

Chinese Bulletin of Botany ›› 2023, Vol. 58 ›› Issue (6): 857-860.DOI: 10.11983/CBB23138 cstr: 32102.14.CBB23138
Special Issue: 大食物观
• COMMENTARIES • Next Articles
Received:2023-10-10
Accepted:2023-10-17
Online:2023-11-01
Published:2023-12-01
Contact:
* E-mail: xhhuang@shnu.edu.cn
Xiting Yu, Xuehui Huang. New Insights Into the Origin of Modern Maize-hybridization of Two Teosintes[J]. Chinese Bulletin of Botany, 2023, 58(6): 857-860.
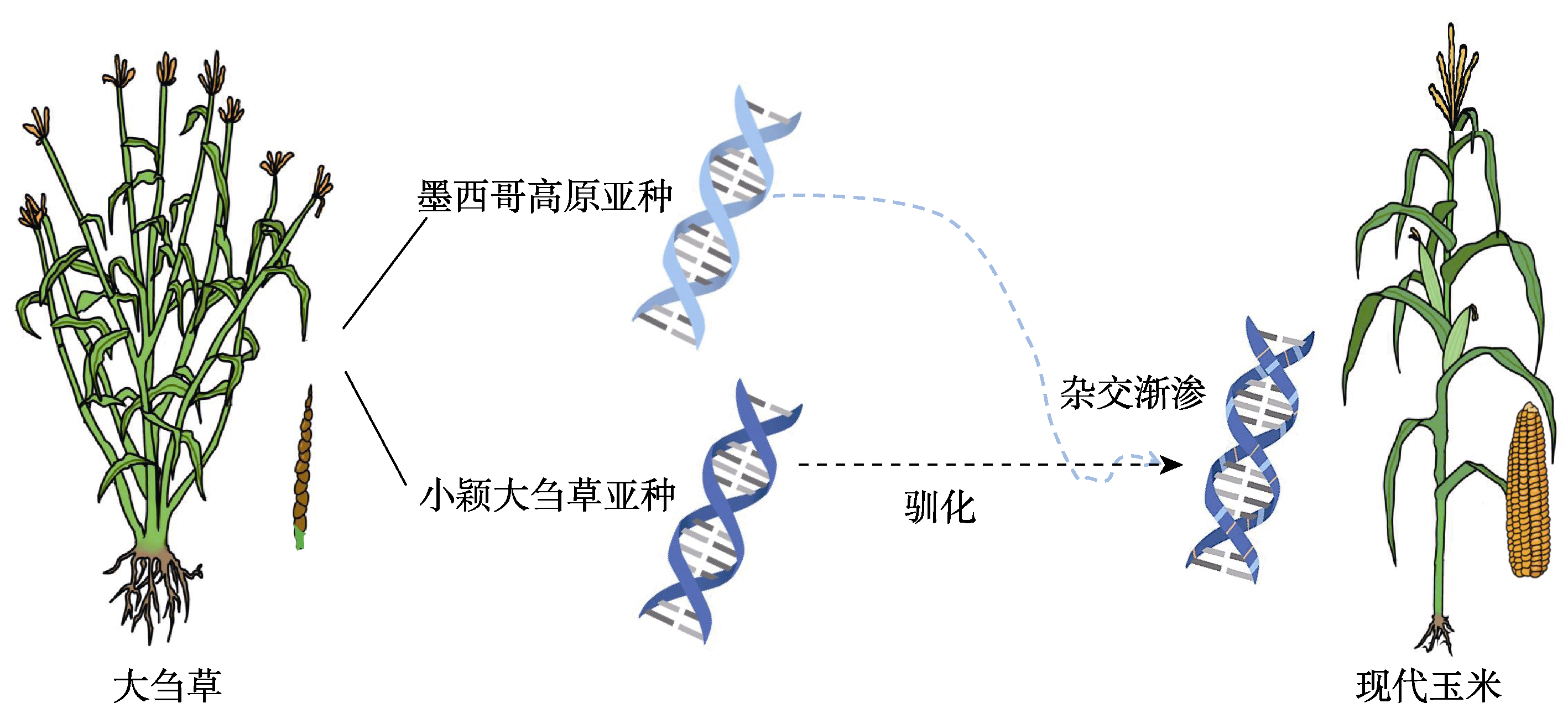
Figure 1 A simplified diagram of the origin process of modern maize Researchers speculate that modern maize first originated from the parviglumis type of teosinte in the Mexican lowlands, and then introgression with the mexicana type of teosinte in the Mexican highlands occurred. The deep blue, light blue, and orange colors in the DNA double helix represent the haplotypes of parviglumis-type teosinte, mexicana-type teosinte, and new allelic variation during domestication, respectively.
| [1] |
Calfee E, Gates D, Lorant A, Perkins MT, Coop G, Ross-Ibarra J (2021). Selective sorting of ancestral introgression in maize and teosinte along an elevational cline. PLoS Genet 17, e1009810.
