

植物学报 ›› 2021, Vol. 56 ›› Issue (3): 245-261.DOI: 10.11983/CBB20209 cstr: 32102.14.CBB20209
• 研究论文 • 下一篇
范凯, 叶方婷, 毛志君, 潘鑫峰, 李兆伟, 林文雄*( )
)
收稿日期:2020-12-25
接受日期:2021-03-25
出版日期:2021-05-01
发布日期:2021-04-30
通讯作者:
林文雄
作者简介:*E-mail: lwx@fafu.edu.cn基金资助:
Kai Fan, Fangting Ye, Zhijun Mao, Xinfeng Pan, Zhaowei Li, Wenxiong Lin*( )
)
Received:2020-12-25
Accepted:2021-03-25
Online:2021-05-01
Published:2021-04-30
Contact:
Wenxiong Lin
摘要: 小热激蛋白(sHSP)是一类重要的响应外界环境变化以及调控植物生长发育的蛋白家族。基于在睡莲(Nymphaea colorata)、水稻(Oryza sativa)、拟南芥(Arabidopsis thaliana)和葡萄(Vitis vinifera)中分别鉴定到的33个NcsHSPs、24个OssHSPs、17个AtsHSPs和47个VvsHSPs, 表明sHSP家族可分为12个亚家族, 不同亚家族包含不同的sHSP成员数目、保守基序、基因结构以及复制基因数目。在4种模式被子植物的sHSP成员中共鉴定到12个基因复制事件, 片段复制事件和串联复制事件均与sHSP成员的扩增有关, 且片段复制事件发生的时间早于串联复制事件。在所有sHSP成员中, 拟南芥和葡萄的sHSP成员的同源性最高, 其次为睡莲和葡萄的sHSP成员。sHSP家族在被子植物中可能向更短的氨基酸长度、更小的分子量、更简单的基因结构以及更集中的染色体分布进化。此外, 在睡莲、水稻、拟南芥和葡萄中鉴定了一些可能与调控植物生长发育相关的候选基因。研究结果为4种模式被子植物sHSP家族的比较基因组学研究奠定了重要基础, 并为其它被子植物sHSP家族的研究提供重要参考。
范凯, 叶方婷, 毛志君, 潘鑫峰, 李兆伟, 林文雄. 被子植物小热激蛋白家族的比较基因组学分析. 植物学报, 2021, 56(3): 245-261.
Kai Fan, Fangting Ye, Zhijun Mao, Xinfeng Pan, Zhaowei Li, Wenxiong Lin. Comparative Genomics of the Small Heat Shock Protein Family in Angiosperms. Chinese Bulletin of Botany, 2021, 56(3): 245-261.
| Protein name | Locus name | Subfamily | Protein length (aa) | Molecular weight (kDa) | Theoretical pI | Instability index | Aliphatic index | GRAVY |
|---|---|---|---|---|---|---|---|---|
| NcsHSP01 | GWHPAAYW001658 | CI | 60 | 6.64 | 9.16 | 41.10 | 99.00 | -0.218 |
| NcsHSP02 | GWHPAAYW001878 | CII | 308 | 34.77 | 9.03 | 44.31 | 74.64 | -0.624 |
| NcsHSP03 | GWHPAAYW002745 | CV | 276 | 30.85 | 4.88 | 44.78 | 77.36 | -0.291 |
| NcsHSP04 | GWHPAAYW004666 | PX | 1189 | 132.46 | 5.86 | 44.51 | 85.53 | -0.300 |
| NcsHSP05 | GWHPAAYW006130 | ER | 218 | 24.32 | 5.46 | 64.70 | 95.14 | -0.411 |
| NcsHSP06 | GWHPAAYW006158 | CII | 378 | 43.02 | 8.36 | 50.40 | 74.76 | -0.780 |
| NcsHSP07 | GWHPAAYW006159 | CII | 662 | 75.02 | 9.13 | 55.90 | 79.64 | -0.665 |
| NcsHSP08 | GWHPAAYW006161 | CII | 314 | 35.79 | 8.86 | 47.40 | 79.39 | -0.709 |
| NcsHSP09 | GWHPAAYW006746 | CIII | 164 | 18.56 | 5.75 | 53.09 | 81.28 | -0.489 |
| NcsHSP10 | GWHPAAYW007213 | CIV | 132 | 14.36 | 6.83 | 58.83 | 82.80 | -0.117 |
| NcsHSP11 | GWHPAAYW009125 | CII | 165 | 19.09 | 5.48 | 54.83 | 69.03 | -0.921 |
| NcsHSP12 | GWHPAAYW010630 | MTII | 235 | 26.36 | 6.79 | 56.35 | 67.57 | -0.687 |
| NcsHSP13 | GWHPAAYW010715 | CI | 129 | 15.42 | 9.12 | 60.26 | 59.53 | -1.343 |
| NcsHSP14 | GWHPAAYW013759 | CVI | 154 | 17.47 | 5.57 | 46.32 | 73.96 | -0.573 |
| NcsHSP15 | GWHPAAYW014339 | CI | 156 | 17.60 | 5.24 | 55.71 | 73.72 | -0.552 |
| NcsHSP16 | GWHPAAYW014344 | CI | 158 | 18.29 | 8.92 | 54.19 | 74.05 | -0.682 |
| NcsHSP17 | GWHPAAYW015449 | MTI/CP | 208 | 23.47 | 6.01 | 36.92 | 78.75 | -0.667 |
| NcsHSP18 | GWHPAAYW018459 | CI | 833 | 94.56 | 6.14 | 53.05 | 69.45 | -0.668 |
| NcsHSP19 | GWHPAAYW019072 | CI | 163 | 18.55 | 6.19 | 62.44 | 71.04 | -0.601 |
| NcsHSP20 | GWHPAAYW019077 | CI | 337 | 37.62 | 7.83 | 57.71 | 78.66 | -0.457 |
表1 睡莲、水稻、拟南芥和葡萄中sHSP成员的鉴定及结构分析
Table 1 The identification and structural analysis of the sHSP members in waterlily, rice, Arabidopsis and grape
| Protein name | Locus name | Subfamily | Protein length (aa) | Molecular weight (kDa) | Theoretical pI | Instability index | Aliphatic index | GRAVY |
|---|---|---|---|---|---|---|---|---|
| NcsHSP01 | GWHPAAYW001658 | CI | 60 | 6.64 | 9.16 | 41.10 | 99.00 | -0.218 |
| NcsHSP02 | GWHPAAYW001878 | CII | 308 | 34.77 | 9.03 | 44.31 | 74.64 | -0.624 |
| NcsHSP03 | GWHPAAYW002745 | CV | 276 | 30.85 | 4.88 | 44.78 | 77.36 | -0.291 |
| NcsHSP04 | GWHPAAYW004666 | PX | 1189 | 132.46 | 5.86 | 44.51 | 85.53 | -0.300 |
| NcsHSP05 | GWHPAAYW006130 | ER | 218 | 24.32 | 5.46 | 64.70 | 95.14 | -0.411 |
| NcsHSP06 | GWHPAAYW006158 | CII | 378 | 43.02 | 8.36 | 50.40 | 74.76 | -0.780 |
| NcsHSP07 | GWHPAAYW006159 | CII | 662 | 75.02 | 9.13 | 55.90 | 79.64 | -0.665 |
| NcsHSP08 | GWHPAAYW006161 | CII | 314 | 35.79 | 8.86 | 47.40 | 79.39 | -0.709 |
| NcsHSP09 | GWHPAAYW006746 | CIII | 164 | 18.56 | 5.75 | 53.09 | 81.28 | -0.489 |
| NcsHSP10 | GWHPAAYW007213 | CIV | 132 | 14.36 | 6.83 | 58.83 | 82.80 | -0.117 |
| NcsHSP11 | GWHPAAYW009125 | CII | 165 | 19.09 | 5.48 | 54.83 | 69.03 | -0.921 |
| NcsHSP12 | GWHPAAYW010630 | MTII | 235 | 26.36 | 6.79 | 56.35 | 67.57 | -0.687 |
| NcsHSP13 | GWHPAAYW010715 | CI | 129 | 15.42 | 9.12 | 60.26 | 59.53 | -1.343 |
| NcsHSP14 | GWHPAAYW013759 | CVI | 154 | 17.47 | 5.57 | 46.32 | 73.96 | -0.573 |
| NcsHSP15 | GWHPAAYW014339 | CI | 156 | 17.60 | 5.24 | 55.71 | 73.72 | -0.552 |
| NcsHSP16 | GWHPAAYW014344 | CI | 158 | 18.29 | 8.92 | 54.19 | 74.05 | -0.682 |
| NcsHSP17 | GWHPAAYW015449 | MTI/CP | 208 | 23.47 | 6.01 | 36.92 | 78.75 | -0.667 |
