

Chinese Bulletin of Botany ›› 2023, Vol. 58 ›› Issue (1): 1-5.DOI: 10.11983/CBB22271 cstr: 32102.14.CBB22271
Special Issue: 杂粮生物学专辑 (2023年58卷1期)
• COMMENTARY • Next Articles
Li Guo, Xuehan Wang, Feng Tian*( )
)
Received:2022-12-05
Accepted:2022-12-13
Online:2023-01-01
Published:2023-01-05
Contact:
*E-mail: ft55@cau.edu.cn
Li Guo, Xuehan Wang, Feng Tian. Multi-omics Integrative Network Map, a Key to Accurately Deco-ding the Maize Functional Genomics[J]. Chinese Bulletin of Botany, 2023, 58(1): 1-5.
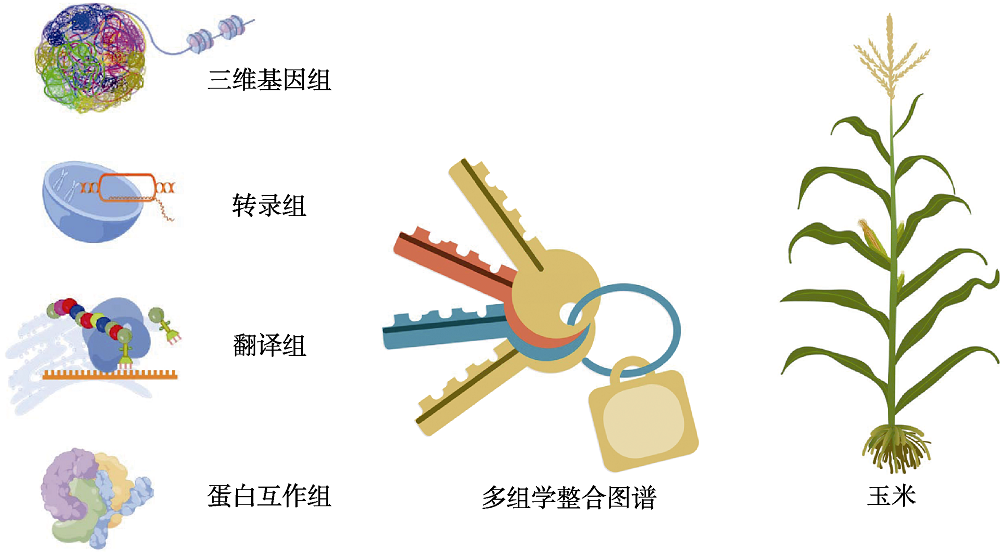
Figure 1 Multi-omics integrative network map is a new key to accurately decode the maize functional genomics (this figure is drawn by figdraw and the plant picture is adapted from Chen et al., 2021)
| [1] |
汪海, 赖锦盛, 王海洋, 李新海 (2022). 作物智能设计育种——自然变异的智能组合和人工变异的智能创制. 中国农业科技导报 24(6), 1-8.
DOI |
| [2] | 王向峰, 才卓 (2019). 中国种业科技创新的智能时代——“玉米育种4.0”. 玉米科学 27, 1-9. |
| [3] |
Chen Q, Li W, Tan L, Tian F (2021). Harnessing knowledge from maize and rice domestication for new crop breeding. Mol Plant 14, 9-26.
DOI PMID |
| [4] |
Doebley J, Stec A, Hubbard L (1997). The evolution of apical dominance in maize. Nature 386, 485-488.
DOI |
| [5] | Dong ZB, Li W, Unger-Wallace E, Yang JL, Vollbrecht E, Chuck G (2017). Ideal crop plant architecture is mediated by tassels replace upper ears1, a BTB/POZ ankyrin repeat gene directly targeted by TEOSINTE BRANCHED1. Proc Natl Acad Sci USA 114, E8656-E8664. |
| [6] |
Gallavotti A, Zhao Q, Kyozuka J, Meeley RB, Ritter MK, Doebley JF, Pè ME, Schmidt RJ (2004). The role of barren stalk1 in the architecture of maize. Nature 432, 630-635.
DOI |
| [7] |
Gälweiler L, Guan CH, Müller A, Wisman E, Mendgen K, Yephremov A, Palme K (1998). Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science 282, 2226-2230.
DOI PMID |
| [8] |
Gui ST, Wei WJ, Jiang CL, Luo JY, Chen L, Wu SS, Li WQ, Wang YB, Li SY, Yang N, Li Q, Fernie AR, Yan JB (2022). A pan-Zea genome map for enhancing maize improvement. Genome Biol 23, 178.
