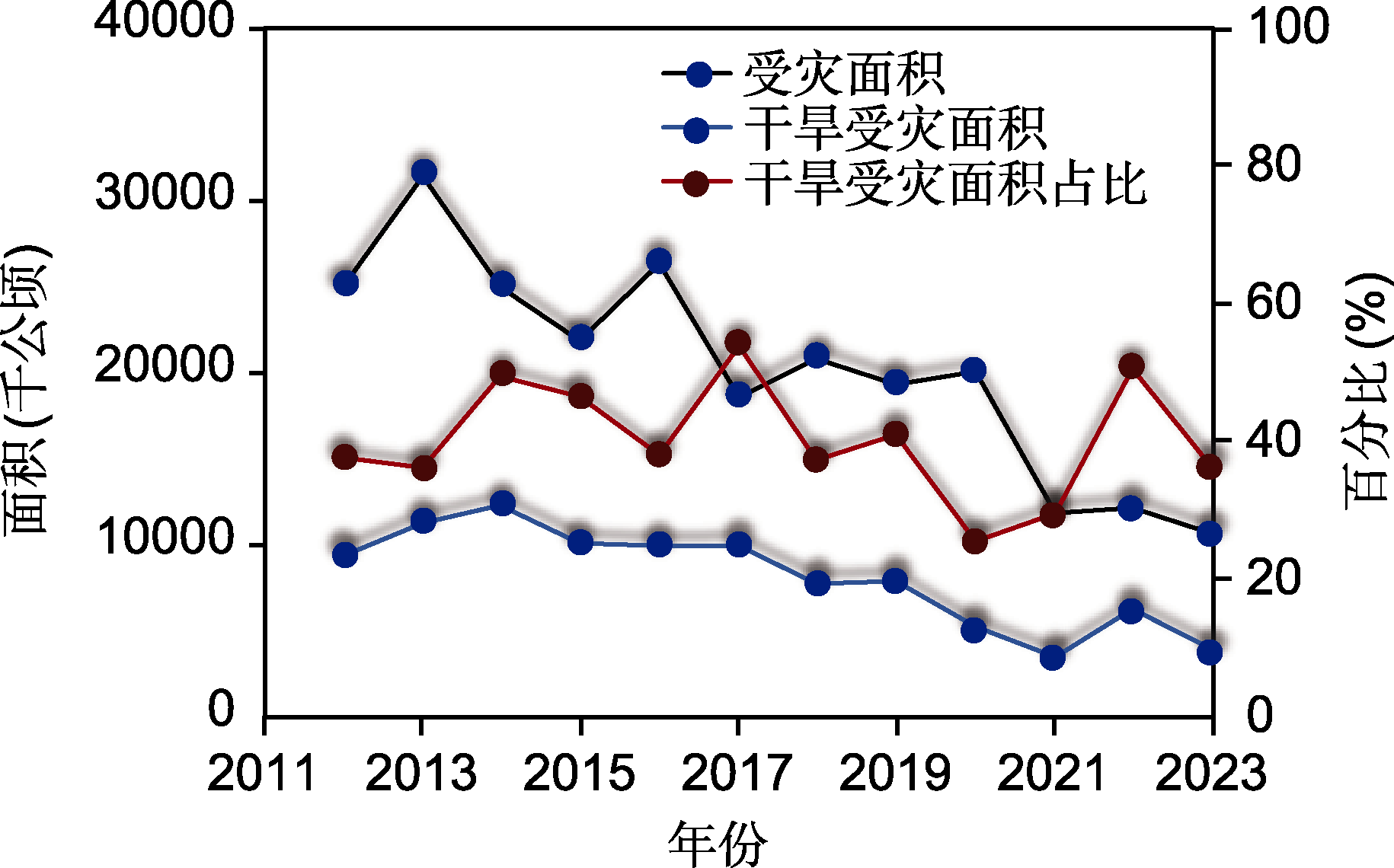
Figure 1 Percentage of crop affected by drought out of all abiotic stress in China from 2012 to 2023 (data were obtained from China Statistical Abstract 2023)
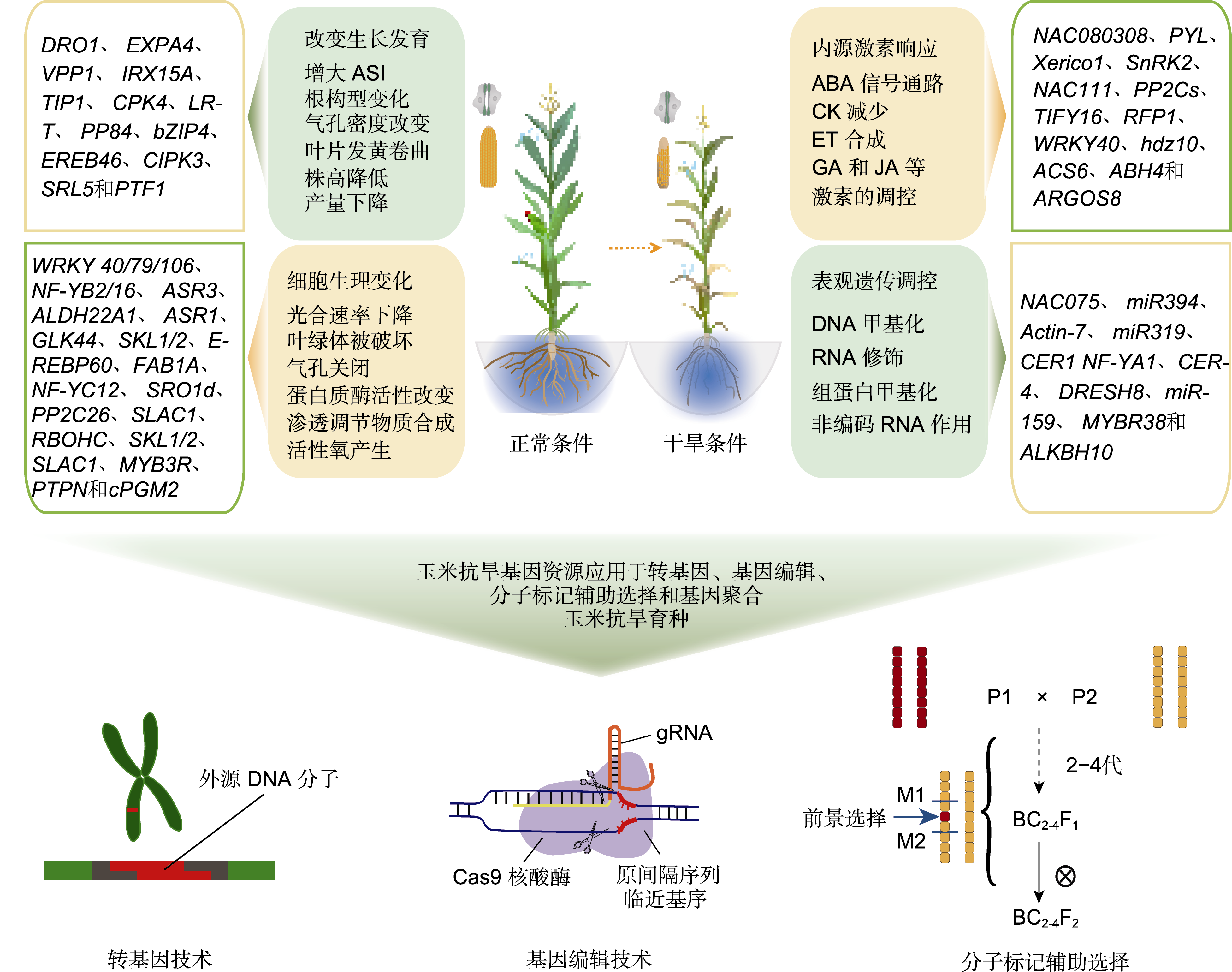
Figure 2 Different ways of maize response to drought stress and potential approaches of genetic improvement of drought resistance P1: Donor parent with drought resistant genes; P2: The recurrent parent with excellent agronomic traits but drought resistance that needs improvement. All the genes are cloned from Zea mays. For simplicity, ‘Zm’ and ‘Zea mays’ are omitted in gene names. ABA: Abscisic acid; CK: Cytokinin; ET: Endothelin; GA: Gibberellin; JA: Jasmonic acid; ASI: Anthesis and silking interval; M1, M2: Molecular marker
| 抗旱基因 | 基因功能 | 参考文献 |
|---|---|---|
| ABH4 | 编码脱落酸8'-羟化酶, 调控脱落酸的水平 | Blankenagel et al., |
| ACS6 | 编码ACC合酶 | Young et al., |
| ALDH22A1 | 编码醛脱氢酶 | Huang et al., |
| ARGOS8 | 乙烯反应的负调控因子 | Shi et al., |
| ASR1 | 调控支链氨基酸的生物合成 | Virlouvet et al., |
| ALKBH10 | 编码m6A脱甲基酶 | Miao et al., |
| bZIP4 | 促进侧根数量增加和主根伸长 | Ma et al., |
| CIPK3 | 提高干旱胁迫下种子根的长度 | Li et al., |
| cPGM2 | 编码磷酸葡萄糖变位酶, 参与糖代谢 | Wu et al., |
| CPK4 | 参与钙信号转导及脱落酸介导的气孔关闭 | Jiang et al., |
| DRESH8 | 产生siRNA, 调控干旱应答和种子大小基因的表达 | Sun et al., |
| DRO1 | 根尖向重力性和根系构型 | Feng et al., |
| EREB46 | 叶片表皮蜡质积累 | Yang et al., |
| EXPA4 | 促进雌穂生长, 减小干旱诱导产生的散粉和吐丝时间间隔 | Liu et al., |
| FAB1A | 编码1-磷脂酰肌醇-4-磷酸-5-激酶, 参与磷酸肌醇代谢 | Wu et al., |
| GLK44 | 调控玉米的色氨酸合成 | Zhang et al., |
| IRX15A | 负调控叶片的气孔密度 | Zhang et al., |
| LRT | 抑制玉米侧根的起始和伸长 | Zhang et al., |
| NAC111 | 调控气孔关闭, 提高水分利用效率 | Mao et al., |
| NF-YA1 | 茉莉酸信号转导、组蛋白修饰和染色质重塑 | Yang et al., |
| PP2C26 | 负调控脱落酸信号转导 | Lu et al., |
| PP84 | 抑制气孔关闭 | Guo et al., |
| PTF1 | 促进根系发育和脱落酸合成 | Li et al., |
| PTPN | 编码新型核苷酸酶 | Zhang et al., |
| RBOHC | 编码NADPH氧化酶 | Gao et al., |
| RFP1 | 编码E3连接酶 | Xia et al., |
| Rtn16 | 编码网状蛋白, 增强液泡H+-ATPase活性 | Tian et al., |
| SKL1/2 | 减少干旱下活性氧(reactive oxygen species, ROS)的积累 | Liu et al., |
| SRL5 | 参与角质层蜡质结构的形成 | Pan et al., |
| SRO1d | 提高气孔的ROS含量, 促进气孔关闭 | Gao et al., |