DOI URL |
| [2] |
Doebley JF, Gaut BS, Smith BD (2006). The molecular genetics of crop domestication. Cell 127, 1309-1321.
DOI PMID |
| [3] |
Huang XH, Huang SW, Han B, Li JY (2022a). The integrated genomics of crop domestication and breeding. Cell 185, 2828-2839.
DOI URL |
| [4] |
Huang XH, Kurata N, Wei XH, Wang ZX, Wang AH, Zhao Q, Zhao Y, Liu KY, Lu HY, Li WJ, Guo YL, Lu YQ, Zhou CC, Fan DL, Weng QJ, Zhu CR, Huang T, Zhang L, Wang YC, Feng L, Furuumi H, Kubo T, Miyabayashi T, Yuan XP, Xu Q, Dong GJ, Zhan QL, Li CY, Fujiyama A, Toyoda A, Lu TT, Feng Q, Qian Q, Li JY, Han B (2012). A map of rice genome variation reveals the origin of cultivated rice. Nature 490, 497-501.
DOI |
| [5] | Huang YC, Wang HH, Zhu YD, Huang X, Li S, Wu XG, Zhao Y, Bao ZG, Qin L, Jin YB, Cui YH, Ma GJ, Xiao Q, Wang Q, Wang JC, Yang XR, Liu HJ, Lu XD, Larkins BA, Wang WQ, Wu YR (2022b). THP9 enhances seed protein content and nitrogen-use efficiency in maize. Nature 612, 292-300. |
| [6] |
Hufford MB, Lubinksy P, Pyhäjärvi T, Devengenzo MT, Ellstrand NC, Ross-Ibarra J (2013). The genomic signature of crop-wild introgression in maize. PLoS Genet 9, e1003477.
DOI URL |
| [7] |
Jing CY, Zhang FM, Wang XH, Wang MX, Zhou L, Cai Z, Han JD, Geng MF, Yu WH, Jiao ZH, Huang L, Liu R, Zheng XM, Meng QL, Ren NN, Zhang HX, Du YS, Wang X, Qiang CG, Zou XH, Gaut BS, Ge S (2023). Multiple domestications of Asian rice. Nat Plants 9, 1221-1235.
DOI |
| [8] |
Li CB, Zhou AL, Sang T (2006). Rice domestication by reducing shattering. Science 311, 1936-1939.
DOI PMID |
| [9] |
Matsuoka Y, Vigouroux Y, Goodman MM, Sanchez GJ, Buckler E, Doebley J (2002). A single domestication for maize shown by multilocus microsatellite genotyping. Proc Natl Acad Sci USA 99, 6080-6084.
DOI PMID |
| [10] |
Pääbo S (2015). The diverse origins of the human gene pool. Nat Rev Genet 16, 313-314.
DOI PMID |
| [11] |
Wang H, Studer AJ, Zhao Q, Meeley R, Doebley JF (2015). Evidence that the origin of naked kernels during maize domestication was caused by a single amino acid substitution in tga1. Genetics 200, 965-974.
DOI |
| [12] | Yang N, Wang YB, Liu XG, Jin ML, Vallebueno-Estrada M, Calfee E, Chen L, Dilkes BP, Gui ST, Fan XM, Harper TK, Kennett DJ, Li WQ, Lu YL, Luo JY, Mambak-kam S, Menon M, Snodgrass S, Veller C, Wu SS, Wu SY, Xiao YJ, Yang XH, Stitzer MC, Runcie D, Yan JB, Ross-Ibarra J (2023). Two teosintes made modern maize. Science doi: 10.1126/science.adg8940 |
| [13] |
Yu H, Lin T, Meng XB, Du HL, Zhang JK, Liu GF, Chen ML, Jing YH, Kou LQ, Li XX, Gao Q, Liang Y, Liu XD, Fan ZL, Liang YT, Cheng ZK, Chen MS, Tian ZX, Wang YH, Chu CC, Zuo JR, Wan JM, Qian Q, Han B, Zuccolo A, Wing RA, Gao CX, Liang CZ, Li JY (2021). A route to de novo domestication of wild allotetraploid rice. Cell 184, 1156-1170.