| NcsHSP18 | GWHPAAYW018459 | CI | 833 | 94.56 | 6.14 | 53.05 | 69.45 | -0.668 |
| NcsHSP19 | GWHPAAYW019072 | CI | 163 | 18.55 | 6.19 | 62.44 | 71.04 | -0.601 |
| NcsHSP20 | GWHPAAYW019077 | CI | 337 | 37.62 | 7.83 | 57.71 | 78.66 | -0.457 |
| Protein name | Locus name | Subfamily | Protein length (aa) | Molecular weight (kDa) | Theoretical pI | Instability index | Aliphatic index | GRAVY |
|---|---|---|---|---|---|---|---|---|
| NcsHSP21 | GWHPAAYW019078 | CI | 156 | 17.38 | 5.69 | 54.61 | 75.00 | -0.449 |
| NcsHSP22 | GWHPAAYW019481 | CP | 222 | 25.65 | 9.17 | 54.03 | 60.63 | -0.880 |
| NcsHSP23 | GWHPAAYW022038 | CI | 159 | 18.24 | 5.60 | 65.94 | 69.18 | -0.648 |
| NcsHSP24 | GWHPAAYW025914 | CVI | 160 | 18.57 | 5.82 | 41.67 | 70.00 | -0.914 |
| NcsHSP25 | GWHPAAYW027820 | ER | 192 | 21.54 | 6.54 | 31.35 | 102.08 | -0.202 |
| NcsHSP26 | GWHPAAYW027821 | ER | 676 | 74.58 | 5.31 | 41.90 | 85.41 | -0.292 |
| NcsHSP27 | GWHPAAYW027823 | ER | 180 | 20.20 | 9.21 | 61.79 | 73.67 | -0.636 |
| NcsHSP28 | GWHPAAYW027825 | ER | 191 | 21.24 | 7.93 | 29.89 | 91.41 | -0.376 |
| NcsHSP29 | GWHPAAYW027826 | ER | 191 | 21.48 | 6.00 | 34.01 | 98.53 | -0.246 |
| NcsHSP30 | GWHPAAYW027987 | CP | 226 | 25.18 | 6.65 | 50.88 | 75.88 | -0.522 |
| NcsHSP31 | GWHPAAYW030961 | ER | 133 | 15.78 | 10.21 | 41.23 | 77.59 | -0.929 |
| NcsHSP32 | GWHPAAYW030963 | ER | 126 | 14.05 | 5.63 | 37.53 | 109.13 | -0.024 |
| NcsHSP33 | GWHPAAYW030964 | ER | 143 | 15.67 | 6.08 | 33.52 | 109.16 | 0.075 |
| OssHSP01 | LOC_Os01g04340.1 | CI | 150 | 16.65 | 6.31 | 34.230 | 77.200 | -0.536 |
| OssHSP02 | LOC_Os01g04350.1 | CI | 166 | 17.89 | 4.96 | 55.210 | 87.590 | 0.051 |
| OssHSP03 | LOC_Os01g04360.1 | CI | 149 | 16.90 | 6.76 | 47.050 | 69.870 | -0.648 |
| OssHSP04 | LOC_Os01g04370.1 | CI | 150 | 16.94 | 6.18 | 52.240 | 76.530 | -0.605 |
| OssHSP05 | LOC_Os01g04380.1 | CI | 150 | 16.96 | 6.18 | 52.240 | 73.930 | -0.617 |
| OssHSP06 | LOC_Os01g08860.1 | CII | 166 | 18.03 | 5.61 | 45.580 | 78.130 | -0.386 |
| OssHSP07 | LOC_Os02g03570.1 | CI | 177 | 18.87 | 6.93 | 52.450 | 71.750 | -0.406 |
| OssHSP08 | LOC_Os02g10710.1 | MTII | 219 | 23.59 | 7.74 | 47.270 | 86.030 | -0.320 |
| OssHSP09 | LOC_Os02g12610.1 | CII | 175 | 19.02 | 5.73 | 39.010 | 84.060 | -0.486 |
| OssHSP10 | LOC_Os02g48140.1 | CVII | 164 | 17.77 | 6.97 | 46.260 | 67.320 | -0.534 |
| OssHSP11 | LOC_Os02g52150.1 | MTI/CP | 221 | 24.15 | 7.96 | 48.750 | 77.330 | -0.519 |
| OssHSP12 | LOC_Os02g54140.1 | CIII | 172 | 18.60 | 7.85 | 40.550 | 77.150 | -0.538 |
| OssHSP13 | LOC_Os03g14180.1 | CP | 240 | 26.66 | 6.78 | 53.350 | 75.120 | -0.541 |
| OssHSP14 | LOC_Os03g15960.1 | CI | 161 | 17.91 | 5.79 | 57.860 | 64.780 | -0.680 |
| OssHSP15 | LOC_Os03g16020.1 | CI | 154 | 17.37 | 6.18 | 49.280 | 66.430 | -0.706 |
| OssHSP16 | LOC_Os03g16030.1 | CI | 161 | 18.08 | 6.77 | 58.690 | 61.680 | -0.746 |
| OssHSP17 | LOC_Os03g16040.1 | CI | 159 | 17.66 | 6.18 | 55.510 | 66.790 | -0.719 |
| OssHSP18 | LOC_Os04g36750.1 | ER | 215 | 23.23 | 5.34 | 35.060 | 82.650 | -0.365 |
| OssHSP19 | LOC_Os05g23140.1 | CP | 251 | 27.64 | 9.42 | 39.120 | 69.240 | -0.488 |
| OssHSP20 | LOC_Os05g42120.1 | CV | 203 | 22.28 | 4.82 | 54.030 | 69.800 | -0.330 |
| OssHSP21 | LOC_Os06g11610.1 | MTI/CP | 248 | 26.23 | 5.27 | 43.470 | 79.270 | -0.323 |
| OssHSP22 | LOC_Os06g14240.1 | PX | 146 | 16.02 | 8.13 | 36.730 | 84.790 | -0.292 |
| OssHSP23 | LOC_Os07g33350.1 | CIV | 219 | 23.76 | 11.74 | 93.800 | 71.320 | -0.642 |
| OssHSP24 | LOC_Os11g13980.1 | ER | 206 | 21.87 | 6.01 | 48.890 | 81.500 | -0.126 |
| AtsHSP01 | AT1G07400.1 | CI | 157 | 17.83 | 5.99 | 43.380 | 70.060 | -0.680 |
| AtsHSP02 | AT1G52560.1 | MTII | 232 | 26.54 | 6.86 | 45.660 | 68.880 | -0.869 |
| AtsHSP03 | AT1G53540.1 | CI | 157 | 17.60 | 5.36 | 53.800 | 71.970 | -0.578 |
| AtsHSP04 | AT1G54050.1 | CIII | 155 | 17.36 | 7.88 | 58.630 | 93.680 | -0.504 |
| AtsHSP05 | AT1G59860.1 | CI | 155 | 17.62 | 6.85 | 50.240 | 70.970 | -0.678 |
| AtsHSP06 | AT2G29500.1 | CI | 153 | 17.56 | 6.33 | 47.820 | 71.900 | -0.720 |
| AtsHSP07 | AT3G46230.1 | CI | 156 | 17.44 | 5.20 | 46.980 | 68.590 | -0.569 |
表1 (续)
Table 1 (continued)
| Protein name | Locus name | Subfamily | Protein length (aa) | Molecular weight (kDa) | Theoretical pI | Instability index | Aliphatic index | GRAVY |
|---|---|---|---|---|---|---|---|---|
| NcsHSP21 | GWHPAAYW019078 | CI | 156 | 17.38 | 5.69 | 54.61 | 75.00 | -0.449 |
| NcsHSP22 | GWHPAAYW019481 | CP | 222 | 25.65 | 9.17 | 54.03 | 60.63 | -0.880 |
| NcsHSP23 | GWHPAAYW022038 | CI | 159 | 18.24 | 5.60 | 65.94 | 69.18 | -0.648 |
| NcsHSP24 | GWHPAAYW025914 | CVI | 160 | 18.57 | 5.82 | 41.67 | 70.00 | -0.914 |
| NcsHSP25 | GWHPAAYW027820 | ER | 192 | 21.54 | 6.54 | 31.35 | 102.08 | -0.202 |
| NcsHSP26 | GWHPAAYW027821 | ER | 676 | 74.58 | 5.31 | 41.90 | 85.41 | -0.292 |