| TIFY16 | 与茉莉酸信号途径的MYC2互作 | Zhang et al., |
| TIP1 | 编码S-酰基转移酶, 促进根毛伸长 | Zhang et al., |
| VPP1 | 编码质膜H+-ATPase, 促进根系发育 | Wang et al., |
| WRKY106 | 增强抗氧化相关酶活性, 降低ROS含量 | Wang et al., |
| WRKY40 | 增强抗氧化相关酶活性, 降低ROS含量 | Wang et al., |
| WRKY79 | 促进脱落酸的生物合成 | Gulzar et al., |
| Xerico1 | 调控脱落酸的代谢稳态 | Brugière et al., |
Table 1 Drought-resistant genes identified in maize and the main functions
| 抗旱基因 | 基因功能 | 参考文献 |
|---|---|---|
| ABH4 | 编码脱落酸8'-羟化酶, 调控脱落酸的水平 | Blankenagel et al., |
| ACS6 | 编码ACC合酶 | Young et al., |
| ALDH22A1 | 编码醛脱氢酶 | Huang et al., |
| ARGOS8 | 乙烯反应的负调控因子 | Shi et al., |
| ASR1 | 调控支链氨基酸的生物合成 | Virlouvet et al., |
| ALKBH10 | 编码m6A脱甲基酶 | Miao et al., |
| bZIP4 | 促进侧根数量增加和主根伸长 | Ma et al., |
| CIPK3 | 提高干旱胁迫下种子根的长度 | Li et al., |
| cPGM2 | 编码磷酸葡萄糖变位酶, 参与糖代谢 | Wu et al., |
| CPK4 | 参与钙信号转导及脱落酸介导的气孔关闭 | Jiang et al., |
| DRESH8 | 产生siRNA, 调控干旱应答和种子大小基因的表达 | Sun et al., |
| DRO1 | 根尖向重力性和根系构型 | Feng et al., |
| EREB46 | 叶片表皮蜡质积累 | Yang et al., |
| EXPA4 | 促进雌穂生长, 减小干旱诱导产生的散粉和吐丝时间间隔 | Liu et al., |
| FAB1A | 编码1-磷脂酰肌醇-4-磷酸-5-激酶, 参与磷酸肌醇代谢 | Wu et al., |
| GLK44 | 调控玉米的色氨酸合成 | Zhang et al., |
| IRX15A | 负调控叶片的气孔密度 | Zhang et al., |
| LRT | 抑制玉米侧根的起始和伸长 | Zhang et al., |
| NAC111 | 调控气孔关闭, 提高水分利用效率 | Mao et al., |
| NF-YA1 | 茉莉酸信号转导、组蛋白修饰和染色质重塑 | Yang et al., |
| PP2C26 | 负调控脱落酸信号转导 | Lu et al., |
| PP84 | 抑制气孔关闭 | Guo et al., |
| PTF1 | 促进根系发育和脱落酸合成 | Li et al., |
| PTPN | 编码新型核苷酸酶 | Zhang et al., |
| RBOHC | 编码NADPH氧化酶 | Gao et al., |
| RFP1 | 编码E3连接酶 | Xia et al., |
| Rtn16 | 编码网状蛋白, 增强液泡H+-ATPase活性 | Tian et al., |
| SKL1/2 | 减少干旱下活性氧(reactive oxygen species, ROS)的积累 | Liu et al., |
| SRL5 | 参与角质层蜡质结构的形成 | Pan et al., |
| SRO1d | 提高气孔的ROS含量, 促进气孔关闭 | Gao et al., |
| TIFY16 | 与茉莉酸信号途径的MYC2互作 | Zhang et al., |
| TIP1 | 编码S-酰基转移酶, 促进根毛伸长 | Zhang et al., |
| VPP1 | 编码质膜H+-ATPase, 促进根系发育 | Wang et al., |
| WRKY106 | 增强抗氧化相关酶活性, 降低ROS含量 | Wang et al., |
| WRKY40 | 增强抗氧化相关酶活性, 降低ROS含量 | Wang et al., |
| WRKY79 | 促进脱落酸的生物合成 | Gulzar et al., |
| Xerico1 | 调控脱落酸的代谢稳态 | Brugière et al., |
| [1] |
高冠龙, 冯起, 张小由, 司建华, 鱼腾飞 (2018). 植物叶片光合作用的气孔与非气孔限制研究综述. 干旱区研究 35, 929-937.