DOI |
| [9] | Han LQ, Zhong WS, Qian J, Jin ML, Tian P, Zhu WC, Zhang HW, Sun YH, Feng JW, Liu XG, Chen G, Farid B, Li RN, Xiong ZM, Tian ZH, Li J, Luo Z, Du DX, Chen SJ, Jin QX, Li JX, Li Z, Liang Y, Jin XM, Peng Y, Zheng C, Ye XN, Yin YJ, Chen H, Li WF, Chen LL, Li Q, Yan JB, Yang F, Li L (2022). A multi-omics integrative network map of maize. Nat Genet https://www.nature.com/articles/s41588-022-01262-1 |
| [10] | Hufford MB, Seetharam AS, Woodhouse MR, Chougule KM, Ou SJ, Liu JN, Ricci WA, Guo TT, Olson A, Qiu YJ, Della Coletta R, Tittes S, Hudson AI, Marand AP, Wei S, Lu ZY, Wang B, Tello-Ruiz MK, Piri RD, Wang N, Kim DW, Zeng YB, O’Connor CH, Li XR, Gilbert AM, Baggs E, Krasileva KV, Portwood JL II, Cannon EKS, Andorf CM, Manchanda N, Snodgrass SJ, Hufnagel DE, Jiang QH, Pedersen S, Syring ML, Kudrna DA, Llaca V, Fengler K, Schmitz RJ, Ross-Ibarra J, Yu JM, Gent JI, Hirsch CN, Ware D, Dawe RK (2021). De novo assembly, annotation, and comparative analysis of 26 diverse maize genomes. Science 373, 655-662. |
| [11] |
Liang YM, Liu HJ, Yan JB, Tian F (2021). Natural variation in crops: realized understanding, continuing promise. Annu Rev Plant Biol 72, 357-385.
DOI PMID |
| [12] |
Matsuoka Y, Vigouroux Y, Goodman MM, Sanchez GJ, Buckler E, Doebley J (2002). A single domestication for maize shown by multilocus microsatellite genotyping. Proc Natl Acad Sci USA 99, 6080-6084.
DOI PMID |
| [13] |
Peng Y, Xiong D, Zhao L, Ouyang WZ, Wang SQ, Sun J, Zhang Q, Guan PP, Xie L, Li WQ, Li GL, Yan JB, Li XW (2019). Chromatin interaction maps reveal genetic regulation for quantitative traits in maize. Nat Commun 10, 2632.
DOI PMID |
| [14] | Schnable JC, Springer NM, Freeling M (2011). Differentiation of the maize subgenomes by genome dominance and both ancient and ongoing gene loss. Proc Natl Acad Sci USA 108, 4069-4074. |
| [15] |
Tu XY, Mejía-Guerra MK, Valdes Franco JA, Tzeng D, Chu PY, Shen W, Wei YY, Dai XR, Li PH, Buckler ES, Zhong SL (2020). Reconstructing the maize leaf regulatory network using ChIP-seq data of 104 transcription factors. Nat Commun 11, 5089.
DOI PMID |
| [16] |
Walley JW, Sartor RC, Shen ZX, Schmitz RJ, Wu KJ, Urich MA, Nery JR, Smith LG, Schnable JC, Ecker JR, Briggs SP (2016). Integration of omic networks in a developmental atlas of maize. Science 353, 814-818.
DOI PMID |
| [17] |
Wen WW, Jin M, Li K, Liu HJ, Xiao YJ, Zhao MC, Alseekh S, Li WQ, de Abreu e Lima F, Brotman Y, Willmitzer L, Fernie AR, Yan JB (2018). An integrated multi-layered analysis of the metabolic networks of different tissues uncovers key genetic components of primary metabolism in maize. Plant J 93, 1116-1128.
DOI URL |
| [18] | Whipple CJ, Kebrom TH, Weber AL, Yang F, Hall D, Meeley R, Schmidt R, Doebley J, Brutnell TP, Jackson DP (2011). grassy tillers1 promotes apical dominance in maize and responds to shade signals in the grasses. Proc Natl Acad Sci USA 108, E506-E512. |
| [19] | Xiao YG, Guo JY, Dong ZB, Richardson A, Patterson E, Mangrum S, Bybee S, Bertolini E, Bartlett M, Chuck G, Eveland AL, Scanlon MJ, Whipple C (2022). Boundary domain genes were recruited to suppress bract growth and promote branching in maize. Sci Adv 8, m6835. |
| [20] |
Xu XS, Crow M, Rice BR, Li F, Harris B, Liu L, Demesa-Arevalo E, Lu ZF, Wang LY, Fox N, Wang XF, Drenkow J, Luo AD, Char SN, Yang B, Sylvester AW, Gingeras TR, Schmitz RJ, Ware D, Lipka AE, Gillis J, Jackson D (2021). Single-cell RNA sequencing of developing maize ears facilitates functional analysis and trait candidate gene discovery. Dev Cell 56, 557-568.
DOI PMID |
| [21] |
Yang F, Lei YY, Zhou ML, Yao QL, Han YC, Wu X, Zhong WS, Zhu CH, Xu WZ, Tao R, Chen X, Lin D, Rahman K, Tyagi R, Habib Z, Xiao SB, Wang D, Yu Y, Chen HC, Fu ZF, Cao G (2018). Development and application of a recombination-based library versus library high-throughput yeast two-hybrid (RLL-Y2H) screening system. Nucleic Acids Res 46, e17.