| NcsHSP27 | GWHPAAYW027823 | ER | 180 | 20.20 | 9.21 | 61.79 | 73.67 | -0.636 |
| NcsHSP28 | GWHPAAYW027825 | ER | 191 | 21.24 | 7.93 | 29.89 | 91.41 | -0.376 |
| NcsHSP29 | GWHPAAYW027826 | ER | 191 | 21.48 | 6.00 | 34.01 | 98.53 | -0.246 |
| NcsHSP30 | GWHPAAYW027987 | CP | 226 | 25.18 | 6.65 | 50.88 | 75.88 | -0.522 |
| NcsHSP31 | GWHPAAYW030961 | ER | 133 | 15.78 | 10.21 | 41.23 | 77.59 | -0.929 |
| NcsHSP32 | GWHPAAYW030963 | ER | 126 | 14.05 | 5.63 | 37.53 | 109.13 | -0.024 |
| NcsHSP33 | GWHPAAYW030964 | ER | 143 | 15.67 | 6.08 | 33.52 | 109.16 | 0.075 |
| OssHSP01 | LOC_Os01g04340.1 | CI | 150 | 16.65 | 6.31 | 34.230 | 77.200 | -0.536 |
| OssHSP02 | LOC_Os01g04350.1 | CI | 166 | 17.89 | 4.96 | 55.210 | 87.590 | 0.051 |
| OssHSP03 | LOC_Os01g04360.1 | CI | 149 | 16.90 | 6.76 | 47.050 | 69.870 | -0.648 |
| OssHSP04 | LOC_Os01g04370.1 | CI | 150 | 16.94 | 6.18 | 52.240 | 76.530 | -0.605 |
| OssHSP05 | LOC_Os01g04380.1 | CI | 150 | 16.96 | 6.18 | 52.240 | 73.930 | -0.617 |
| OssHSP06 | LOC_Os01g08860.1 | CII | 166 | 18.03 | 5.61 | 45.580 | 78.130 | -0.386 |
| OssHSP07 | LOC_Os02g03570.1 | CI | 177 | 18.87 | 6.93 | 52.450 | 71.750 | -0.406 |
| OssHSP08 | LOC_Os02g10710.1 | MTII | 219 | 23.59 | 7.74 | 47.270 | 86.030 | -0.320 |
| OssHSP09 | LOC_Os02g12610.1 | CII | 175 | 19.02 | 5.73 | 39.010 | 84.060 | -0.486 |
| OssHSP10 | LOC_Os02g48140.1 | CVII | 164 | 17.77 | 6.97 | 46.260 | 67.320 | -0.534 |
| OssHSP11 | LOC_Os02g52150.1 | MTI/CP | 221 | 24.15 | 7.96 | 48.750 | 77.330 | -0.519 |
| OssHSP12 | LOC_Os02g54140.1 | CIII | 172 | 18.60 | 7.85 | 40.550 | 77.150 | -0.538 |
| OssHSP13 | LOC_Os03g14180.1 | CP | 240 | 26.66 | 6.78 | 53.350 | 75.120 | -0.541 |
| OssHSP14 | LOC_Os03g15960.1 | CI | 161 | 17.91 | 5.79 | 57.860 | 64.780 | -0.680 |
| OssHSP15 | LOC_Os03g16020.1 | CI | 154 | 17.37 | 6.18 | 49.280 | 66.430 | -0.706 |
| OssHSP16 | LOC_Os03g16030.1 | CI | 161 | 18.08 | 6.77 | 58.690 | 61.680 | -0.746 |
| OssHSP17 | LOC_Os03g16040.1 | CI | 159 | 17.66 | 6.18 | 55.510 | 66.790 | -0.719 |
| OssHSP18 | LOC_Os04g36750.1 | ER | 215 | 23.23 | 5.34 | 35.060 | 82.650 | -0.365 |
| OssHSP19 | LOC_Os05g23140.1 | CP | 251 | 27.64 | 9.42 | 39.120 | 69.240 | -0.488 |
| OssHSP20 | LOC_Os05g42120.1 | CV | 203 | 22.28 | 4.82 | 54.030 | 69.800 | -0.330 |
| OssHSP21 | LOC_Os06g11610.1 | MTI/CP | 248 | 26.23 | 5.27 | 43.470 | 79.270 | -0.323 |
| OssHSP22 | LOC_Os06g14240.1 | PX | 146 | 16.02 | 8.13 | 36.730 | 84.790 | -0.292 |
| OssHSP23 | LOC_Os07g33350.1 | CIV | 219 | 23.76 | 11.74 | 93.800 | 71.320 | -0.642 |
| OssHSP24 | LOC_Os11g13980.1 | ER | 206 | 21.87 | 6.01 | 48.890 | 81.500 | -0.126 |
| AtsHSP01 | AT1G07400.1 | CI | 157 | 17.83 | 5.99 | 43.380 | 70.060 | -0.680 |
| AtsHSP02 | AT1G52560.1 | MTII | 232 | 26.54 | 6.86 | 45.660 | 68.880 | -0.869 |
| AtsHSP03 | AT1G53540.1 | CI | 157 | 17.60 | 5.36 | 53.800 | 71.970 | -0.578 |
| AtsHSP04 | AT1G54050.1 | CIII | 155 | 17.36 | 7.88 | 58.630 | 93.680 | -0.504 |
| AtsHSP05 | AT1G59860.1 | CI | 155 | 17.62 | 6.85 | 50.240 | 70.970 | -0.678 |
| AtsHSP06 | AT2G29500.1 | CI | 153 | 17.56 | 6.33 | 47.820 | 71.900 | -0.720 |
| AtsHSP07 | AT3G46230.1 | CI | 156 | 17.44 | 5.20 | 46.980 | 68.590 | -0.569 |
| Protein name | Locus name | Subfamily | Protein length (aa) | Molecular weight (kDa) | Theoretical pI | Instability index | Aliphatic index | GRAVY |
|---|---|---|---|---|---|---|---|---|
| AtsHSP08 | AT4G10250.1 | ER | 195 | 22.00 | 5.58 | 36.450 | 93.950 | -0.469 |
| AtsHSP09 | AT4G21870.1 | CIV | 134 | 15.39 | 5.17 | 73.930 | 93.810 | -0.325 |
| AtsHSP10 | AT4G25200.1 | MTI/CP | 210 | 23.61 | 6.46 | 58.790 | 80.760 | -0.649 |
| AtsHSP11 | AT4G27670.1 | CP | 227 | 25.34 | 8.49 | 46.860 | 72.070 | -0.641 |
| AtsHSP12 | AT5G12020.1 | CII | 155 | 17.62 | 6.32 | 38.840 | 80.390 | -0.617 |
| AtsHSP13 | AT5G12030.1 | CII | 156 | 17.69 | 5.59 | 44.370 | 75.000 | -0.596 |
| AtsHSP14 | AT5G37670.1 | PX | 137 | 15.70 | 7.94 | 50.680 | 86.640 | -0.512 |
| AtsHSP15 | AT5G51440.1 | MTI/CP | 210 | 23.47 | 8.96 | 52.510 | 84.000 | -0.558 |
| AtsHSP16 | AT5G54660.1 | CV | 192 | 21.66 | 5.47 | 38.030 | 71.560 | -0.500 |
| AtsHSP17 | AT5G59720.1 | CI | 161 | 18.13 | 6.77 | 48.090 | 67.200 | -0.657 |
| VvsHSP01 | VIT_200s0707g00010.1 | CIV | 136 | 15.69 | 4.89 | 53.490 | 82.430 | -0.315 |
| VvsHSP02 | VIT_200s0992g00020.1 | CIV | 136 | 15.70 | 5.01 | 54.190 | 82.430 | -0.312 |
| VvsHSP03 | VIT_201s0010g02290.1 | CP | 226 | 25.56 | 6.77 | 48.710 | 62.170 | -0.706 |
| VvsHSP04 | VIT_202s0154g00480.1 | MTI/CP | 201 | 22.45 | 9.24 | 45.150 | 77.060 | -0.531 |
| VvsHSP05 | VIT_202s0154g00490.1 | MTI/CP | 201 | 22.55 | 9.11 | 51.710 | 75.670 | -0.561 |
| VvsHSP06 | VIT_204s0008g01490.1 | CII | 156 | 17.34 | 5.94 | 41.520 | 82.500 | -0.444 |
| VvsHSP07 | VIT_204s0008g01500.1 | CII | 152 | 16.69 | 6.84 | 42.090 | 79.540 | -0.390 |