DOI |
| [2] | 高辉, 张红芳, 袁思安, 李小婷 (2013). 植物内源激素对干旱胁迫的响应研究. 绿色科技 (11), 5-7. |
| [3] | 高世斌, 冯质雷, 李晚忱, 荣廷昭 (2005). 干旱胁迫下玉米根系性状和产量的QTLs分析. 作物学报 31, 718-722. |
| [4] | 国家统计局 (2023). 中国统计摘要-2023. 北京: 中国统计出版社. pp. 1-212. |
| [5] | 郭莹, 化青春, 虎梦霞, 王勇, 袁俊秀, 杨芳萍 (2023). 分子标记辅助选择在作物育种中的应用及展望. 寒旱农业科学 2, 785-790. |
| [6] | 金锐, 张从合, 朱全贵, 苏法 (2021). 分子标记辅助选择在玉米抗病和抗虫育种上的应用. 安徽农业科学 49(16), 10-15. |
| [7] | 宋博研, 朱卫国 (2011). 组蛋白甲基化修饰效应分子的研究进展. 遗传 33, 285-292. |
| [8] | 宋凤斌, 戴俊英 (2004). 干旱胁迫对玉米花粉和花丝表面超微结构及两者活力的影响. 吉林农业大学学报 26, 1-5. |
| [9] | 尹立林, 马云龙, 项韬, 朱猛进, 余梅, 李新云, 刘小磊, 赵书红 (2019). 全基因组选择模型研究进展及展望. 畜牧兽医学报 50, 233-242. |
| [10] | 赵坤 (2019). 我国玉米生产现状及发展趋势. 新农业 (12), 49. |
| [11] | 周文期, 刘忠祥, 王晓娟, 何海军, 周玉乾, 杨彦忠, 连晓荣, 李永生 (2022). 利用BSA-Seq方法快速定位作物农艺性状QTL/基因概述. 甘肃农业科技 53(4), 1-10. |
| [12] |
周文期, 周玉乾, 李永生, 何海军, 杨彦忠, 王晓娟, 连晓荣, 刘忠祥, 胡筑兵 (2023). 玉米ZmICE2基因调控气孔发育. 植物学报 58, 866-881.
DOI |
| [13] |
Apel K, Hirt H (2004). Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu Rev Plant Biol 55, 373-399.
PMID |
| [14] |
Armstrong F, Leung J, Grabov A, Brearley J, Giraudat J, Blatt MR (1995). Sensitivity to abscisic acid of guard-cell K+ channels is suppressed by abi1-1, a mutant Arabidopsis gene encoding a putative protein phosphatase. Proc Natl Acad Sci USA 92, 9520-9524.
PMID |
| [15] | Blankenagel S, Eggels S, Frey M, Grill E, Bauer E, Dawid C, Fernie AR, Haberer G, Hammerl R, Barbosa Medeiros D, Ouzunova M, Presterl T, Ruß V, Schäufele R, Schlüter U, Tardieu F, Urbany C, Urzinger S, Weber APM, Schön CC, Avramova V (2022). Natural alleles of the abscisic acid catabolism gene ZmAbh4 modulate water use efficiency and carbon isotope discrimination in maize. Plant Cell 34, 3860-3872. |
| [16] |
Brugière N, Zhang WJ, Xu QZ, Scolaro EJ, Lu C, Kahsay RY, Kise R, Trecker L, Williams RW, Hakimi S, Niu XP, Lafitte R, Habben JE (2017). Overexpression of RING domain E3 ligase ZmXerico1 confers drought tolerance through regulation of ABA homeostasis. Plant Physiol 175, 1350-1369.
DOI PMID |
| [17] |
Caine RS, Yin XJ, Sloan J, Harrison EL, Mohammed U, Fulton T, Biswal AK, Dionora J, Chater CC, Coe RA, Bandyopadhyay A, Murchie EH, Swarup R, Quick WP, Gray JE (2019). Rice with reduced stomatal density conserves water and has improved drought tolerance under future climate conditions. New Phytol 221, 371-384.
DOI PMID |
| [18] | Cao LR, Ma CC, Ye FY, Pang YY, Wang GR, Fahim AM, Lu XM (2023). Genome-wide identification of NF-Y gene family in maize (Zea mays L.) and the positive role of ZmNF-YC12 in drought resistance and recovery ability. Front Plant Sci 14, 1159955. |
| [19] |
Castorina G, Domergue F, Chiara M, Zilio M, Persico M, Ricciardi V, Horner DS, Consonni G (2020). Drought- responsive ZmFDL1/MYB94 regulates cuticle biosynthesis and cuticle-dependent leaf permeability. Plant Physiol 184, 266-282.
DOI PMID |
| [20] |
de Carvalho MHC (2008). Drought stress and reactive oxygen species: production, scavenging and signaling. Plant Signal Behav 3, 156-165.
DOI PMID |
| [21] | Doudna JA, Charpentier E (2014). Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 346, 1258096. |
| [22] | Du LY, Huang XL, Ding L, Wang ZX, Tang DL, Chen B, Ao LJY, Liu YL, Kang ZS, Mao HD (2023). TaERF87 and TaAKS1 synergistically regulate TaP5CS1/TaP5CR1-mediated proline biosynthesis to enhance drought tolerance in wheat. New Phytol 237, 232-250. |
| [23] | Feng XJ, Jia L, Cai YT, Guan HR, Zheng D, Zhang WX, Xiong H, Zhou HM, Wen Y, Hu Y, Zhang XM, Wang QJ, Wu FK, Xu J, Lu YL (2022). ABA-inducible DEEPER ROOTING 1 improves adaptation of maize to water deficiency. Plant Biotechnol J 20, 2077-2088. |
| [24] |
Flint-Garcia SA, Thornsberry JM, Buckler ES (2003). Structure of linkage disequilibrium in plants. Annu Rev Plant Biol 54, 357-374.
