

Chinese Bulletin of Botany ›› 2023, Vol. 58 ›› Issue (6): 866-881.DOI: 10.11983/CBB22261 cstr: 32102.14.CBB22261
• EXPERIMENTAL COMMUNICATIONS • Previous Articles Next Articles
Wenqi Zhou1,*( ), Yuqian Zhou1, Yongsheng Li1, Haijun He1, Yanzhong Yang1, Xiaojuan Wang1, Xiaorong Lian1, Zhongxiang Liu1, Zhubing Hu2,*(
), Yuqian Zhou1, Yongsheng Li1, Haijun He1, Yanzhong Yang1, Xiaojuan Wang1, Xiaorong Lian1, Zhongxiang Liu1, Zhubing Hu2,*( )
)
Received:2022-11-14
Accepted:2023-04-18
Online:2023-11-01
Published:2023-11-27
Contact:
* E-mail: zhouwenqi850202@163.com;zhubinghu@henu.edu.cn
Wenqi Zhou, Yuqian Zhou, Yongsheng Li, Haijun He, Yanzhong Yang, Xiaojuan Wang, Xiaorong Lian, Zhongxiang Liu, Zhubing Hu. ZmICE2 Regulates Stomatal Development in Maize[J]. Chinese Bulletin of Botany, 2023, 58(6): 866-881.
| Primer name | Forward primer (5′-3′) | Reverse primer (5′-3′) |
|---|---|---|
| ZmICE2 | ACCCCAACAAAACCAGGAC | GTCTCTTTTTCAGCGGTCTTG |
| T-ice2-1 | GAGGAGGACGACGACAAGAAG | GCTAATTGCTACCGAAAACGC |
| T-ice1-1 | GCGACCCATCAACCCATAGC | CGGTGTTGATGGGTTGAAGC |
| T-ice2-c9 | ACGAGAATGGGTGAGTGTGG | AAGAGCGAGAACATCTGCGA |
| GAPDH | CCATCACTGCCACACAGAAAAC | AGGAACACGGAAGGACATACCAG |
| RT-ZmICE2 | CTTCCTCGGGCGGCGGCGGTG | ACCGTGCTGTTGGCGTTGGAG |
Table 1 Primer sequences used in this study
| Primer name | Forward primer (5′-3′) | Reverse primer (5′-3′) |
|---|---|---|
| ZmICE2 | ACCCCAACAAAACCAGGAC | GTCTCTTTTTCAGCGGTCTTG |
| T-ice2-1 | GAGGAGGACGACGACAAGAAG | GCTAATTGCTACCGAAAACGC |
| T-ice1-1 | GCGACCCATCAACCCATAGC | CGGTGTTGATGGGTTGAAGC |