| VvsHSP08 | VIT_204s0008g01510.1 | CII | 156 | 17.40 | 5.77 | 44.190 | 76.220 | -0.462 |
| VvsHSP09 | VIT_204s0008g01520.1 | CII | 156 | 17.58 | 5.58 | 40.180 | 78.080 | -0.513 |
| VvsHSP10 | VIT_204s0008g01530.1 | CII | 480 | 53.16 | 8.89 | 39.780 | 68.600 | -0.713 |
| VvsHSP11 | VIT_204s0008g01550.1 | CII | 156 | 17.41 | 5.94 | 39.580 | 71.790 | -0.533 |
| VvsHSP12 | VIT_204s0008g01560.1 | CII | 113 | 12.66 | 8.66 | 38.840 | 83.540 | -0.473 |
| VvsHSP13 | VIT_204s0008g01570.1 | CII | 166 | 18.60 | 5.95 | 40.640 | 68.670 | -0.542 |
| VvsHSP14 | VIT_204s0008g01580.1 | CII | 156 | 17.42 | 6.62 | 42.220 | 76.790 | -0.510 |
| VvsHSP15 | VIT_204s0008g01590.1 | CII | 155 | 17.29 | 5.94 | 37.610 | 81.030 | -0.466 |
| VvsHSP16 | VIT_204s0008g01610.1 | CII | 158 | 18.14 | 6.33 | 40.180 | 75.820 | -0.617 |
| VvsHSP17 | VIT_204s0008g01620.1 | CII | 159 | 18.42 | 8.46 | 42.080 | 83.900 | -0.459 |
| VvsHSP18 | VIT_206s0004g05770.1 | CI | 144 | 16.31 | 6.93 | 33.000 | 75.070 | -0.642 |
| VvsHSP19 | VIT_208s0058g00210.1 | CI | 148 | 16.88 | 5.81 | 64.560 | 66.490 | -0.629 |
| VvsHSP20 | VIT_209s0002g00640.1 | CIII | 160 | 17.89 | 6.30 | 49.690 | 81.000 | -0.516 |
| VvsHSP21 | VIT_209s0002g06790.1 | MTII | 233 | 26.31 | 7.78 | 53.870 | 82.060 | -0.750 |
| VvsHSP22 | VIT_212s0035g01910.1 | ER | 250 | 28.39 | 7.94 | 49.940 | 89.400 | -0.388 |
| VvsHSP23 | VIT_213s0019g00860.1 | PX | 142 | 15.81 | 6.75 | 51.850 | 85.000 | -0.311 |
| VvsHSP24 | VIT_213s0019g02740.1 | CI | 151 | 17.17 | 5.81 | 46.350 | 74.110 | -0.553 |
| VvsHSP25 | VIT_213s0019g02760.1 | CI | 140 | 15.80 | 6.77 | 42.530 | 72.290 | -0.589 |
| VvsHSP26 | VIT_213s0019g02770.1 | CI | 151 | 17.10 | 5.81 | 48.460 | 72.850 | -0.554 |
| VvsHSP27 | VIT_213s0019g02780.1 | CI | 151 | 17.02 | 5.80 | 45.690 | 70.260 | -0.587 |
| VvsHSP28 | VIT_213s0019g02820.1 | CI | 151 | 17.12 | 5.81 | 47.080 | 70.260 | -0.591 |
| VvsHSP29 | VIT_213s0019g02840.1 | CI | 151 | 17.09 | 5.54 | 50.220 | 72.850 | -0.551 |
| VvsHSP30 | VIT_213s0019g02850.1 | CI | 151 | 17.05 | 5.80 | 45.440 | 70.260 | -0.590 |
| VvsHSP31 | VIT_213s0019g02900.1 | CI | 108 | 12.64 | 6.18 | 52.990 | 80.190 | -0.798 |
| VvsHSP32 | VIT_213s0019g02920.1 | CI | 136 | 15.27 | 5.70 | 46.020 | 79.490 | -0.552 |
| VvsHSP33 | VIT_213s0019g02930.1 | CI | 160 | 18.17 | 6.78 | 53.430 | 76.690 | -0.644 |
| VvsHSP34 | VIT_213s0019g03000.1 | CI | 160 | 18.15 | 7.93 | 55.700 | 76.690 | -0.644 |
表1 (续)
Table 1 (continued)
| Protein name | Locus name | Subfamily | Protein length (aa) | Molecular weight (kDa) | Theoretical pI | Instability index | Aliphatic index | GRAVY |
|---|---|---|---|---|---|---|---|---|
| AtsHSP08 | AT4G10250.1 | ER | 195 | 22.00 | 5.58 | 36.450 | 93.950 | -0.469 |
| AtsHSP09 | AT4G21870.1 | CIV | 134 | 15.39 | 5.17 | 73.930 | 93.810 | -0.325 |
| AtsHSP10 | AT4G25200.1 | MTI/CP | 210 | 23.61 | 6.46 | 58.790 | 80.760 | -0.649 |
| AtsHSP11 | AT4G27670.1 | CP | 227 | 25.34 | 8.49 | 46.860 | 72.070 | -0.641 |
| AtsHSP12 | AT5G12020.1 | CII | 155 | 17.62 | 6.32 | 38.840 | 80.390 | -0.617 |
| AtsHSP13 | AT5G12030.1 | CII | 156 | 17.69 | 5.59 | 44.370 | 75.000 | -0.596 |
| AtsHSP14 | AT5G37670.1 | PX | 137 | 15.70 | 7.94 | 50.680 | 86.640 | -0.512 |
| AtsHSP15 | AT5G51440.1 | MTI/CP | 210 | 23.47 | 8.96 | 52.510 | 84.000 | -0.558 |
| AtsHSP16 | AT5G54660.1 | CV | 192 | 21.66 | 5.47 | 38.030 | 71.560 | -0.500 |
| AtsHSP17 | AT5G59720.1 | CI | 161 | 18.13 | 6.77 | 48.090 | 67.200 | -0.657 |
| VvsHSP01 | VIT_200s0707g00010.1 | CIV | 136 | 15.69 | 4.89 | 53.490 | 82.430 | -0.315 |
| VvsHSP02 | VIT_200s0992g00020.1 | CIV | 136 | 15.70 | 5.01 | 54.190 | 82.430 | -0.312 |
| VvsHSP03 | VIT_201s0010g02290.1 | CP | 226 | 25.56 | 6.77 | 48.710 | 62.170 | -0.706 |
| VvsHSP04 | VIT_202s0154g00480.1 | MTI/CP | 201 | 22.45 | 9.24 | 45.150 | 77.060 | -0.531 |
| VvsHSP05 | VIT_202s0154g00490.1 | MTI/CP | 201 | 22.55 | 9.11 | 51.710 | 75.670 | -0.561 |
| VvsHSP06 | VIT_204s0008g01490.1 | CII | 156 | 17.34 | 5.94 | 41.520 | 82.500 | -0.444 |
| VvsHSP07 | VIT_204s0008g01500.1 | CII | 152 | 16.69 | 6.84 | 42.090 | 79.540 | -0.390 |
| VvsHSP08 | VIT_204s0008g01510.1 | CII | 156 | 17.40 | 5.77 | 44.190 | 76.220 | -0.462 |
| VvsHSP09 | VIT_204s0008g01520.1 | CII | 156 | 17.58 | 5.58 | 40.180 | 78.080 | -0.513 |
| VvsHSP10 | VIT_204s0008g01530.1 | CII | 480 | 53.16 | 8.89 | 39.780 | 68.600 | -0.713 |
| VvsHSP11 | VIT_204s0008g01550.1 | CII | 156 | 17.41 | 5.94 | 39.580 | 71.790 | -0.533 |
| VvsHSP12 | VIT_204s0008g01560.1 | CII | 113 | 12.66 | 8.66 | 38.840 | 83.540 | -0.473 |
| VvsHSP13 | VIT_204s0008g01570.1 | CII | 166 | 18.60 | 5.95 | 40.640 | 68.670 | -0.542 |
| VvsHSP14 | VIT_204s0008g01580.1 | CII | 156 | 17.42 | 6.62 | 42.220 | 76.790 | -0.510 |
| VvsHSP15 | VIT_204s0008g01590.1 | CII | 155 | 17.29 | 5.94 | 37.610 | 81.030 | -0.466 |