PMID |
| [25] | Fujii H, Chinnusamy V, Rodrigues A, Rubio S, Antoni R, Park SY, Cutler SR, Sheen J, Rodriguez PL, Zhu JK (2009). In vitro reconstitution of an abscisic acid signaling pathway. Nature 462, 660-664. |
| [26] | Gaffney J, Schussler J, Löffler C, Cai WG, Paszkiewicz S, Messina C, Groeteke J, Keaschall J, Cooper M (2015). Industry-scale evaluation of maize hybrids selected for increased yield in drought-stress conditions of the US corn belt. Crop Sci 55, 1608-1618. |
| [27] | Gao HJ, Cui JJ, Liu SX, Wang SH, Lian YY, Bai YT, Zhu TF, Wu HH, Wang YJ, Yang SP, Li XF, Zhuang JH, Chen LM, Gong ZZ, Qin F (2022). Natural variations of ZmSRO1d modulate the trade-off between drought resistance and yield by affecting ZmRBOHC-mediated stomatal ROS production in maize. Mol Plant 15, 1558-1574. |
| [28] | Gill SS, Tuteja N (2010). Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol Biochem 48, 909-930. |
| [29] | Gulzar F, Fu JY, Zhu CY, Yan J, Li XL, Meraj TA, Shen QQ, Hassan B, Wang Q (2021). Maize WRKY transcription factor ZmWRKY79 positively regulates drought tolerance through elevating ABA biosynthesis. Int J Mol Sci 22, 10080. |
| [30] | Guo YZ, Shi YB, Wang YL, Liu F, Li Z, Qi JS, Wang Y, Zhang JB, Yang SH, Wang Y, Gong ZZ (2023). The clade F PP2C phosphatase ZmPP84 negatively regulates drought tolerance by repressing stomatal closure in maize. New Phytol 237, 1728-1744. |
| [31] |
Gupta A, Rico-Medina A, Caño-Delgado AI (2020). The physiology of plant responses to drought. Science 368, 266-269.
DOI PMID |
| [32] |
Gusev A, Ko A, Shi H, Bhatia G, Chung W, Penninx BW, Jansen R, de Geus EJ, Boomsma DI, Wright FA, Sullivan PF, Nikkola E, Alvarez M, Civelek M, Lusis AJ, Lehtimäki T, Raitoharju E, Kähönen M, Seppälä I, Raitakari OT, Kuusisto J, Laakso M, Price AL, Pajukanta P, Pasaniuc B (2016). Integrative approaches for large-scale transcriptome-wide association studies. Nat Genet 48, 245-252.
DOI PMID |
| [33] |
He ZH, Zhong JW, Sun XP, Wang BC, Terzaghi W, Dai MQ (2018). The maize ABA receptors ZmPYL8, 9, and 12 facilitate plant drought resistance. Front Plant Sci 9, 422.
DOI PMID |
| [34] | Heffner EL, Sorrells ME, Jannink JL (2009). Genomic selection for crop improvement. Crop Sci 49, 1-12. |
| [35] | Hetherington AM, Woodward FI (2003). The role of stomata in sensing and driving environmental change. Nature 424, 901-908. |
| [36] |
Hou X, Xie KB, Yao JL, Qi ZY, Xiong LZ (2009). A homolog of human ski-interacting protein in rice positively regulates cell viability and stress tolerance. Proc Natl Acad Sci USA 106, 6410-6415.
DOI PMID |
| [37] | Huang WZ, Ma XR, Wang QL, Gao YF, Xue Y, Niu XL, Yu GR, Liu YS (2008). Significant improvement of stress tolerance in tobacco plants by overexpressing a stress- responsive aldehyde dehydrogenase gene from maize (Zea mays). Plant Mol Biol 68, 451-463. |
| [38] | Jiang SS, Zhang D, Wang L, Pan JW, Liu Y, Kong XP, Zhou Y, Li DQ (2013). A maize calcium-dependent protein kinase gene, ZmCPK4, positively regulated abscisic acid signaling and enhanced drought stress tolerance in transgenic Arabidopsis. Plant Physiol Biochem 71, 112-120. |
| [39] | Jiao P, Jiang ZZ, Miao M, Wei XT, Wang CL, Liu SY, Guan SY, Ma YY (2024). Zmhdz9, an HD-Zip transcription factor, promotes drought stress resistance in maize by modulating ABA and lignin accumulation. Int J Biol Macromol 258, 128849. |
| [40] |
Kazan K, Manners JM (2013). MYC2: the master in action. Mol Plant 6, 686-703.
DOI PMID |
| [41] | Kim TH, Böhmer M, Hu HH, Nishimura N, Schroeder JI (2010). Guard cell signal transduction network: advances in understanding abscisic acid, CO2, and Ca2+ signaling. Annu Rev Plant Biol 61, 561-591. |
| [42] |
Kooyers NJ (2015). The evolution of drought escape and avoidance in natural herbaceous populations. Plant Sci 234, 155-162.
DOI PMID |
| [43] | Kramp RE, Liancourt P, Herberich MM, Saul L, Weides S, Tielbörger K, Májeková M (2022). Functional traits and their plasticity shift from tolerant to avoidant under extreme drought. Ecology 103, e3826. |
| [44] |
Krannich CT, Maletzki L, Kurowsky C, Horn R (2015). Network candidate genes in breeding for drought tolerant crops. Int J Mol Sci 16, 16378-16400.
DOI PMID |
| [45] |
Lebaudy A, Pascaud F, Véry AA, Alcon C, Dreyer I, Thibaud JB, Lacombe B (2010). Preferential KAT1-KAT2 heteromerization determines inward K+ current properties in Arabidopsis guard cells. J Biol Chem 285, 6265-6274.