| T-ice2-c9 | ACGAGAATGGGTGAGTGTGG | AAGAGCGAGAACATCTGCGA |
| GAPDH | CCATCACTGCCACACAGAAAAC | AGGAACACGGAAGGACATACCAG |
| RT-ZmICE2 | CTTCCTCGGGCGGCGGCGGTG | ACCGTGCTGTTGGCGTTGGAG |
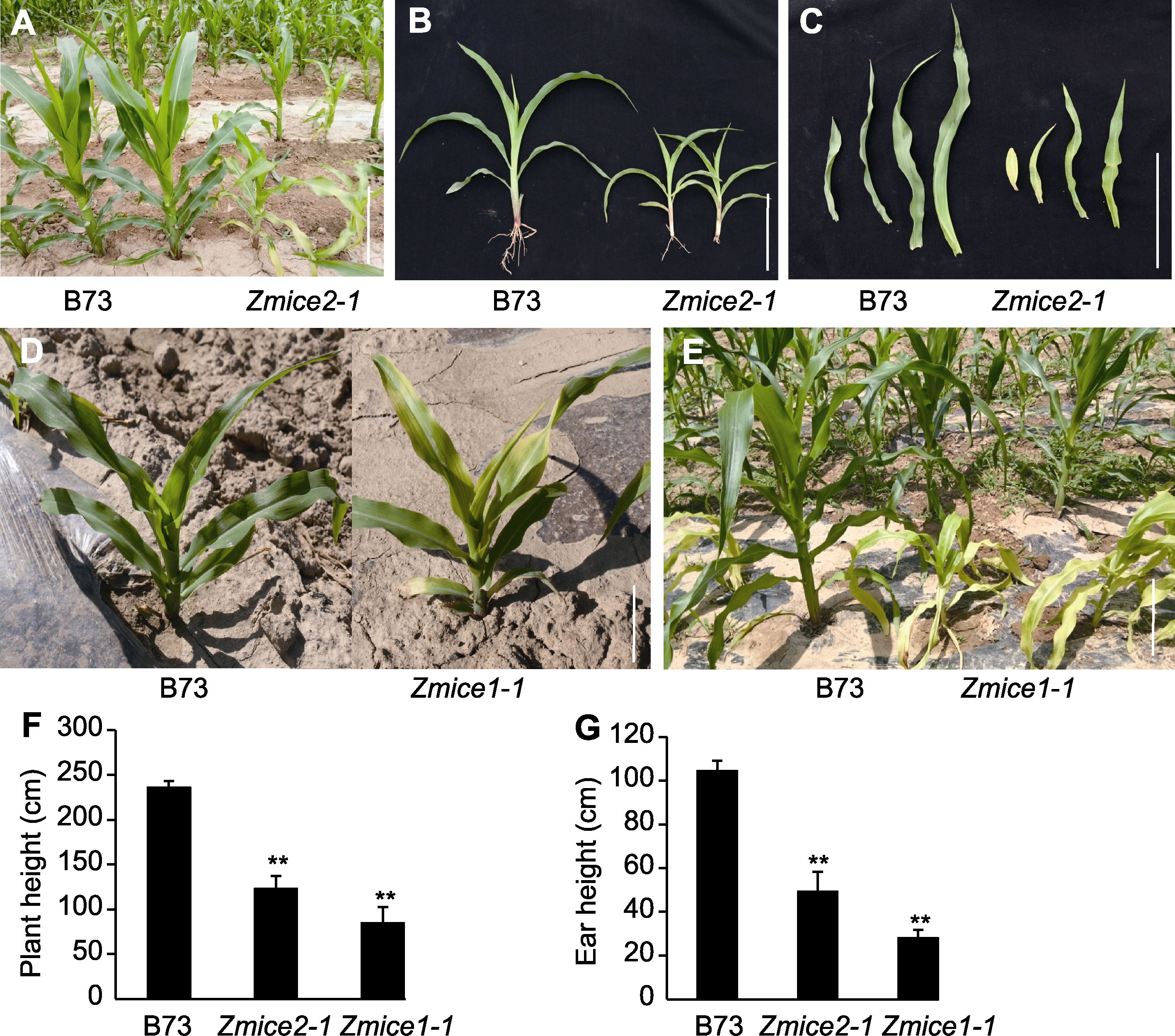
Figure 1 Field phenotypes of Zmice1-1 and Zmice2-1 mutants at seedling stage (A)-(C) Zmice2-1 mutant showed yellow leaf phenotype and short plant type at seedling stage; (D), (E) From the five-leaf and one-heart stage, the Zmice1-1 leaves gradually began to turn yellow, and the phenotype became more obvious at the later stage; (F), (G) The plant height and ear height of Zmice2-1 and Zmice1-1 were significantly decreased compared with the control (** indicate extremely significant differences (P<0.01)). In the figure (G), since the homozygote of Zmice1-1 had not developed a female spike, the spike height was calculated as the height from the ground of the fallen 6 leaves at maturity stage. (B), (C), (D) Bars=10 cm; (A), (E) Bars=20 cm
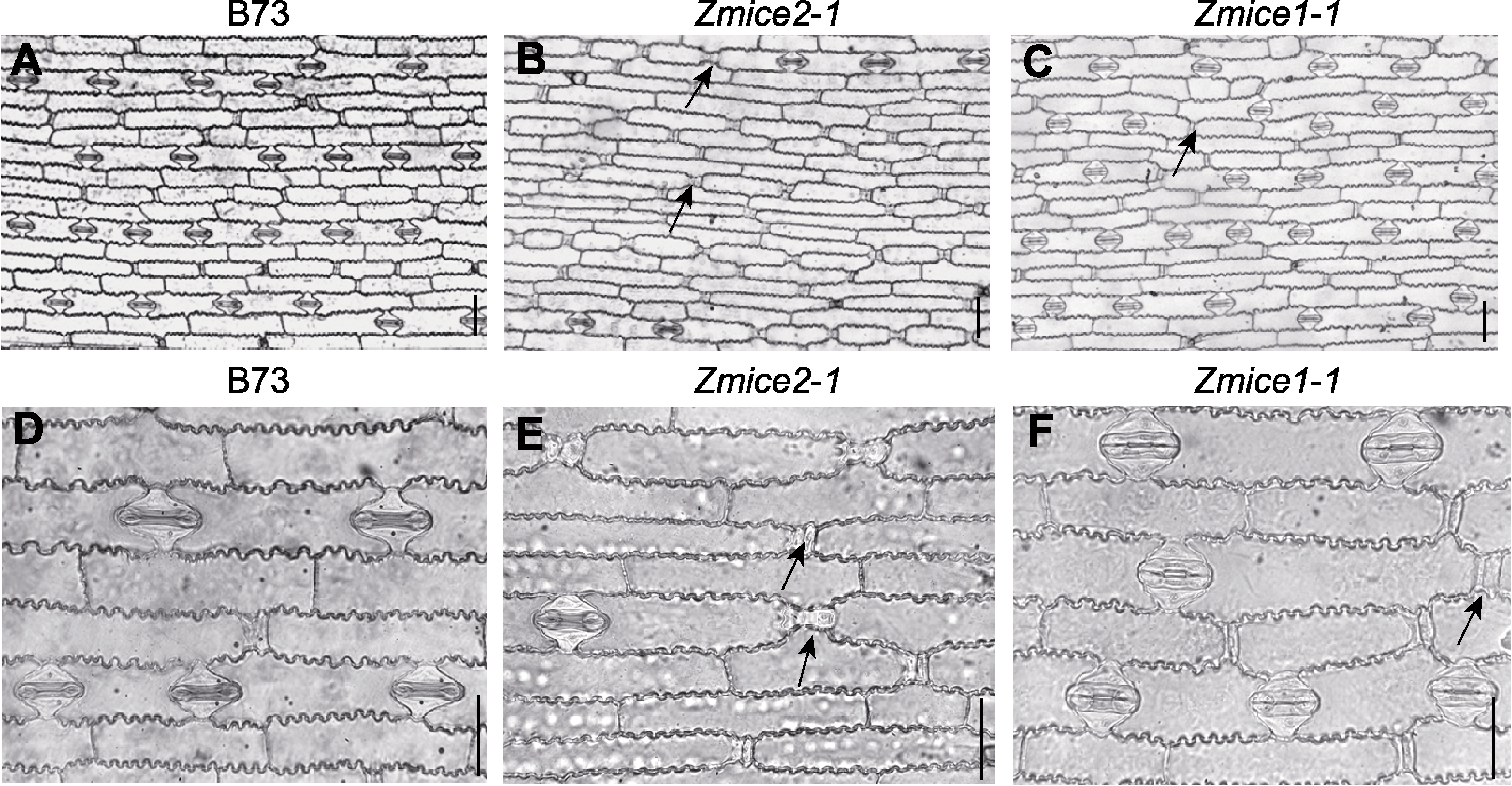
Figure 2 Morphology and stomatal distribution of Zmice1-1 and Zmice2-1 mutants (A) Schematic diagram of stomata and epidermal cells in leaves of B73 seedlings (with one stomata spaced by one epidermal long cell in a monolinear regular arrangement); (B) Stomata and epidermal cell schema of young leaves of Zmice2-1 (there were only a few stomata in the same size field of vision, and stomatal density decreased significantly); (C) Stomata and epidermal cell pattern of Zmice1-1 young leaves (showed no significant difference from the control); (D) Epidermal