| VvsHSP16 | VIT_204s0008g01610.1 | CII | 158 | 18.14 | 6.33 | 40.180 | 75.820 | -0.617 |
| VvsHSP17 | VIT_204s0008g01620.1 | CII | 159 | 18.42 | 8.46 | 42.080 | 83.900 | -0.459 |
| VvsHSP18 | VIT_206s0004g05770.1 | CI | 144 | 16.31 | 6.93 | 33.000 | 75.070 | -0.642 |
| VvsHSP19 | VIT_208s0058g00210.1 | CI | 148 | 16.88 | 5.81 | 64.560 | 66.490 | -0.629 |
| VvsHSP20 | VIT_209s0002g00640.1 | CIII | 160 | 17.89 | 6.30 | 49.690 | 81.000 | -0.516 |
| VvsHSP21 | VIT_209s0002g06790.1 | MTII | 233 | 26.31 | 7.78 | 53.870 | 82.060 | -0.750 |
| VvsHSP22 | VIT_212s0035g01910.1 | ER | 250 | 28.39 | 7.94 | 49.940 | 89.400 | -0.388 |
| VvsHSP23 | VIT_213s0019g00860.1 | PX | 142 | 15.81 | 6.75 | 51.850 | 85.000 | -0.311 |
| VvsHSP24 | VIT_213s0019g02740.1 | CI | 151 | 17.17 | 5.81 | 46.350 | 74.110 | -0.553 |
| VvsHSP25 | VIT_213s0019g02760.1 | CI | 140 | 15.80 | 6.77 | 42.530 | 72.290 | -0.589 |
| VvsHSP26 | VIT_213s0019g02770.1 | CI | 151 | 17.10 | 5.81 | 48.460 | 72.850 | -0.554 |
| VvsHSP27 | VIT_213s0019g02780.1 | CI | 151 | 17.02 | 5.80 | 45.690 | 70.260 | -0.587 |
| VvsHSP28 | VIT_213s0019g02820.1 | CI | 151 | 17.12 | 5.81 | 47.080 | 70.260 | -0.591 |
| VvsHSP29 | VIT_213s0019g02840.1 | CI | 151 | 17.09 | 5.54 | 50.220 | 72.850 | -0.551 |
| VvsHSP30 | VIT_213s0019g02850.1 | CI | 151 | 17.05 | 5.80 | 45.440 | 70.260 | -0.590 |
| VvsHSP31 | VIT_213s0019g02900.1 | CI | 108 | 12.64 | 6.18 | 52.990 | 80.190 | -0.798 |
| VvsHSP32 | VIT_213s0019g02920.1 | CI | 136 | 15.27 | 5.70 | 46.020 | 79.490 | -0.552 |
| VvsHSP33 | VIT_213s0019g02930.1 | CI | 160 | 18.17 | 6.78 | 53.430 | 76.690 | -0.644 |
| VvsHSP34 | VIT_213s0019g03000.1 | CI | 160 | 18.15 | 7.93 | 55.700 | 76.690 | -0.644 |
| Protein name | Locus name | Subfamily | Protein length (aa) | Molecular weight (kDa) | Theoretical pI | Instability index | Aliphatic index | GRAVY |
|---|---|---|---|---|---|---|---|---|
| VvsHSP35 | VIT_213s0019g03010.1 | CI | 144 | 16.37 | 9.21 | 49.720 | 69.030 | -0.785 |
| VvsHSP36 | VIT_213s0019g03030.1 | CI | 108 | 12.67 | 6.76 | 37.260 | 77.500 | -0.817 |
| VvsHSP37 | VIT_213s0019g03050.1 | CI | 165 | 19.23 | 6.46 | 61.350 | 79.090 | -0.721 |
| VvsHSP38 | VIT_213s0019g03090.1 | CI | 160 | 18.17 | 5.43 | 62.670 | 73.690 | -0.673 |
| VvsHSP39 | VIT_213s0019g03160.1 | CI | 160 | 18.02 | 7.94 | 64.610 | 73.120 | -0.631 |
| VvsHSP40 | VIT_213s0019g03170.1 | CI | 159 | 18.19 | 6.17 | 51.140 | 71.010 | -0.655 |
| VvsHSP41 | VIT_216s0022g00510.1 | MTI/CP | 208 | 23.74 | 5.61 | 60.250 | 75.870 | -0.721 |
| VvsHSP42 | VIT_216s0098g01060.1 | CP | 227 | 25.03 | 6.35 | 44.280 | 72.600 | -0.505 |
| VvsHSP43 | VIT_218s0001g01570.1 | CVI | 163 | 18.28 | 6.33 | 48.000 | 71.720 | -0.790 |
| VvsHSP44 | VIT_218s0001g01610.1 | CVI | 159 | 18.00 | 5.74 | 44.410 | 72.890 | -0.731 |
| VvsHSP45 | VIT_218s0089g01270.1 | ER | 186 | 21.13 | 5.89 | 35.370 | 90.590 | -0.567 |
| VvsHSP46 | VIT_219s0014g05050.1 | CV | 192 | 22.39 | 5.35 | 48.680 | 70.470 | -0.754 |
| VvsHSP47 | VIT_219s0085g01050.1 | CVII | 146 | 16.45 | 5.90 | 50.360 | 72.740 | -0.530 |
表1 (续)
Table 1 (continued)
| Protein name | Locus name | Subfamily | Protein length (aa) | Molecular weight (kDa) | Theoretical pI | Instability index | Aliphatic index | GRAVY |
|---|---|---|---|---|---|---|---|---|
| VvsHSP35 | VIT_213s0019g03010.1 | CI | 144 | 16.37 | 9.21 | 49.720 | 69.030 | -0.785 |
| VvsHSP36 | VIT_213s0019g03030.1 | CI | 108 | 12.67 | 6.76 | 37.260 | 77.500 | -0.817 |
| VvsHSP37 | VIT_213s0019g03050.1 | CI | 165 | 19.23 | 6.46 | 61.350 | 79.090 | -0.721 |
| VvsHSP38 | VIT_213s0019g03090.1 | CI | 160 | 18.17 | 5.43 | 62.670 | 73.690 | -0.673 |
| VvsHSP39 | VIT_213s0019g03160.1 | CI | 160 | 18.02 | 7.94 | 64.610 | 73.120 | -0.631 |
| VvsHSP40 | VIT_213s0019g03170.1 | CI | 159 | 18.19 | 6.17 | 51.140 | 71.010 | -0.655 |
| VvsHSP41 | VIT_216s0022g00510.1 | MTI/CP | 208 | 23.74 | 5.61 | 60.250 | 75.870 | -0.721 |
| VvsHSP42 | VIT_216s0098g01060.1 | CP | 227 | 25.03 | 6.35 | 44.280 | 72.600 | -0.505 |
| VvsHSP43 | VIT_218s0001g01570.1 | CVI | 163 | 18.28 | 6.33 | 48.000 | 71.720 | -0.790 |
| VvsHSP44 | VIT_218s0001g01610.1 | CVI | 159 | 18.00 | 5.74 | 44.410 | 72.890 | -0.731 |
| VvsHSP45 | VIT_218s0089g01270.1 | ER | 186 | 21.13 | 5.89 | 35.370 | 90.590 | -0.567 |
| VvsHSP46 | VIT_219s0014g05050.1 | CV | 192 | 22.39 | 5.35 | 48.680 | 70.470 | -0.754 |
| VvsHSP47 | VIT_219s0085g01050.1 | CVII | 146 | 16.45 | 5.90 | 50.360 | 72.740 | -0.530 |
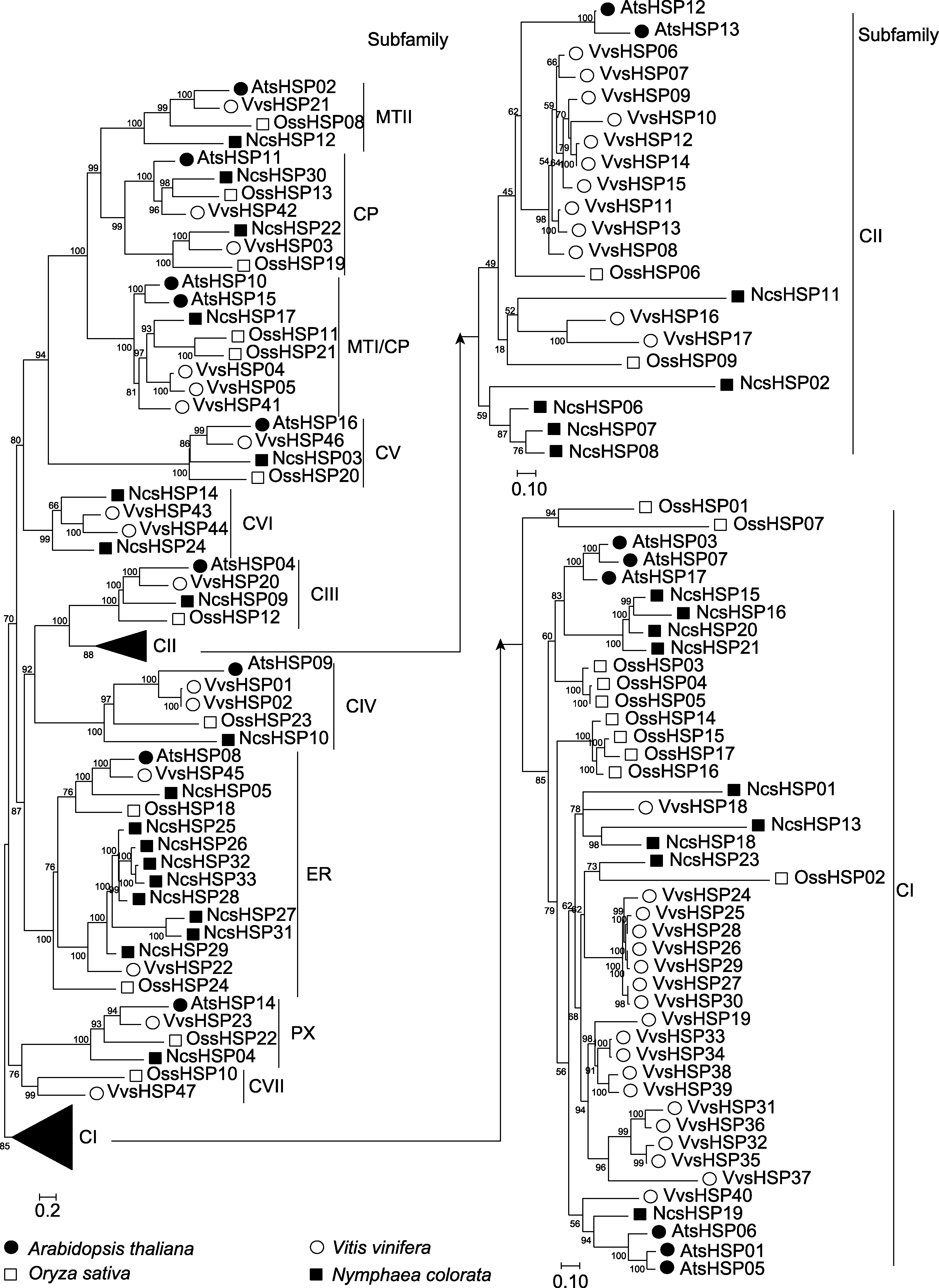
图1 睡莲、水稻、拟南芥和葡萄中sHSP成员的系统进化分析 通过IQ-tree软件构建系统进化树, 分支上的数字代表bootstrap值。
Figure 1 The phylogenetic analysis of sHSP members in waterlily, rice, Arabidopsis and grape The phylogenetic tree was constructed by IQ-tree software. The numbers in the clades stood for the bootstrap values.