DOI PMID |
| [46] | Li CH, Guo J, Wang DM, Chen XJ, Guan HH, Li YX, Zhang DF, Liu XY, He GH, Wang TY, Li Y (2023). Genomic insight into changes of root architecture under drought stress in maize. Plant Cell Environ 46, 1860-1872. |
| [47] | Li DL, Liu Q, Schnable PS (2021a). TWAS results are complementary to and less affected by linkage disequilibrium than GWAS. Plant Physiol 186, 1800-1811. |
| [48] | Li DX, Wang MW, Zhang TP, Chen X, Li CY, Liu Y, Brestic M, Chen THH, Yang XH (2021b). Glycinebetaine mitigated the photoinhibition of photosystem II at high temperature in transgenic tomato plants. Photosynth Res 147, 301-315. |
| [49] | Li L, Du YC, He C, Dietrich CR, Li JK, Ma XL, Wang R, Liu Q, Liu SZ, Wang GY, Schnable PS, Zheng J (2019a). Maize glossy6 is involved in cuticular wax deposition and drought tolerance. J Exp Bot 70, 3089-3099. |
| [50] | Li ZX, Liu C, Zhang Y, Wang BM, Ran QJ, Zhang JR (2019b). The bHLH family member ZmPTF1 regulates drought tolerance in maize by promoting root development and abscisic acid synthesis. J Exp Bot 70, 5471-5486. |
| [51] | Liang YN, Jiang YL, Du M, Li BY, Chen L, Chen MC, Jin DM, Wu JD (2019). ZmASR3 from the maize ASR gene family positively regulates drought tolerance in transgenic Arabidopsis. Int J Mol Sci 20, 2278. |
| [52] | Liu BX, Zhang B, Yang ZR, Liu Y, Yang SP, Shi YL, Jiang CF, Qin F (2021a). Manipulating ZmEXPA4 expression ameliorates the drought-induced prolonged anthesis and silking interval in maize. Plant Cell 33, 2058-2071. |
| [53] | Liu L, Gallagher J, Arevalo ED, Chen R, Skopelitis T, Wu QY, Bartlett M, Jackson D (2021b). Enhancing grain- yield-related traits by CRISPR-Cas9 promoter editing of maize CLE genes. Nat Plants 7, 287-294. |
| [54] |
Liu SX, Li CP, Wang HW, Wang SH, Yang SP, Liu XH, Yan JB, Li BL, Beatty M, Zastrow-Hayes G, Song SH, Qin F (2020). Mapping regulatory variants controlling gene expression in drought response and tolerance in maize. Genome Biol 21, 163.
DOI PMID |
| [55] | Liu SX, Liu XH, Zhang XM, Chang SJ, Ma C, Qin F (2022). Co-expression of ZmVPP1 with ZmNAC111 confers robust drought resistance in maize. Genes (Basel) 14, 8. |
| [56] | Liu YQ, Li AQ, Liang MN, Zhang Q, Wu JD (2023). Overexpression of the maize genes ZmSKL1 and ZmSKL2 positively regulates drought stress tolerance in transgenic Arabidopsis. Plant Cell Rep 42, 521-533. |
| [57] | Lu FZ, Li WC, Peng YL, Cao Y, Qu JT, Sun FA, Yang QQ, Lu YL, Zhang XH, Zheng LJ, Fu FL, Yu HQ (2022). ZmPP2C26 alternative splicing variants negatively regulate drought tolerance in maize. Front Plant Sci 13, 85-1531. |
| [58] | Lu HF, Lin T, Klein J, Wang SH, Qi JJ, Zhou Q, Sun JJ, Zhang ZH, Weng YQ, Huang SW (2014). QTL-seq identifies an early flowering QTL located near Flowering Locus T in cucumber. Theor Appl Genet 127, 1491-1499. |
| [59] |
Lynch J (1995). Root architecture and plant productivity. Plant Physiol 109, 7-13.
DOI PMID |
| [60] |
Ma HZ, Liu C, Li ZX, Ran QJ, Xie GN, Wang BM, Fang S, Chu JF, Zhang JR (2018). ZmbZIP4 contributes to stress resistance in maize by regulating ABA synthesis and root development. Plant Physiol 178, 753-770.
DOI PMID |
| [61] |
Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, Christmann A, Grill E (2009). Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324, 1064-1068.
DOI PMID |
| [62] | Mao HD, Wang HW, Liu SX, Li ZG, Yang XH, Yan JB, Li JS, Tran LSP, Qin F (2015). A transposable element in a NAC gene is associated with drought tolerance in maize seedlings. Nat Commun 6, 8326. |
| [63] |
Massman JM, Gordillo A, Lorenzana RE, Bernardo R (2013). Genomewide predictions from maize single-cross data. Theor Appl Genet 126, 13-22.
DOI PMID |
| [64] | Mei FM, Chen B, Du LY, Li SM, Zhu DH, Chen N, Zhang YF, Li FF, Wang ZX, Cheng XX, Ding L, Kang ZS, Mao HD (2022). A gain-of-function allele of a DREB transcription factor gene ameliorates drought tolerance in wheat. Plant Cell 34, 4472-4494. |
| [65] | Miao ZY, Zhang T, Qi YH, Song J, Han ZX, Ma C (2020). Evolution of the RNA N6-methyladenosine methylome mediated by genomic duplication. Plant Physiol 182, 345-360. |
| [66] |
Mittler R, Vanderauwera S, Suzuki N, Miller G, Tognetti VB, Vandepoele K, Gollery M, Shulaev V, Van Breusegem F (2011). ROS signaling: the new wave? Trends Plant Sci 16, 300-309.
DOI PMID |
| [67] | Nakaya A, Isobe SN (2012). Will genomic selection be a practical method for plant breeding? Ann Bot 110, 1303-1316. |
| [68] |
Nelson DE, Repetti PP, Adams TR, Creelman RA, Wu JR, Warner DC, Anstrom DC, Bensen RJ, Castiglioni PP, Donnarummo MG, Hinchey BS, Kumimoto RW, Maszle DR, Canales RD, Krolikowski KA, Dotson SB, Gutterson N, Ratcliffe OJ, Heard JE (2007). Plant nuclear factor Y (NF-Y) B subunits confer drought tolerance and lead to improved corn yields on water-limited acres. Proc Natl Acad Sci USA 104, 16450-16455.
DOI PMID |
| [69] | Nemali KS, Bonin C, Dohleman FG, Stephens M, Reeves WR, Nelson DE, Castiglioni P, Whitsel JE, Sammons B, Silady RA, Anstrom D, Sharp RE, Patharkar OR, Clay D, Coffin M, Nemeth MA, Leibman ME, Luethy M, Lawson M (2015). Physiological responses related to increased grain yield under drought in the first biotechnology-derived drought-tolerant maize. Plant Cell Environ 38, 1866-1880. |
| [70] | Nica AC, Dermitzakis ET (2013). Expression quantitative trait loci: present and future. Philos Trans R Soc Lond B Biol Sci 368, 20120362. |
| [71] | Nuccio ML, Wu J, Mowers R, Zhou HP, Meghji M, Primavesi LF, Paul MJ, Chen X, Gao Y, Haque E, Basu SS, Lagrimini LM (2015). Expression of trehalose- 6-phosphate phosphatase in maize ears improves yield in well-watered and drought conditions. Nat Biotechnol 33, 862-869. |
| [72] | Ogura T, Goeschl C, Filiault D, Mirea M, Slovak R, Wolhrab B, Satbhai SB, Busch W (2019). Root system depth in Arabidopsis is shaped by EXOCYST70A3 via the dynamic modulation of auxin transport. Cell 178, 400-412. |
| [73] | Pan ZY, Liu M, Zhao HL, Tan ZD, Liang K, Sun Q, Gong DM, He HJ, Zhou WQ, Qiu FZ (2020). ZmSRL5 is involved in drought tolerance by maintaining cuticular wax structure in maize. J Integr Plant Biol 62, 1895-1909. |
| [74] | Park SY, Fung P, Nishimura N, Jensen DR, Fujii H, Zhao Y, Lumba S, Santiago J, Rodrigues A, Chow TFF, Alfred SE, Bonetta D, Finkelstein R, Provart NJ, Desveaux D, Rodriguez PL, McCourt P, Zhu JK, Schroeder JI, Volkman BF, Cutler SR (2009). Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324, 1068-1071. |
| [75] | Pourghayoumi M, Bakhshi D, Rahemi M, Kamgar- Haghighi AA, Aalami A (2017). The physiological responses of various pomegranate cultivars to drought stress and recovery in order to screen for drought tolerance. Sci Hortic 217, 164-172. |
| [76] |
Qi JS, Song CP, Wang BS, Zhou JM, Kangasjärvi J, Zhu JK, Gong ZZ (2018). Reactive oxygen species signaling and stomatal movement in plant responses to drought stress and pathogen attack. J Integr Plant Biol 60, 805-826.