cell morphology of B73; (E) The epidermal cell morphology of Zmice2-1 (in the setting stage number of stomata reduced); (F) Stomatal morphology and epidermal flat cell morphology of leaves at Zmice1-1 setting stage (with increased stomatal opening). Arrows indicate immature stomatas. Bars=20 μm
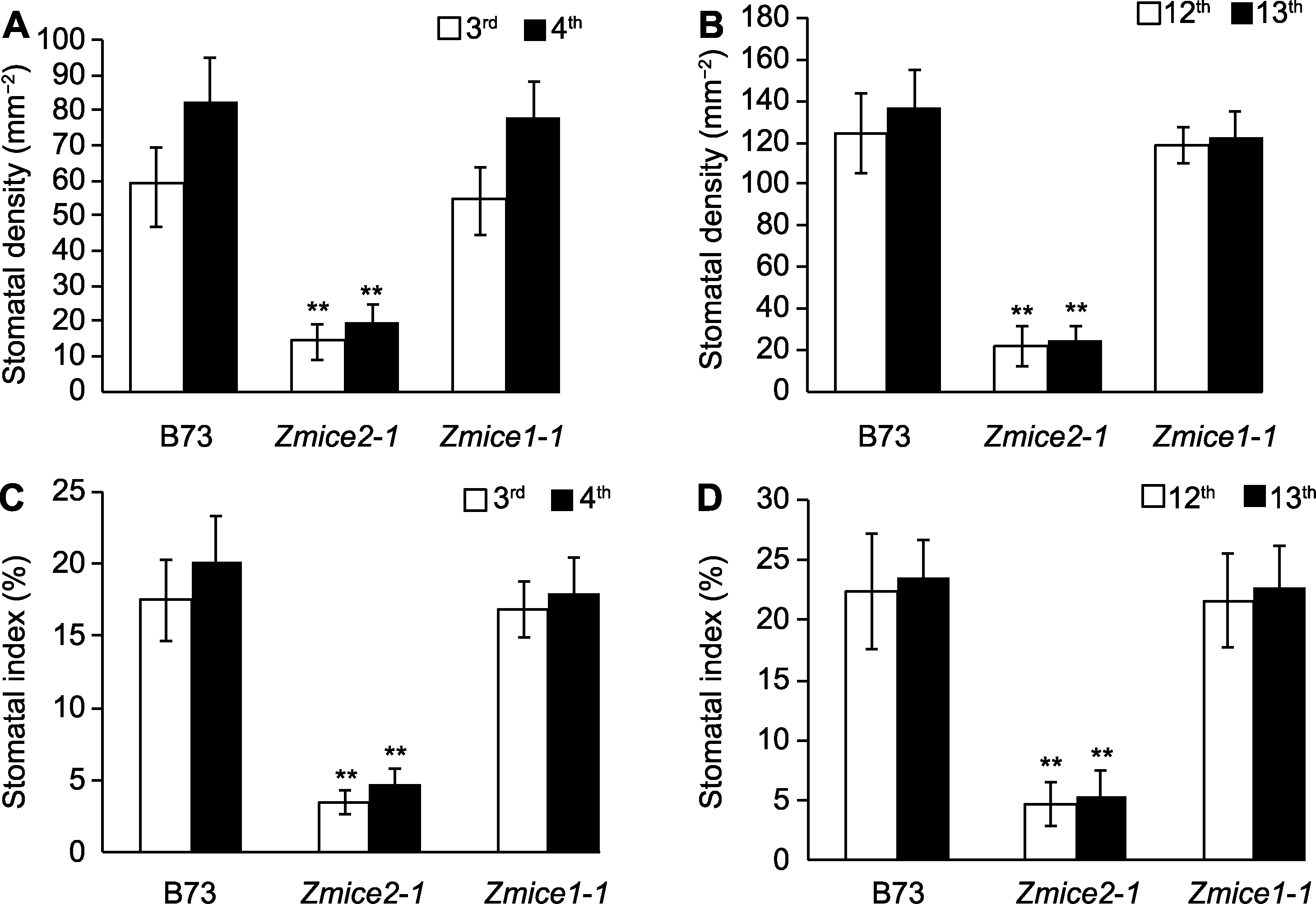
Figure 3 Stomatal density and stomatal index of B73 and Zmice mutants (A), (B) The stomatal density of the 3rd, 4th, 12th and 13th leaves was calculated, the stomatal density of Zmice2-1 was significantly lower than that of the control, while the stomatal density of Zmice1-1 showed no significant change (n=200); (C), (D) The stomatal indexes of the 3rd, 4th, 12th and 13th leaves were counted, Zmice2-1 stomatal indexes were significantly lower than that of the control, while Zmice1-1 stomatal indexes had no significant change (n=200). n=200 represented the statistical number of stomata (≥15 microscopic fields) and 3 biological replicates. ** P<0.01
| Species | Homologous gene | The function of prediction |
|---|---|---|
| Arabidopsis thaliana | AT3G26744 | Basic helix-loop-helix (bHLH) DNA-binding superfamily protein |
| Oryza sativa | LOC_Os11g32100 | Inducer of CBF expression 1, putative, expressed |
| Brachypodium distachyum | Bradi4g17460 | ICE87 |
| Zea mays | GRMZM2G033356 | Helix-loop-helix DNA-binding domain containing protein |
| Sorghum bicolor | Sb05g019530 | Helix-loop-helix DNA-binding domain containing protein |
| Vitis vinifera | GSVIVG00008637001 | Helix-loop-helix DNA-binding domain containing protein |