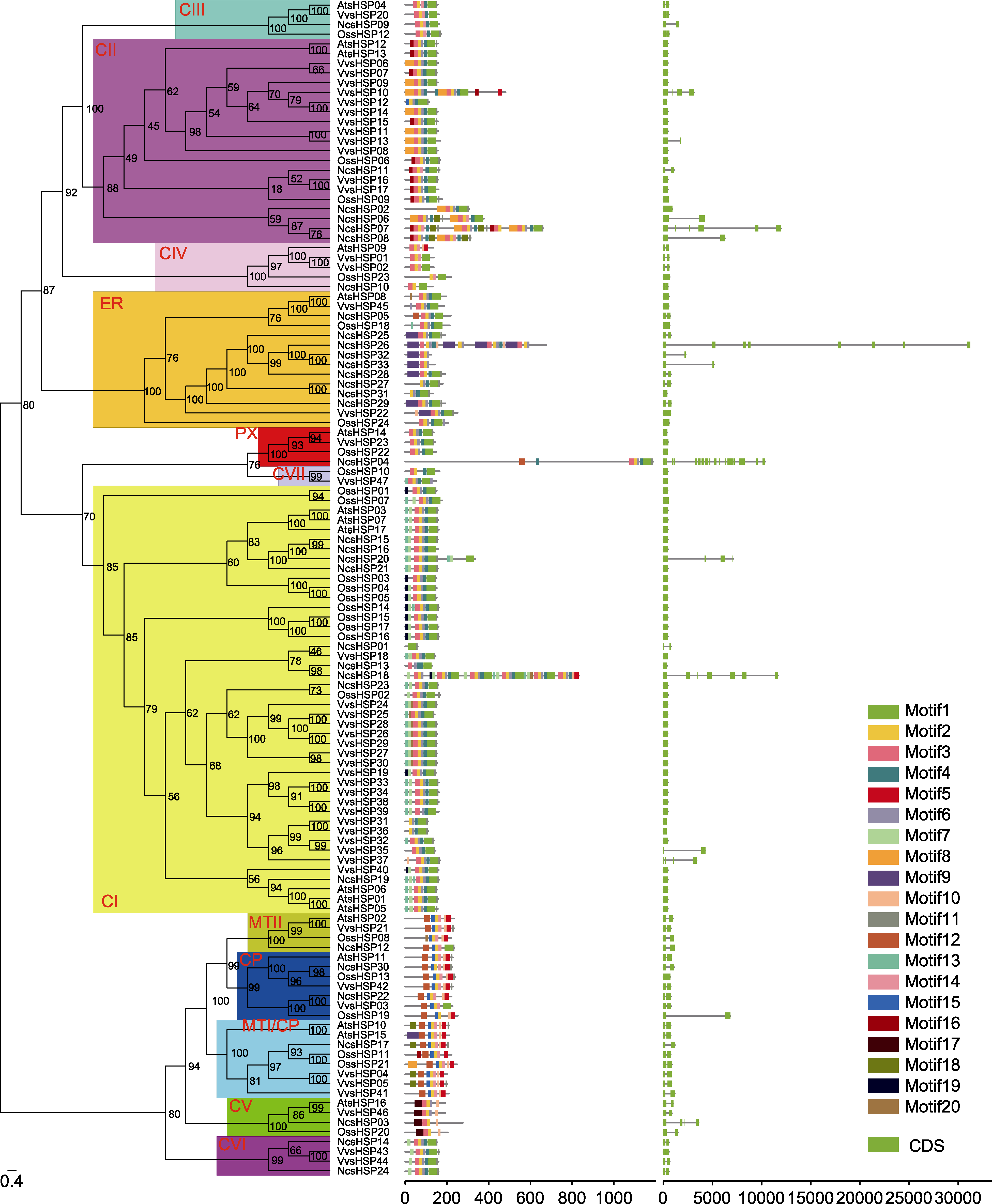
图2 睡莲、水稻、拟南芥和葡萄中sHSP家族的系统进化(左)、保守基序(中)和基因结构(右)分析 通过IQ-tree软件构建系统进化树(左), 分支上的数字代表bootstrap值, sHSP亚家族用红色字母表示。保守基序(中)通过MEME程序识别, 不同颜色代表不同的保守基序, 保守基序的位置使用下方标尺进行估计。根据内含子和外显子的位置构建基因结构示意图(右), 绿色方框表示外显子, 黑色线表示内含子, 内含子和外显子的位置使用下方标尺进行估计。
Figure 2 The phylogenetic analysis (left), conserved motif (middle), and gene structure (right) of the sHSF family in waterlily, rice, Arabidopsis and grape The phylogenetic tree (left) was constructed by IQ-tree software. The numbers in the clades stood for the bootstrap values, and sHSP subfamilies were indicated by red letters. The conserved motifs (middle) were identified by MEME software. Each conserved motif was marked by a specific color, and the location of motifs can be estimated using the scale at the bottom. The gene structures (right) were visualized according to the location of exons and introns. The exons and introns were represented by green box and black line, respectively. The location of exons and introns can be estimated using the scale at the bottom.
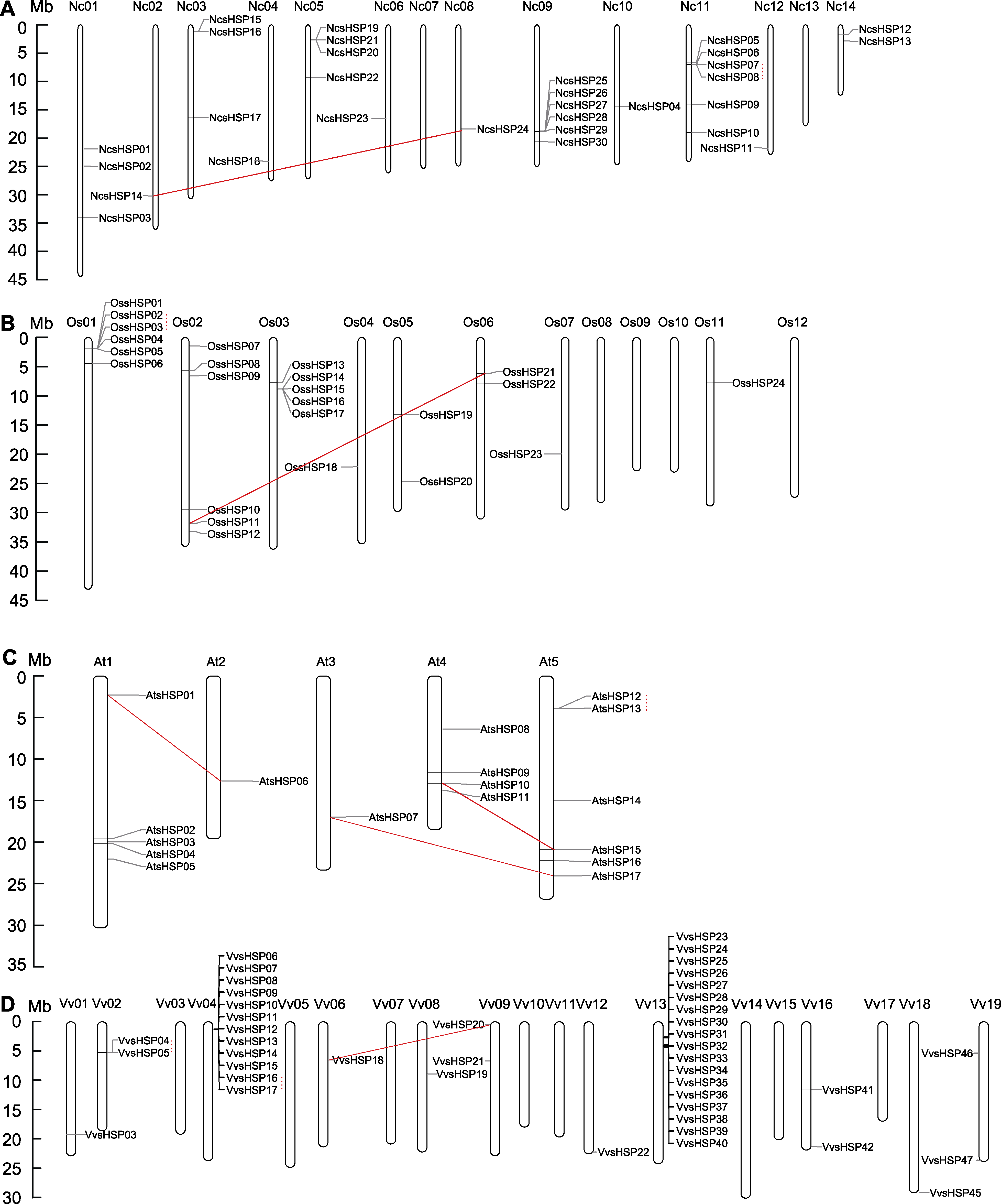
图3 睡莲、水稻、拟南芥和葡萄中NcsHSP (A)、OssHSP (B)、AtsHSP (C)和VvsHSP成员(D)的染色体分布 实线连线和虚线连线分别代表片段复制事件和串联复制事件。
Figure 3 Chromosomal locations of NcsHSP (A), OssHSP (B), AtsHSP (C) and VvsHSP (D) on the waterlily, rice, Arabidopsis and grape chromosomes The solid lines and dot lines represented the segmental and tandem duplication events, respectively.