DOI URL |
| [1] |
Juan Cui, Xiaoyu Yu, Yuejiao Yu, Chengwei Liang, Jian Sun, Wenfu Chen.
Analysis of Texture Factors and Genetic Basis Influencing the Differences in Eating Quality between Northeast China and Japanese Japonica Rice [J]. Chinese Bulletin of Botany, 2025, 60(4): 1-0. |
| [2] | Chuanyong Wang, Dian Zhuang, Zhengda Song, Henghua Zhai, Naiwei Li, Fan Zhang. Structural and Comparative Analysis of the Complete Chloroplast Genome and Phylogenetic Inference of the Aronia melanocarpa [J]. Chinese Bulletin of Botany, 2025, 60(4): 1-0. |
| [3] | Zhang Ruli, Li Dezhu, Zhang Yuxiao. Population Genetic Structure and Climate Adaptation Analysis of Brachystachyum densiflorum [J]. Chinese Bulletin of Botany, 2025, 60(3): 407-424. |
| [4] | Linfeng Xia, Rui Li, Haizheng Wang, Daling Feng, Chunyang Wang. Research Advances and Prospects in Charophytes Genomics [J]. Chinese Bulletin of Botany, 2025, 60(2): 271-282. |
| [5] | Lin Zhen, Xiang Jiabao, Cai Hejiayi, Gao Bei, Yang Jintao, Li Junyi, Zhou Qingsong, Huang Xiaolei, Deng Jun. Mitochondrial genomic data for seven Hemipteran species [J]. Biodiv Sci, 2025, 33(2): 24434-. |
| [6] | Cao Dong, Li Huanlong, Peng Yang, Wei Cunzheng. Progresses in the study of the relationship between plant genome size and traits [J]. Biodiv Sci, 2025, 33(2): 24192-. |
| [7] | Hong Deng, Zhanyou Zhong, Chunni Kou, Shuli Zhu, Yuefei Li, Yuguo Xia, Zhi Wu, Jie Li, Weitao Chen. Population genetic structure and evolutionary history of Hemibagrus guttatus based on mitochondrial genomes [J]. Biodiv Sci, 2025, 33(1): 24241-. |
| [8] | Xiangtan Yao, Xinyi Zhang, Yang Chen, Ye Yuan, Wangda Cheng, Tianrui Wang, Yingxiong Qiu. Genomic resequencing reveals the genetic diversity of the cultivated water caltrop, and the origin and domestication of ‘Nanhuling’ [J]. Biodiv Sci, 2024, 32(9): 24212-. |
| [9] | Qiang Zhang, Zhenyu Zhao, Pinghua Li. Research Progress of Gene Editing Technology in Maize [J]. Chinese Bulletin of Botany, 2024, 59(6): 978-998. |
| [10] | Qingguo Du, Wenxue Li. Research Progress in the Regulation of Development and Stress Responses by Long Non-coding RNAs in Maize [J]. Chinese Bulletin of Botany, 2024, 59(6): 950-962. |
| [11] | Ziyang Wang, Shengxue Liu, Zhirui Yang, Feng Qin. Genetic Dissection of Drought Resistance in Maize [J]. Chinese Bulletin of Botany, 2024, 59(6): 883-902. |
| [12] | Juan Yang, Yuelei Zhao, Xiaoyuan Chen, Baobao Wang, Haiyang Wang. Regulation Mechanism and Breeding Application of Flowering Time in Maize [J]. Chinese Bulletin of Botany, 2024, 59(6): 912-931. |
| [13] | Hengyu Yan, Zhaoxia Li, Yubin Li. Research Progress on Heat Stress Impact on Maize Growth and Heat-Tolerant Maize Screening in China [J]. Chinese Bulletin of Botany, 2024, 59(6): 1007-1023. |
| [14] | Yuan Li, Kaijian Fan, Tai An, Cong Li, Junxia Jiang, Hao Niu, Weiwei Zeng, Yanfang Heng, Hu Li, Junjie Fu, Huihui Li, Liang Li. Study on Multi-environment Genome-wide Prediction of Inbred Agronomic Traits in Maize Natural Populations [J]. Chinese Bulletin of Botany, 2024, 59(6): 1041-1053. |
| [15] | Wenli Yang, Zhao Li, Zhiming Liu, Zhihua Zhang, Jinsheng Yang, Yanjie Lü, Yongjun Wang. Senescence Characteristics of Maize Leaves at Different Maturity Stages and Their Effect on Phyllosphere Bacteria [J]. Chinese Bulletin of Botany, 2024, 59(6): 1024-1040. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