| V. vinifera | GSVIVG00032998001 | Inducer of CBF expression 1 |
| Populus trichocarpa | POPTR_0012s10780 | ICE1; DNA binding/transcription activator/transcription factor |
| P. trichocarpa | POPTR_0015s11650 | ICE1; DNA binding/transcription activator/transcription factor |
Table 2 Gene code and function annotation of ZmICE2 in different species
| Species | Homologous gene | The function of prediction |
|---|---|---|
| Arabidopsis thaliana | AT3G26744 | Basic helix-loop-helix (bHLH) DNA-binding superfamily protein |
| Oryza sativa | LOC_Os11g32100 | Inducer of CBF expression 1, putative, expressed |
| Brachypodium distachyum | Bradi4g17460 | ICE87 |
| Zea mays | GRMZM2G033356 | Helix-loop-helix DNA-binding domain containing protein |
| Sorghum bicolor | Sb05g019530 | Helix-loop-helix DNA-binding domain containing protein |
| Vitis vinifera | GSVIVG00008637001 | Helix-loop-helix DNA-binding domain containing protein |
| V. vinifera | GSVIVG00032998001 | Inducer of CBF expression 1 |
| Populus trichocarpa | POPTR_0012s10780 | ICE1; DNA binding/transcription activator/transcription factor |
| P. trichocarpa | POPTR_0015s11650 | ICE1; DNA binding/transcription activator/transcription factor |
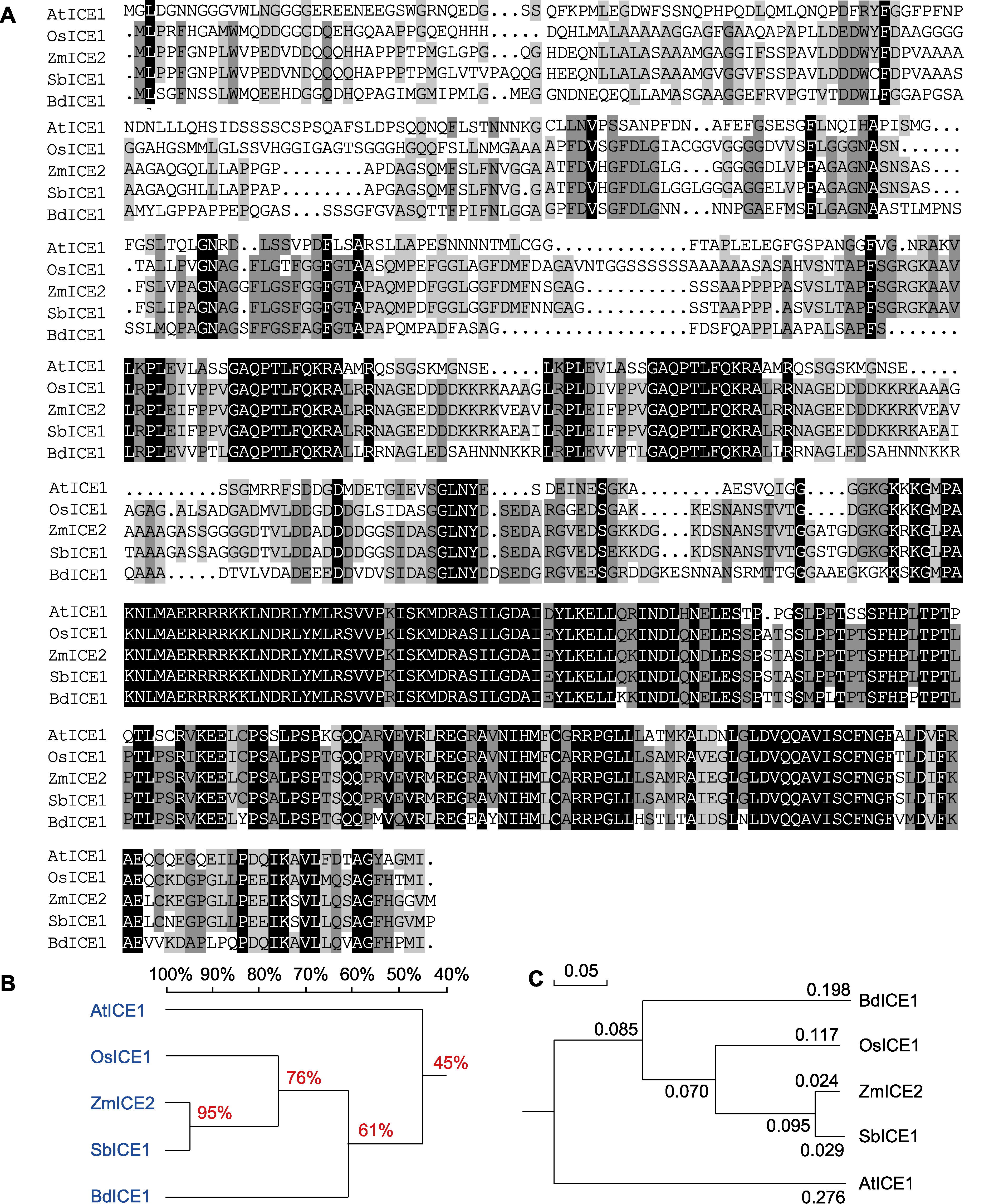
Figure 4 Sequence alignment of amino acid sequence of ZmICE2 with its homologues and phylogenetic analysis (A) Conservative analysis of ZmICE2 protein sequences (with black indicating 100% identical amino acid sequences; dark gray means 75% the same amino acid sequences; light gray means 50% the same amino acid sequences); (B) Homologous tree analysis of ZmICE2 protein; (C) Phylogenetic tree analysis of ZmICE2 protein