DOI URL |
| [22] |
Zhu WC, Xu J, Chen SJ, Chen J, Liang Y, Zhang CJ, Li Q, Lai JS, Li L (2021). Large-scale translatome profiling annotates the functional genome and reveals the key role of genic 3' untranslated regions in translatomic variation in plants. Plant Commun 2, 100181.
DOI URL |
| [1] | FU Zhao-Qi, HU Xu, TIAN Qin-Rui, GE Yan-Ling, ZHOU Hong-Juan, WU Xiao-Yun, CHEN Li-Xin. Nocturnal sap flow characteristics of two typical forest tree species and responses to environmental factors in the loess region of West Shanxi, China [J]. Chin J Plant Ecol, 2024, 48(9): 1128-1142. |
| [2] | Qiang Zhang, Zhenyu Zhao, Pinghua Li. Research Progress of Gene Editing Technology in Maize [J]. Chinese Bulletin of Botany, 2024, 59(6): 978-998. |
| [3] | Juan Yang, Yuelei Zhao, Xiaoyuan Chen, Baobao Wang, Haiyang Wang. Regulation Mechanism and Breeding Application of Flowering Time in Maize [J]. Chinese Bulletin of Botany, 2024, 59(6): 912-931. |
| [4] | Hengyu Yan, Zhaoxia Li, Yubin Li. Research Progress on Heat Stress Impact on Maize Growth and Heat-Tolerant Maize Screening in China [J]. Chinese Bulletin of Botany, 2024, 59(6): 1007-1023. |
| [5] | Yuan Li, Kaijian Fan, Tai An, Cong Li, Junxia Jiang, Hao Niu, Weiwei Zeng, Yanfang Heng, Hu Li, Junjie Fu, Huihui Li, Liang Li. Study on Multi-environment Genome-wide Prediction of Inbred Agronomic Traits in Maize Natural Populations [J]. Chinese Bulletin of Botany, 2024, 59(6): 1041-1053. |
| [6] | Qingguo Du, Wenxue Li. Research Progress in the Regulation of Development and Stress Responses by Long Non-coding RNAs in Maize [J]. Chinese Bulletin of Botany, 2024, 59(6): 950-962. |
| [7] | Ziyang Wang, Shengxue Liu, Zhirui Yang, Feng Qin. Genetic Dissection of Drought Resistance in Maize [J]. Chinese Bulletin of Botany, 2024, 59(6): 883-902. |
| [8] | Wenli Yang, Zhao Li, Zhiming Liu, Zhihua Zhang, Jinsheng Yang, Yanjie Lü, Yongjun Wang. Senescence Characteristics of Maize Leaves at Different Maturity Stages and Their Effect on Phyllosphere Bacteria [J]. Chinese Bulletin of Botany, 2024, 59(6): 1024-1040. |
| [9] | Tao Wang, Jinglei Feng, Cui Zhang. Research Progress on Molecular Mechanisms of Heat Stress Affecting the Growth and Development of Maize [J]. Chinese Bulletin of Botany, 2024, 59(6): 963-977. |
| [10] | Suowei Wu, Xueli An, Xiangyuan Wan. Molecular Mechanisms of Male Sterility and their Applications in Biotechnology-based Male-sterility Hybrid Seed Production in Maize [J]. Chinese Bulletin of Botany, 2024, 59(6): 932-949. |
| [11] | Mingmin Zheng, Qiang Huang, Peng Zhang, Xiaowei Liu, Zhuofan Zhao, Hongyang Yi, Tingzhao Rong, Moju Cao. Research Progress on Cytoplasmic Male Sterility and Fertility Restoration in Maize [J]. Chinese Bulletin of Botany, 2024, 59(6): 999-1006. |
| [12] | CHENG Ke-Xin, DU Yao, LI Kai-Hang, WANG Hao-Chen, YANG Yan, JIN Yi, HE Xiao-Qing. Genetic mechanism of interaction between maize and phyllospheric microbiome [J]. Chin J Plant Ecol, 2024, 48(2): 215-228. |
| [13] | WANG Xiu-Ying, CHEN Qi, DU Hua-Li, ZHANG Rui, MA Hong-Lu. Evapotranspiration interpolation in alpine marshes wetland on the Qingzang Plateau based on machine learning [J]. Chin J Plant Ecol, 2023, 47(7): 912-921. |
| [14] | Wenqi Zhou, Yuqian Zhou, Yongsheng Li, Haijun He, Yanzhong Yang, Xiaojuan Wang, Xiaorong Lian, Zhongxiang Liu, Zhubing Hu. ZmICE2 Regulates Stomatal Development in Maize [J]. Chinese Bulletin of Botany, 2023, 58(6): 866-881. |
| [15] | Xiting Yu, Xuehui Huang. New Insights Into the Origin of Modern Maize-hybridization of Two Teosintes [J]. Chinese Bulletin of Botany, 2023, 58(6): 857-860. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||