DOI |
| [77] | Qin LM, Sun L, Wei L, Yuan JR, Kong FF, Zhang Y, Miao X, Xia GM, Liu SW (2021). Maize SRO1e represses anthocyanin synthesis through regulating the MBW complex in response to abiotic stress. Plant J 105, 1010-1025. |
| [78] |
Reguera M, Peleg Z, Blumwald E (2012). Targeting metabolic pathways for genetic engineering abiotic stress-tolerance in crops. Biochim Biophys Acta 1819, 186-194.
DOI PMID |
| [79] |
Ren W, Zhao LF, Liang JX, Wang LF, Chen LM, Li PC, Liu ZG, Li XJ, Zhang ZH, Li JP, He KH, Zhao Z, Ali F, Mi GH, Yan JB, Zhang FS, Chen FJ, Yuan LX, Pan QC (2022). Genome-wide dissection of changes in maize root system architecture during modern breeding. Nat Plants 8, 1408-1422.
DOI PMID |
| [80] | Ribaut JM, Ragot M (2007). Marker-assisted selection to improve drought adaptation in maize: the backcross approach, perspectives, limitations, and alternatives. J Exp Bot 58, 351-360. |
| [81] | Schneider KA, Brothers ME, Kelly JD (1997). Marker- assisted selection to improve drought resistance in common bean. Crop Sci 37, 51-60. |
| [82] |
Shavrukov Y, Kurishbayev A, Jatayev S, Shvidchenko V, Zotova L, Koekemoer F, de Groot S, Soole K, Langridge P (2017). Early flowering as a drought escape mechanism in plants: how can it aid wheat production? Front Plant Sci 8, 1950.
DOI PMID |
| [83] |
Shi JR, Gao HR, Wang HY, Lafitte HR, Archibald RL, Yang MZ, Hakimi SM, Mo H, Habben JE (2017). ARGOS8 variants generated by CRISPR-Cas9 improve maize grain yield under field drought stress conditions. Plant Biotechnol J 15, 207-216.
DOI PMID |
| [84] |
Singh D, Laxmi A (2015). Transcriptional regulation of drought response: a tortuous network of transcriptional factors. Front Plant Sci 6, 895.
DOI PMID |
| [85] |
Soma F, Takahashi F, Suzuki T, Shinozaki K, Yamaguchi-Shinozaki K (2020). Plant Raf-like kinases regulate the mRNA population upstream of ABA-unresponsive SnRK2 kinases under drought stress. Nat Commun 11, 1373.
DOI PMID |
| [86] | Sun XP, Xiang YL, Dou NN, Zhang H, Pei SR, Franco AV, Menon M, Monier B, Ferebee T, Liu T, Liu SY, Gao YC, Wang JB, Terzaghi W, Yan JB, Hearne S, Li L, Li F, Dai MQ (2023). The role of transposon inverted repeats in balancing drought tolerance and yield-related traits in maize. Nat Biotechnol 41, 120-127. |
| [87] | Takagi H, Abe A, Yoshida K, Kosugi S, Natsume S, Mitsuoka C, Uemura A, Utsushi H, Tamiru M, Takuno S, Innan H, Cano LM, Kamoun S, Terauchi R (2013). QTL-seq: rapid mapping of quantitative trait loci in rice by whole genome resequencing of DNA from two bulked populations. Plant J 74, 174-183. |
| [88] | Tang Q, Lv HZ, Li QM, Zhang XY, Li L, Xu J, Wu FK, Wang QJ, Feng XJ, Lu YL (2022). Characteristics of microRNAs and target genes in maize root under drought stress. Int J Mol Sci 23, 4968. |
| [89] |
Tian T, Wang SH, Yang SP, Yang ZR, Liu SX, Wang YJ, Gao HJ, Zhang SS, Yang XH, Jiang CF, Qin F (2023). Genome assembly and genetic dissection of a prominent drought-resistant maize germplasm. Nat Genet 55, 496-506.
DOI PMID |
| [90] | Uga Y, Sugimoto K, Ogawa S, Rane J, Ishitani M, Hara N, Kitomi Y, Inukai Y, Ono K, Kanno N, Inoue H, Takehisa H, Motoyama R, Nagamura Y, Wu JZ, Matsumoto T, Takai T, Okuno K, Yano M (2013). Control of root system architecture by DEEPER ROOTING 1 increases rice yield under drought conditions. Nat Genet 45, 1097-1102. |
| [91] |
Van Dooren TJM, Silveira AB, Gilbault E, Jiménez-Gómez JM, Martin A, Bach L, Tisné S, Quadrana L, Loudet O, Colot V (2020). Mild drought in the vegetative stage induces phenotypic, gene expression, and DNA methylation plasticity in Arabidopsis but no transgenerational effects. J Exp Bot 71, 3588-3602.
DOI PMID |
| [92] |
Virlouvet L, Jacquemot MP, Gerentes D, Corti H, Bouton S, Gilard F, Valot B, Trouverie J, Tcherkez G, Falque M, Damerval C, Rogowsky P, Perez P, Noctor G, Zivy M, Coursol S (2011). The ZmASR1 protein influences branched-chain amino acid biosynthesis and maintains kernel yield in maize under water-limited conditions. Plant Physiol 157, 917-936.
DOI PMID |
| [93] |
Wainberg M, Sinnott-Armstrong N, Mancuso N, Barbeira AN, Knowles DA, Golan D, Ermel R, Ruusalepp A, Quertermous T, Hao K, Björkegren JLM, Im HK, Pa-saniuc B, Rivas MA, Kundaje A (2019). Opportunities and challenges for transcriptome-wide association stu-dies. Nat Genet 51, 592-599.