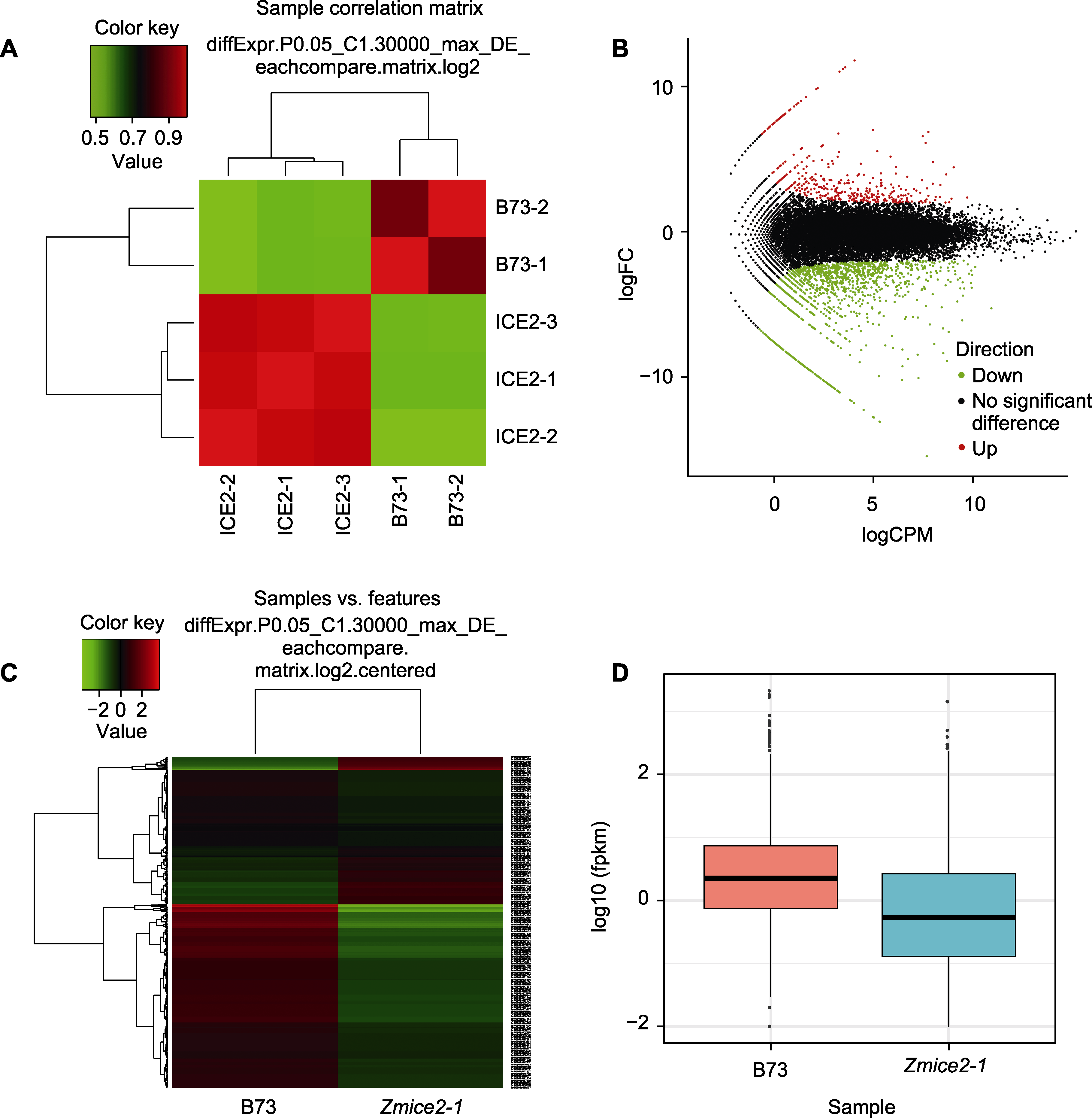
Figure 5 Cluster analysis of differentially expressed genes in B73 and Zmice2-1 (A) Sample correlation analysis based on differential expression gene; (B) MA map of differentially expressed genes (each dot represents a gene); (C) Cluster analysis diagram of differentially expressed genes (each row represents a gene and each column represents a sample); (D) Boxplot of transcript expression (ZmICE2 expression decreased in Zmice2-1).

Figure 7 Phenotype of ZmICE2 gene editing mutant (A) ZmICE2 transgenic test-tube seedlings; (B), (C) Phenotype of ZmICE2 gene editing T0 generation transgenic plant in field; (D) Phenotype of T2 generation transgenic plant in field (leaf yellowing); (E), (F) The epidermal stomatal schema of control (CK) and Zmice2-2 (Zmice2-2 lacked a large number of stomata and could not form normal stomata). Arrows indicate immature stomata. (A)-(D) Bars=10 cm; (E), (F) Bars=20 μm
| [1] | 陈亮, 侯岁稳 (2017). 植物气孔发育的分子遗传调控. 中国科学: 生命科学 47, 798-807. |
| [2] | 陈青云 (2017). 玉米中STOMAGEN-Like基因调控气孔发育的功能研究. 硕士论文. 南宁: 广西大学. pp. 35-60. |
| [3] | 刘延波, 项阳, 秦利军, 赵德刚 (2014). 转玉米ZmSDD1基因烟草降低气孔密度提高抗旱性. 植物生理学报 50, 1889-1898. |
| [4] |
牛艳丽, 柏胜龙, 王麒云, 刘凌云 (2017). 单细胞组学技术及其在植物保卫细胞研究中的应用. 植物学报 52, 788-796.
DOI |
| [5] |
商业绯, 李明, 丁博, 牛浩, 杨振宁, 陈小强, 曹高燚, 谢晓东 (2017). 生长素调控植物气孔发育的研究进展. 植物学报 52, 235-240.
DOI |
| [6] |
王宏亮, 郭思义, 王棚涛, 宋纯鹏 (2018). 植物气孔发育机制研究进展. 植物学报 53, 164-174.