| Duplicated gene 1 | Duplicated gene 2 | Subfamily | Ka | Ks | Ka/Ks | Purifying selection | Duplicated type |
|---|---|---|---|---|---|---|---|
| NcsHSP07 | NcsHSP08 | CII | 0.067786 | 0.358961 | 0.188839 | Yes | Tandem |
| NcsHSP14 | NcsHSP24 | CVI | 0.346841 | 0.789231 | 0.439467 | Yes | Segmental |
| OssHSP03 | OssHSP04 | CI | 0.02811 | 0.129535 | 0.217006 | Yes | Tandem |
| OssHSP11 | OssHSP21 | MTI/CP | 0.303273 | 0.454245 | 0.66764 | Yes | Segmental |
| AtsHSP01 | AtsHSP06 | CI | 0.08714 | 1.101078 | 0.079141 | Yes | Segmental |
| AtsHSP07 | AtsHSP17 | CI | 0.110454 | 1.291145 | 0.085547 | Yes | Segmental |
| AtsHSP10 | AtsHSP15 | MTI/CP | 0.175047 | 1.17229 | 0.14932 | Yes | Segmental |
| AtsHSP12 | AtsHSP13 | CII | 0.060844 | 0.53554 | 0.113613 | Yes | Tandem |
| VvsHSP04 | VvsHSP05 | MTI/CP | 0.077421 | 0.10198 | 0.759181 | Yes | Tandem |
| VvsHSP16 | VvsHSP17 | CII | 0.212704 | 0.880614 | 0.241541 | Yes | Tandem |
| VvsHSP18 | VvsHSP40 | CI | 0.206071 | 1.642523 | 0.12546 | Yes | Segmental |
| VvsHSP43 | VvsHSP44 | CVI | 0.21995 | 0.81194 | 0.270894 | Yes | Tandem |
表2 在睡莲、水稻、拟南芥和葡萄中复制的sHSP成员的Ka和Ks分析
Table 2 Ka and Ks analysis for the duplicated sHSP members in waterlily, rice, Arabidopsis and grape
| Duplicated gene 1 | Duplicated gene 2 | Subfamily | Ka | Ks | Ka/Ks | Purifying selection | Duplicated type |
|---|---|---|---|---|---|---|---|
| NcsHSP07 | NcsHSP08 | CII | 0.067786 | 0.358961 | 0.188839 | Yes | Tandem |
| NcsHSP14 | NcsHSP24 | CVI | 0.346841 | 0.789231 | 0.439467 | Yes | Segmental |
| OssHSP03 | OssHSP04 | CI | 0.02811 | 0.129535 | 0.217006 | Yes | Tandem |
| OssHSP11 | OssHSP21 | MTI/CP | 0.303273 | 0.454245 | 0.66764 | Yes | Segmental |
| AtsHSP01 | AtsHSP06 | CI | 0.08714 | 1.101078 | 0.079141 | Yes | Segmental |
| AtsHSP07 | AtsHSP17 | CI | 0.110454 | 1.291145 | 0.085547 | Yes | Segmental |
| AtsHSP10 | AtsHSP15 | MTI/CP | 0.175047 | 1.17229 | 0.14932 | Yes | Segmental |
| AtsHSP12 | AtsHSP13 | CII | 0.060844 | 0.53554 | 0.113613 | Yes | Tandem |
| VvsHSP04 | VvsHSP05 | MTI/CP | 0.077421 | 0.10198 | 0.759181 | Yes | Tandem |
| VvsHSP16 | VvsHSP17 | CII | 0.212704 | 0.880614 | 0.241541 | Yes | Tandem |
| VvsHSP18 | VvsHSP40 | CI | 0.206071 | 1.642523 | 0.12546 | Yes | Segmental |
| VvsHSP43 | VvsHSP44 | CVI | 0.21995 | 0.81194 | 0.270894 | Yes | Tandem |
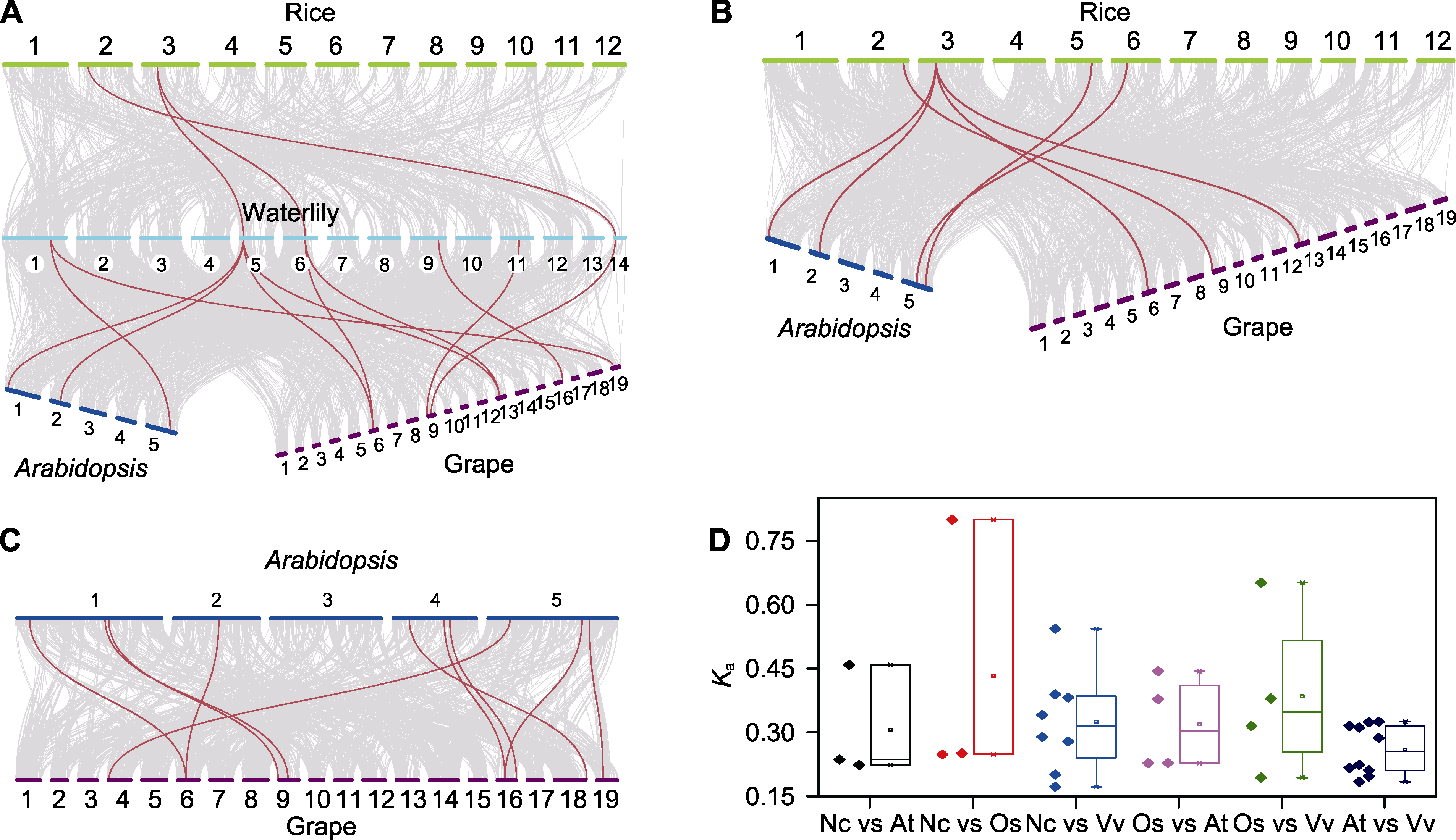
图4 睡莲、水稻、拟南芥和葡萄中sHSP成员的共线性分析 (A) 睡莲与水稻、拟南芥和葡萄中sHSP成员的共线性分析; (B) 水稻与拟南芥和葡萄sHSP成员的共线性分析; (C) 拟南芥与葡萄sHSP成员的共线性分析; (D) 在不同物种中sHSP同源基因对的Ka值分析。(A)-(C) 灰线代表所有的同源基因, 红线代表sHSP同源基因。
Figure 4 Synteny analysis of sHSP members in waterlily, rice, Arabidopsis and grape (A) Synteny analysis of sHSP members between waterlily and rice, Arabidopsis and grape; (B) Synteny analysis of sHSP members between rice and Arabidopsis and grape; (C) Synteny analysis of sHSP members between Arabidopsis and grape; (D) Comparison of Ka values of orthologous sHSPs between different species. (A)-(C) Gray lines connected orthologous genes, and orthologous sHSPs were marked with red lines.
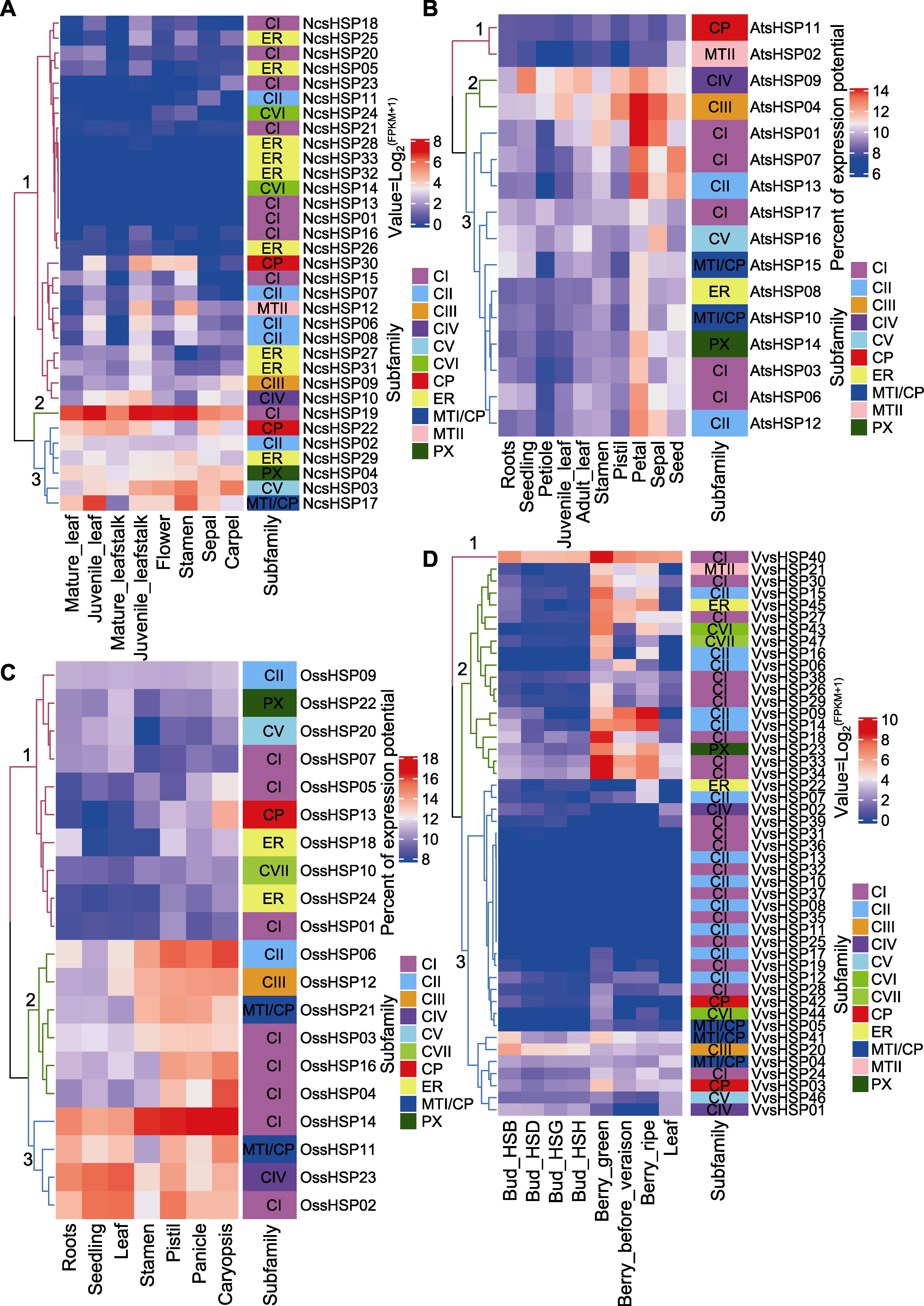
图5 NcsHSP (A)、AtsHSP (B)、OssHSP (C)和VvsHSP (D)在不同组织中的表达分析 两种颜色的标识分别代表表达值和sHSP亚家族。
Figure 5 Expression profiles of NcsHSP (A), AtsHSP (B), OssHSP (C) and VvsHSP (D) in different tissues Two color bars represented expression level and sHSP subfamilies, respectively.