DOI PMID |
| [94] | Wan LY, Zhang JF, Zhang HW, Zhang ZJ, Quan RD, Zhou SR, Huang RF (2011). Transcriptional activation of OsDERF1 in OsERF3 and OsAP2-39 negatively modulates ethylene synthesis and drought tolerance in rice. PLoS One 6, e25216. |
| [95] | Wang BM, Li ZX, Ran QJ, Li P, Peng ZH, Zhang JR (2018a). ZmNF-YB16 overexpression improves drought resistance and yield by enhancing photosynthesis and the antioxidant capacity of maize plants. Front Plant Sci 9, 709. |
| [96] | Wang CT, Ru JN, Liu YW, Li M, Zhao D, Yang JF, Fu JD, Xu ZS (2018b). Maize WRKY transcription factor ZmWRKY106 confers drought and heat tolerance in transgenic plants. Int J Mol Sci 19, 3046. |
| [97] | Wang CT, Ru JN, Liu YW, Yang JF, Li M, Xu ZS, Fu JD (2018c). The maize WRKY transcription factor ZmWRKY40 confers drought resistance in transgenic Arabidopsis. Int J Mol Sci 19, 2580. |
| [98] |
Wang N, Cheng M, Chen Y, Liu BJ, Wang XN, Li GJ, Zhou YH, Luo P, Xi ZY, Yong HJ, Zhang DG, Li MS, Zhang XC, Vicente FS, Hao ZF, Li XH (2021). Natural variations in the non-coding region of ZmNAC080308 contributes maintaining grain yield under drought stress in maize. BMC Plant Biol 21, 305.
DOI PMID |
| [99] | Wang XL, Wang HW, Liu SX, Ferjani A, Li JS, Yan JB, Yang XH, Qin F (2016). Genetic variation in ZmVPP1 contributes to drought tolerance in maize seedlings. Nat Genet 48, 1233-1241. |
| [100] | Wang YG, Fu FL, Yu HQ, Hu T, Zhang YY, Tao Y, Zhu JK, Zhao Y, Li WC (2018d). Interaction network of core ABA signaling components in maize. Plant Mol Biol 96, 245-263. |
| [101] |
Wen WW, Li D, Li X, Gao YQ, Li WQ, Li HH, Liu J, Liu HJ, Chen W, Luo J, Yan JB (2014). Metabolome-based genome-wide association study of maize kernel leads to novel biochemical insights. Nat Commun 5, 3438.
DOI PMID |
| [102] | Wu JD, Jiang YL, Liang YN, Chen L, Chen WJ, Cheng BJ (2019). Expression of the maize MYB transcription factor ZmMYB3R enhances drought and salt stress tolerance in transgenic plants. Plant Physiol Biochem 137, 179-188. |
| [103] |
Wu X, Feng H, Wu D, Yan SJ, Zhang P, Wang WB, Zhang J, Ye JL, Dai GX, Fan Y, Li WK, Song BX, Geng ZD, Yang WL, Chen GX, Qin F, Terzaghi W, Stitzer M, Li L, Xiong LZ, Yan JB, Buckler E, Yang WN, Dai MQ (2021). Using high-throughput multiple optical phenotyping to decipher the genetic architecture of maize drought tolerance. Genome Biol 22, 185.
DOI PMID |
| [104] |
Xia ZL, Liu QJ, Wu JY, Ding JQ (2012). ZmRFP1, the putative ortholog of SDIR1, encodes a RING-H2 E3 ubiquitin ligase and responds to drought stress in an ABA-dependent manner in maize. Gene 495, 146-153.
DOI PMID |
| [105] |
Xiang YL, Sun XP, Gao S, Qin F, Dai MQ (2017). Deletion of an endoplasmic reticulum stress response element in a ZmPP2C-A gene facilitates drought tolerance of maize seedlings. Mol Plant 10, 456-469.
DOI PMID |
| [106] | Xu HX, Cao YW, Xu YF, Ma PT, Ma FF, Song LP, Li LH, An DG (2017). Marker-assisted development and evaluation of near-isogenic lines for broad-spectrum powdery mildew resistance gene Pm2b introgressed into different genetic backgrounds of wheat. Front Plant Sci 8, 1322. |
| [107] | Xu YB, Lu YL, Xie CX, Gao SB, Wan JM, Prasanna BM (2012). Whole-genome strategies for marker-assisted plant breeding. Mol Breed 29, 833-854. |
| [108] | Yang Y, Shi JX, Chen LM, Xiao WH, Yu JJ (2022a). ZmEREB46, a maize ortholog of Arabidopsis WAX INDUCER1/SHINE1, is involved in the biosynthesis of leaf epicuticular very-long-chain waxes and drought tolerance. Plant Sci 321, 111256. |
| [109] | Yang YL, Wang BM, Wang JM, He CM, Zhang DF, Li P, Zhang JR, Li ZX (2022b). Transcription factors ZmNF- YA1 and ZmNF-YB16 regulate plant growth and drought tolerance in maize. Plant Physiol 190, 1506-1525. |
| [110] |
Young TE, Meeley RB, Gallie DR (2004). ACC synthase expression regulates leaf performance and drought tolerance in maize. Plant J 40, 813-825.
PMID |
| [111] |
Yu J, Xu F, Wei ZW, Zhang XX, Chen T, Pu L (2020). Epigenomic landscape and epigenetic regulation in maize. Theor Appl Genet 133, 1467-1489.
DOI PMID |
| [112] | Yu P, Li CH, Li M, He XM, Wang DN, Li HJ, Marcon C, Li Y, Perez-Limón S, Chen XP, Delgado-Baquerizo M, Koller R, Metzner R, van Dusschoten D, Pflugfelder D, Borisjuk L, Plutenko I, Mahon A, Resende MFR, Salvi S, Akale A, Abdalla M, Ahmed MA, Bauer FM, Schnepf A, Lobet G, Heymans A, Suresh K, Schreiber L, McLaughlin CM, Li CJ, Mayer M, Schön CC, Bernau V, von Wirén N, Sawers RJH, Wang TY, Hochholdinger F (2024). Seedling root system adaptation to water availability during maize domestication and global expansion. Nat Genet 56, 1245-1256. |
| [113] | Zhang C, Yang RJ, Zhang TT, Zheng DY, Li XL, Zhang ZB, Li LG, Wu ZY (2023a). ZmTIFY16, a novel maize TIFY transcription factor gene, promotes root growth and development and enhances drought and salt tolerance in Arabidopsis and Zea mays. Plant Growth Regul 100, 149-160. |
| [114] |
Zhang F, Wu JF, Sade N, Wu S, Egbaria A, Fernie AR, Yan JB, Qin F, Chen W, Brotman Y, Dai MQ (2021). Genomic basis underlying the metabolome-mediated drought adaptation of maize. Genome Biol 22, 260.