DOI |
| [7] | 张一弓, 张怡, 张怡, 阿依白合热木·木台力甫, 张道远 (2021). 异源过表达齿肋赤藓ScABI3基因改变拟南芥气孔表型并提高抗旱性. 植物学报 56, 414-421. |
| [8] | 周文期 (2015). 调控水稻叶表皮发育的LPL2和DSP1基因克隆与功能分析. 博士论文. 兰州: 兰州大学. pp. 38-66. |
| [9] | 周文期, 寇思荣, 连晓荣, 杨彦忠, 刘忠祥, 王晓娟, 何海军, 周玉乾 (2020a). 水稻和玉米叶表皮突变体的筛选和鉴定. 植物生理学报 56, 189-199. |
| [10] | 周文期, 连晓荣, 周玉乾, 王兴荣, 杨彦忠, 刘忠祥, 王晓娟, 何海军, 寇思荣 (2020b). EMS诱变玉米自交系种质创新应用. 玉米科学 28(6), 31-38. |
| [11] | 周玉乾, 孟思远, 周文期 (2018). 植物表皮形态建成的分子调控机制. 西北农学报 27, 609-616. |
| [12] |
周文期, 强晓霞, 王森, 江静雯, 卫万荣 (2022). 水稻OsLPL2/PIR基因抗旱耐盐机制研究. 作物学报 48, 1401-1415.
DOI |
| [13] |
周玉萍, 颜嘉豪, 田长恩 (2022). 保卫细胞中ABA信号调控机制研究进展. 植物学报 57, 684-696.
DOI |
| [14] |
Bergmann DC, Sack FD (2007). Stomatal development. Annu Rev Plant Biol 58, 163-181.
PMID |
| [15] |
Caine RS, Yin XJ, Sloan J, Harrison EL, Mohammed U, Fulton T, Biswal AK, Dionora J, Chater CC, Coe RA, Bandyopadhyay A, Murchie EH, Swarup R, Quick WP, Gray JE (2019). Rice with reduced stomatal density conserves water and has improved drought tolerance under future climate conditions. New Phytol 221, 371-384.
DOI PMID |
| [16] |
Chater CCC, Caine RS, Fleming AJ, Gray JE (2017). Origins and evolution of stomatal development. Plant Physiol 174, 624-638.
DOI PMID |
| [17] |
Chen ZH, Chen G, Dai F, Wang YZ, Hills A, Ruan YL, Zhang GP, Franks PJ, Nevo E, Blatt MR (2017). Molecular evolution of grass stomata. Trends Plant Sci 22, 124-139.
DOI URL |
| [18] |
Chinnusamy V, Ohta M, Kanrar S, Lee BH, Hong XH, Agarwal M, Zhu JK (2003). ICE1: a regulator of cold- induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev 17, 1043-1054.
DOI URL |
| [19] | Cuming AC, Cho SH, Kamisugi Y, Graham H, Quatrano RS (2007). Microarray analysis of transcriptional responses to abscisic acid and osmotic, salt, and drought stress in the moss, Physcomitrella patens. New Phytol 176, 275-287. |
| [20] |
Deng CY, Ye HY, Fan M, Pu TL, Yan JB (2017). The rice transcription factors OsICE confer enhanced cold tolerance in transgenic Arabidopsis. Plant Signal Behav 12, e1316442.
DOI URL |
| [21] |
Dong CH, Agarwal M, Zhang YY, Xie Q, Zhu JK (2006). The negative regulator of plant cold responses, HOS1, is a RING E3 ligase that mediates the ubiquitination and degradation of ICE1. Proc Natl Acad Sci USA 103, 8281-8286.
DOI URL |
| [22] |
Edwards D, Kerp H, Hass H (1998). Stomata in early land plants: an anatomical and ecophysiological approach. J Exp Bot 49, 255-278.
DOI URL |
| [23] |
Feng F, Qi WW, Lv YD, Yan SM, Xu LM, Yang WY, Yuan Y, Chen YH, Zhao H, Song RT (2018). OPAQUE11 is a central hub of the regulatory network for maize endosperm development and nutrient metabolism. Plant Cell 30, 375-396.
DOI URL |
| [24] |
Frank MJ, Smith LG (2002). A small, novel protein highly conserved in plants and animals promotes the polarized growth and division of maize leaf epidermal cells. Curr Biol 12, 849-853.
PMID |
| [25] |
Gao Y, Wu MQ, Zhang MJ, Jiang W, Ren XY, Liang EX, Zhang DP, Zhang CQ, Xiao N, Li Y, Dai Y, Chen JM (2018). A maize phytochrome-interacting factors protein ZmPIF1 enhances drought tolerance by inducing stomatal closure and improves grain yield in Oryza sativa. Plant Biotechnol J 16, 1375-1387.
DOI URL |
| [26] |
Han SK, Qi XY, Sugihara K, Dang JH, Endo TA, Miller KL, Kim ED, Miura T, Torii KU (2018). MUTE directly orchestrates cell-state switch and the single symmetric division to create stomata. Dev Cell 45, 303-315.
DOI URL |
| [27] |
Han SK, Torii KU (2019). Linking cell cycle to stomatal differentiation. Curr Opin Plant Biol 51, 66-73.
DOI URL |
| [28] |
Hepworth C, Caine RS, Harrison EL, Sloan J, Gray JE (2018). Stomatal development: focusing on the grasses. Curr Opin Plant Biol 41, 1-7.
DOI PMID |
| [29] |
Jiang HF, Shi YT, Liu JY, Li Z, Fu DY, Wu SF, Li MZ, Yang ZJ, Shi YL, Lai JS, Yang XH, Gong ZZ, Hua J, Yang SH (2022). Natural polymorphism of ZmICE1 contributes to amino acid metabolism that impacts cold tolerance in maize. Nat Plants 8, 1176-1190.
DOI PMID |
| [30] |
Kanaoka MM, Pillitteri LJ, Fujii H, Yoshida Y, Bogenschutz NL, Takabayashi J, Zhu JK, Torii KU (2008). SCREAM/ICE1 and SCREAM2 specify three cell-state transitional steps leading to Arabidopsis stomatal differentiation. Plant Cell 20, 1775-1785.
DOI URL |
| [31] |
Kidokoro S, Kim JS, Ishikawa T, Suzuki T, Shinozaki K, Yamaguchi-Shinozaki K (2020). DREB1A/CBF3 is repressed by transgene-induced DNA methylation in the Arabidopsis ice1-1 mutant. Plant Cell 32, 1035-1048.
DOI URL |
| [32] |
Lau OS, Bergmann DC (2012). Stomatal development: a plant’s perspective on cell polarity, cell fate transitions and intercellular communication. Development 139, 3683-3692.
DOI URL |
| [33] |
Li XM, Han HP, Chen M, Yang W, Liu L, Li N, Ding XH, Chu ZH (2017). Overexpression of OsDT11, which encodes a novel cysteine-rich peptide, enhances drought tolerance and increases ABA concentration in rice. Plant Mol Biol 93, 21-34.
DOI URL |
| [34] |
Liu T, Ohashi-Ito K, Bergmann DC (2009). Orthologs of Arabidopsis thaliana stomatal bHLH genes and regulation of stomatal development in grasses. Development 136, 2265-2276.
DOI URL |
| [35] |
Lu XD, Liu JS, Ren W, Yang Q, Chai ZG, Chen RM, Wang L, Zhao J, Lang ZH, Wang HY, Fan YL, Zhao JR, Zhang CY (2018). Gene-indexed mutations in maize. Mol Plant 11, 496-504.
DOI PMID |
| [36] |
MacAlister CA, Ohashi-Ito K, Bergmann DC (2007). Transcription factor control of asymmetric cell divisions that establish the stomatal lineage. Nature 445, 537-540.
DOI |
| [37] |
Matos JL, Lau OS, Hachez C, Cruz-Ramírez A, Scheres B, Bergmann DC (2014). Irreversible fate commitment in the Arabidopsis stomatal lineage requires a FAMA and RETINOBLASTOMA-RELATED module. eLife 3, e03271.
DOI URL |
| [38] |
McKown KH, Bergmann DC (2020). Stomatal development in the grasses: lessons from models and crops (and crop models). New Phytol 227, 1636-1648.
DOI URL |
| [39] |
Miura K, Jin JB, Lee J, Yoo CY, Stirm V, Miura T, Ashworth EN, Bressan RA, Yun DJ, Hasegawa PM (2007). SIZ1-mediated sumoylation of ICE1 controls CBF3/ DREB1A expression and freezing tolerance in Arabidopsis. Plant Cell 19, 1403-1414.
DOI URL |
| [40] |
Nadeau JA, Sack FD (2002). Control of stomatal distribution on the Arabidopsis leaf surface. Science 296, 1697-1700.
DOI URL |
| [41] |
Ohashi-Ito K, Bergmann DC (2006). Arabidopsis FAMA controls the final proliferation/differentiation switch during stomatal development. Plant Cell 18, 2493-2505.
DOI PMID |
| [42] | Pillitteri LJ, Sloan DB, Bogenschutz NL, Torii KU (2007). Termination of asymmetric cell division and differentiation of stomata. Nature 445, 501-505. |
| [43] |
Pillitteri LJ, Torii KU (2012). Mechanisms of stomatal development. Annu Rev Plant Biol 63, 591-614.
DOI PMID |
| [44] |
Putarjunan A, Ruble J, Srivastava A, Zhao CZ, Rychel AL, Hofstetter AK, Tang XB, Zhu JK, Tama F, Zheng N, Torii KU (2019). Bipartite anchoring of SCREAM enforces stomatal initiation by coupling MAP kinases to SPEECHLESS. Nat Plants 5, 742-754.
DOI PMID |
| [45] | Qu X, Peterson KM, Torii KU (2017). Stomatal development in time: the past and the future. Curr Opin Genes Dev 45, 1-9. |
| [46] |
Raissig MT, Abrash E, Bettadapur A, Vogel JP, Bergmann DC (2016). Grasses use an alternatively wired bHLH transcription factor network to establish stomatal identity. Proc Natl Acad Sci USA 113, 8326-8331.
DOI PMID |
| [47] |
Raissig MT, Matos JL, Ximena Anleu Gil M, Kornfeld A, Bettadapur A, Abrash E, Allison HR, Badgley G, Vogel JP, Berry JA, Bergmann DC (2017). Mobile MUTE specifies subsidiary cells to build physiologically improved grass stomata. Science 355, 1215-1218.
DOI PMID |
| [48] |
Rudall PJ, Hilton J, Bateman RM (2013). Several developmental and morphogenetic factors govern the evolution of stomatal patterning in land plants. New Phytol 200, 598-614.
DOI PMID |
| [49] |
Serna L (2011). Stomatal development in Arabidopsis and grasses: differences and commonalities. Int J Dev Biol 55, 5-10.
DOI URL |
| [50] |
Serna L (2020). The role of grass MUTE orthologues during stomatal development. Front Plant Sci 11, 55.
DOI URL |
| [51] |
Wang HL, Guo SY, Qiao X, Guo JF, Li ZL, Zhou YS, Bai SL, Gao ZY, Wang DJ, Wang PC, Galbraith DW, Song CP (2019). BZU2/ZmMUTE controls symmetrical division of guard mother cell and specifies neighbor cell fate in maize. PLoS Genet 15, e1008377.
DOI URL |
| [52] |
Wei DH, Liu MJ, Chen H, Zheng Y, Liu YX, Wang X, Yang SH, Zhou MQ, Lin J (2018). INDUCER OF CBF EXPRESSION 1 is a male fertility regulator impacting anther dehydration in Arabidopsis. PLoS Genet 14, e1007695.