DOI PMID |
| [115] | Zhang H, Xiang YL, He N, Liu XG, Liu HB, Fang LP, Zhang F, Sun XP, Zhang DL, Li XW, Terzaghi W, Yan JB, Dai MQ (2020a). Enhanced vitamin C production mediated by an ABA-induced PTP-like nucleotidase improves plant drought tolerance in Arabidopsis and maize. Mol Plant 13, 760-776. |
| [116] | Zhang K, Xue M, Qin F, He Y, Zhou YY (2023b). Natural polymorphisms in ZmIRX15A affect water-use efficiency by modulating stomatal density in maize. Plant Biotechnol J 21, 2560-2573. |
| [117] | Zhang M, Chen YH, Xing HY, Ke WS, Shi YL, Sui Z, Xu RB, Gao LL, Guo GG, Li JS, Xing JW, Zhang YR (2023c). Positional cloning and characterization reveal the role of a miRNA precursor gene ZmLRT in the regulation of lateral root number and drought tolerance in maize. J Integr Plant Biol 65, 772-790. |
| [118] | Zhang W, Han ZX, Guo QL, Liu Y, Zheng YX, Wu FL, Jin WB (2014). Identification of maize long non-coding RNAs responsive to drought stress. PLoS One 9, e98958. |
| [119] | Zhang XM, Mi Y, Mao HD, Liu SX, Chen LM, Qin F (2020b). Genetic variation in ZmTIP1 contributes to root hair elongation and drought tolerance in maize. Plant Biotechnol J 18, 1271-1283. |
| [120] | Zhao Y, Ma Q, Jin XL, Peng XJ, Liu JY, Deng L, Yan HW, Sheng L, Jiang HY, Cheng BJ (2014). A novel maize homeodomain-leucine zipper (HD-Zip) I gene, Zmhdz10, positively regulates drought and salt tolerance in both rice and Arabidopsis. Plant Cell Physiol 55, 1142-1156. |
| [121] | Zhao YS, Zeng J, Fernando R, Reif JC (2013). Genomic prediction of hybrid wheat performance. Crop Sci 53, 802-810. |
| [122] | Zhu JK (2002). Salt and drought stress signal transduction in plants. Annu Rev Plant Biol 53, 247-273. |
| [123] | Zhu YQ, Liu Y, Zhou KM, Tian CY, Aslam M, Zhang BL, Liu WJ, Zou HW (2022). Overexpression of ZmEREBP60 enhances drought tolerance in maize. J Plant Physiol 275, 153763. |
| [1] | Suowei Wu, Xueli An, Xiangyuan Wan. Molecular Mechanisms of Male Sterility and their Applications in Biotechnology-based Male-sterility Hybrid Seed Production in Maize [J]. Chinese Bulletin of Botany, 2024, 59(6): 932-949. |
| [2] | Mingmin Zheng, Qiang Huang, Peng Zhang, Xiaowei Liu, Zhuofan Zhao, Hongyang Yi, Tingzhao Rong, Moju Cao. Research Progress on Cytoplasmic Male Sterility and Fertility Restoration in Maize [J]. Chinese Bulletin of Botany, 2024, 59(6): 999-1006. |
| [3] | Wenli Yang, Zhao Li, Zhiming Liu, Zhihua Zhang, Jinsheng Yang, Yanjie Lü, Yongjun Wang. Senescence Characteristics of Maize Leaves at Different Maturity Stages and Their Effect on Phyllosphere Bacteria [J]. Chinese Bulletin of Botany, 2024, 59(6): 1024-1040. |
| [4] | Yuan Li, Kaijian Fan, Tai An, Cong Li, Junxia Jiang, Hao Niu, Weiwei Zeng, Yanfang Heng, Hu Li, Junjie Fu, Huihui Li, Liang Li. Study on Multi-environment Genome-wide Prediction of Inbred Agronomic Traits in Maize Natural Populations [J]. Chinese Bulletin of Botany, 2024, 59(6): 1041-1053. |
| [5] | Qiang Zhang, Zhenyu Zhao, Pinghua Li. Research Progress of Gene Editing Technology in Maize [J]. Chinese Bulletin of Botany, 2024, 59(6): 978-998. |
| [6] | Tao Wang, Jinglei Feng, Cui Zhang. Research Progress on Molecular Mechanisms of Heat Stress Affecting the Growth and Development of Maize [J]. Chinese Bulletin of Botany, 2024, 59(6): 963-977. |
| [7] | Juan Yang, Yuelei Zhao, Xiaoyuan Chen, Baobao Wang, Haiyang Wang. Regulation Mechanism and Breeding Application of Flowering Time in Maize [J]. Chinese Bulletin of Botany, 2024, 59(6): 912-931. |
| [8] | Hengyu Yan, Zhaoxia Li, Yubin Li. Research Progress on Heat Stress Impact on Maize Growth and Heat-Tolerant Maize Screening in China [J]. Chinese Bulletin of Botany, 2024, 59(6): 1007-1023. |
| [9] | Qingguo Du, Wenxue Li. Research Progress in the Regulation of Development and Stress Responses by Long Non-coding RNAs in Maize [J]. Chinese Bulletin of Botany, 2024, 59(6): 950-962. |
| [10] | Xingxin Liao, Yi Niu, Xingwu Duo, Akeyedeli Jumahazi, Marhaba Abdukuyum, Rizwangul Hufur, Haiyan Lan, Jing Cao. Heterologous Expression of Suaeda aralocaspica SaPEPC2 Gene Improves Drought Resistance and Photosynthesis in Transgenic Tobacco [J]. Chinese Bulletin of Botany, 2024, 59(4): 585-599. |
| [11] | Lumei He, Bojun Ma, Xifeng Chen. Advances on the Executor Resistance Genes in Plants [J]. Chinese Bulletin of Botany, 2024, 59(4): 671-680. |
| [12] | CHENG Ke-Xin, DU Yao, LI Kai-Hang, WANG Hao-Chen, YANG Yan, JIN Yi, HE Xiao-Qing. Genetic mechanism of interaction between maize and phyllospheric microbiome [J]. Chin J Plant Ecol, 2024, 48(2): 215-228. |
| [13] | Heping Wang, Zhen Sun, Yuchen Liu, Yanlong Su, Jinyu Du, Yan Zhao, Hongbo Zhao, Zhaoming Wang, Feng Yuan, Yaling Liu, Zhenying Wu, Feng He, Chunxiang Fu. Sequence Identification and Functional Analysis of Cinnamyl Alcohol Dehydrogenase Gene from Agropyron mongolicum [J]. Chinese Bulletin of Botany, 2024, 59(2): 204-216. |
| [14] | Xiting Yu, Xuehui Huang. New Insights Into the Origin of Modern Maize-hybridization of Two Teosintes [J]. Chinese Bulletin of Botany, 2023, 58(6): 857-860. |
| [15] | Wenqi Zhou, Yuqian Zhou, Yongsheng Li, Haijun He, Yanzhong Yang, Xiaojuan Wang, Xiaorong Lian, Zhongxiang Liu, Zhubing Hu. ZmICE2 Regulates Stomatal Development in Maize [J]. Chinese Bulletin of Botany, 2023, 58(6): 866-881. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||