DOI URL |
| [53] |
Wu ZL, Chen L, Yu Q, Zhou WQ, Gou XP, Li J, Hou SW (2019). Multiple transcriptional factors control stomata development in rice. New Phytol 223, 220-232.
DOI PMID |
| [54] | Ye KY, Li H, Ding YL, Shi YT, Song CP, Gong ZZ, Yang SH (2019). BRASSINOSTEROID-INSENSITIVE2 negatively regulates the stability of transcription factor ICE1 in response to cold stress in Arabidopsis. Plant Cell 31, 2682-2696. |
| [55] |
Yoo CY, Pence HE, Jin JB, Miura K, Gosney MJ, Hasegawa PM, Mickelbart MV (2010). The Arabidopsis GTL1 transcription factor regulates water use efficiency and drought tolerance by modulating stomatal density via transrepression of SDD1. Plant Cell 22, 4128-4141.
DOI URL |
| [56] |
Zhou WQ, Wang YC, Wu ZL, Luo L, Liu P, Yan LF, Hou SW (2016). Homologs of SCAR/WAVE complex components are required for epidermal cell morphogenesis in rice. J Exp Bot 67, 4311-4323.
DOI PMID |
| [57] |
Zoulias N, Harrison EL, Casson SA, Gray JE (2018). Molecular control of stomatal development. Biochem J 475, 441-454.
DOI PMID |
| [1] | Juan Yang, Yuelei Zhao, Xiaoyuan Chen, Baobao Wang, Haiyang Wang. Regulation Mechanism and Breeding Application of Flowering Time in Maize [J]. Chinese Bulletin of Botany, 2024, 59(6): 912-931. |
| [2] | Hengyu Yan, Zhaoxia Li, Yubin Li. Research Progress on Heat Stress Impact on Maize Growth and Heat-Tolerant Maize Screening in China [J]. Chinese Bulletin of Botany, 2024, 59(6): 1007-1023. |
| [3] | Qingguo Du, Wenxue Li. Research Progress in the Regulation of Development and Stress Responses by Long Non-coding RNAs in Maize [J]. Chinese Bulletin of Botany, 2024, 59(6): 950-962. |
| [4] | Ziyang Wang, Shengxue Liu, Zhirui Yang, Feng Qin. Genetic Dissection of Drought Resistance in Maize [J]. Chinese Bulletin of Botany, 2024, 59(6): 883-902. |
| [5] | Qiang Zhang, Zhenyu Zhao, Pinghua Li. Research Progress of Gene Editing Technology in Maize [J]. Chinese Bulletin of Botany, 2024, 59(6): 978-998. |
| [6] | Yuan Li, Kaijian Fan, Tai An, Cong Li, Junxia Jiang, Hao Niu, Weiwei Zeng, Yanfang Heng, Hu Li, Junjie Fu, Huihui Li, Liang Li. Study on Multi-environment Genome-wide Prediction of Inbred Agronomic Traits in Maize Natural Populations [J]. Chinese Bulletin of Botany, 2024, 59(6): 1041-1053. |
| [7] | Suowei Wu, Xueli An, Xiangyuan Wan. Molecular Mechanisms of Male Sterility and their Applications in Biotechnology-based Male-sterility Hybrid Seed Production in Maize [J]. Chinese Bulletin of Botany, 2024, 59(6): 932-949. |
| [8] | Mingmin Zheng, Qiang Huang, Peng Zhang, Xiaowei Liu, Zhuofan Zhao, Hongyang Yi, Tingzhao Rong, Moju Cao. Research Progress on Cytoplasmic Male Sterility and Fertility Restoration in Maize [J]. Chinese Bulletin of Botany, 2024, 59(6): 999-1006. |
| [9] | Tao Wang, Jinglei Feng, Cui Zhang. Research Progress on Molecular Mechanisms of Heat Stress Affecting the Growth and Development of Maize [J]. Chinese Bulletin of Botany, 2024, 59(6): 963-977. |
| [10] | Wenli Yang, Zhao Li, Zhiming Liu, Zhihua Zhang, Jinsheng Yang, Yanjie Lü, Yongjun Wang. Senescence Characteristics of Maize Leaves at Different Maturity Stages and Their Effect on Phyllosphere Bacteria [J]. Chinese Bulletin of Botany, 2024, 59(6): 1024-1040. |
| [11] | CHENG Ke-Xin, DU Yao, LI Kai-Hang, WANG Hao-Chen, YANG Yan, JIN Yi, HE Xiao-Qing. Genetic mechanism of interaction between maize and phyllospheric microbiome [J]. Chin J Plant Ecol, 2024, 48(2): 215-228. |
| [12] | Yi Song, Hanghang Chen, Xin Cui, Zhifeng Lu, Shipeng Liao, Yangyang Zhang, Xiaokun Li, Rihuan Cong, Tao Ren, Jianwei Lu. Potassium Nutrient Status-mediated Leaf Growth of Oilseed Rape (Brassica napus) and Its Effect on Phyllosphere Microorganism [J]. Chinese Bulletin of Botany, 2024, 59(1): 54-65. |
| [13] | Xiting Yu, Xuehui Huang. New Insights Into the Origin of Modern Maize-hybridization of Two Teosintes [J]. Chinese Bulletin of Botany, 2023, 58(6): 857-860. |
| [14] | Li Guo, Xuehan Wang, Feng Tian. Multi-omics Integrative Network Map, a Key to Accurately Deco-ding the Maize Functional Genomics [J]. Chinese Bulletin of Botany, 2023, 58(1): 1-5. |
| [15] | Jiahuan Sun, Dong Liu, Jiaqi Zhu, Shuning Zhang, Meixiang Gao. Spatial distribution pattern of soil mite community and body size in wheat- maize rotation farmland [J]. Biodiv Sci, 2022, 30(12): 22292-. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||