| 1 | 何福林, 张斌 (2019). 银杏(Ginkgo biloba) GbHsp20基因家族的鉴定及系统进化分析. 分子植物育种 17, 7368-7376. |
| 2 | 黄祥富, 黄上志, 傅家瑞 (1999). 植物热激蛋白的功能及其基因表达的调控. 植物学通报 16, 530-536. |
| 3 | 栗振义, 龙瑞才, 张铁军, 杨青川, 康俊梅 (2016). 植物热激蛋白研究进展. 生物技术通报 32(2), 7-13. |
| 4 | 刘德立 (1996). 植物热激蛋白及其功能. 植物学通报 13, 14-19. |
| 5 | 张宁, 姜晶, 史洁玮 (2017). 番茄HSP20基因家族的全基因组鉴定、系统进化及表达分析. 沈阳农业大学学报 48, 137-144. |
| 6 | 张奇艳, 雷忠萍, 宋银, 海江波, 贺道华 (2019). 陆地棉扩展蛋白基因的鉴定与特征分析. 中国农业科学 52, 3713-3732. |
| 7 |
Cannon SB, Mitra A, Baumgarten A, Young ND, May G (2004). The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol 4, 10.
DOI URL |
| 8 |
Chen CJ, Chen H, Zhang Y, Thomas HR, Frank MH, He YH, Xia R (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol Plant 13, 1194-1202.
DOI URL |
| 9 |
Dafny-Yelin M, Tzfira T, Vainstein A, Adam Z (2008). Non-redundant functions of sHSP-CIs in acquired thermotolerance and their role in early seed development in Arabidopsis. Plant Mol Biol 67, 363-373.
DOI PMID |
| 10 |
Fan K, Chen YR, Mao ZJ, Fang Y, Li ZW, Lin WW, Zhang YQ, Liu JP, Huang JW, Lin WX (2020a). Pervasive duplication, biased molecular evolution and comprehensive functional analysis of the PP2C family in Glycine max. BMC Genomics 21, 465.
DOI URL |
| 11 |
Fan K, Mao ZJ, Zheng JX, Chen YR, Li ZW, Lin WW, Zhang YQ, Huang JW, Lin WX (2020b). Molecular evolution and expansion of the KUP family in the allopolyploid cotton species Gossypium hirsutum and Gossypium barbadense. Front Plant Sci 11, 545042.
DOI URL |
| 12 |
Fan K, Shen H, Bibi N, Li F, Yuan SN, Wang M, Wang XD (2015). Molecular evolution and species-specific expansion of the NAP members in plants. J Integr Plant Biol 57, 673-687.
DOI URL |
| 13 |
Fan K, Wang M, Miao Y, Ni M, Bibi N, Yuan SN, Li F, Wang XD (2014). Molecular evolution and expansion analysis of the NAC transcription factor in Zea mays. PLoS One 9, e111837.
DOI URL |
| 14 | Guo CL, Guo RR, Xu XZ, Gao M, Li XQ, Song JY, Zheng Y, Wang XP (2014). Evolution and expression analysis of the grape (Vitis vinifera L.) WRKY gene family. J Exp Bot 6, 1513-1528. |
| 15 |
Guo LM, Li J, He J, Liu H, Zhang HM (2020). A class I cytosolic HSP20 of rice enhances heat and salt tolerance in different organisms. Sci Rep 10, 1383.
DOI URL |
| 16 | Guo M, Liu JH, Lu JP, Zhai YF, Wang H, Gong ZH, Wang SB, Lu MH (2015). Genome-wide analysis of the CaHsp20 gene family in pepper: comprehensive sequence and expression profile analysis under heat stress. Front Plant Sci 6, 806. |
| 17 |
Ji XR, Yu YH, Ni PY, Zhang GH, Guo DL (2019). Genome-wide identification of small heat-shock protein (HSP20) gene family in grape and expression profile during berry development. BMC Plant Biol 19, 433.
DOI URL |
| 18 |
Jiao YN, Leebens-Mack J, Ayyampalayam S, Bowers JE, McKain M, McNeal J, Rolf M, Ruzicka DR, Wafula E, Wickett NJ, Wu XL, Zhang Y, Wang J, Zhang YT, Carpenter EJ, Deyholos MK, Kutchan TM, Chanderbali AS, Soltis PS, Stevenson DW, McCombie R, Pires JC, Wong GKS, Soltis DE, DePamphilis DW (2012). A genome triplication associated with early diversification of the core eudicots. Genome Biol 13, R3.
DOI URL |
| 19 |
Key JL, Lin CY, Chen YM (1981). Heat shock proteins of higher plants. Proc Natl Acad Sci USA 78, 3526-3530.
DOI URL |
| 20 |
Lamesch P, Berardini TZ, Li DH, Swarbreck D, Wilks C, Sasidharan R, Muller R, Dreher K, Alexander DL, Garcia-Hernandez M, Karthikeyan AS, Lee CH, Nelson WD, Ploetz L, Singh S, Wensel A, Huala E (2012). The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res 40, D1202-D1210.
DOI URL |
| 21 |
Li JB, Zhang J, Jia HX, Li Y, Xu XD, Wang LJ, Lu MZ (2016). The Populus trichocarpa PtHSP17.8 involved in heat and salt stress tolerances. Plant Cell Rep 35, 1587-1599.
DOI URL |
| 22 |
Li JB, Zhang J, Jia HX, Yue ZQ, Lu MZ, Xin XB, Hu JJ (2018). Genome-wide characterization of the sHsp gene family in Salix suchowensis reveals its functions under different abiotic stresses. Int J Mol Sci 19, 3246.
DOI URL |
| 23 |
Li Q, Zhang N, Zhang LS, Ma H (2015). Differential evolution of members of the rhomboid gene family with conservative and divergent patterns. New Phytol 206, 368-380.
DOI URL |
| 24 |
Ma W, Guan XY, Li J, Pan RH, Wang LY, Liu FJ, Ma HY, Zhu SJ, Hu J, Ruan YL, Chen XY, Zhang TZ (2019). Mitochondrial small heat shock protein mediates seed germination via thermal sensing. Proc Natl Acad Sci USA 116, 4716-4721.
DOI URL |
| 25 |
Ma W, Zhao T, Li J, Liu BL, Fang L, Hu Y, Zhang TZ (2016). Identification and characterization of the GhHsp20 gene family in Gossypium hirsutum. Sci Rep 6, 32517.
DOI URL |
| 26 |
Maere S, de Bodt S, Raes J, Casneuf T, van Montagu M, Kuiper M, van de Peer Y (2005). Modeling gene and genome duplications in eukaryotes. Proc Natl Acad Sci USA 102, 5454-5459.
DOI URL |
| 27 |
Ouyang S, Zhu W, Hamilton J, Lin HN, Campbell M, Childs K, Thibaud-Nissen F, Malek RL, Lee Y, Zheng L, Orvis J, Haas B, Wortman J, Buell CR (2007). The TIGR Rice Genome Annotation Resource: improvements and new features. Nucleic Acids Res 35, D883-D887.
DOI URL |
| 28 |
Park KC, Kim NS (2010). Intron loss mediated structural dynamics and functional differentiation of the polygalacturonase gene family in land plants. Genes Genom 32, 570-577.
DOI URL |
| 29 |
Pertea M, Kim D, Pertea GM, Leek JT, Salzberg SL (2016). Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat Protoc 11, 1650-1667.
DOI URL |
| 30 |
Sarkar NK, Kim YK, Grover A (2009). Rice sHsp genes: genomic organization and expression profiling under stress and development. BMC Genomics 10, 393.
DOI URL |
| 31 |
Scharf KD, Siddique M, Vierling E (2001). The expanding family of Arabidopsis thaliana small heat stress proteins and a new family of proteins containing α-crystallin domains (Acd proteins). Cell Stress Chaperones 6, 225-237.
DOI URL |
| 32 |
Shazadee H, Khan N, Wang JJ, Wang CC, Zeng JG, Huang ZY, Wang XY (2019). Identification and expression profiling of protein phosphatases (PP2C) gene family in Gossypium hirsutum L. Int J Mol Sci 20, 1395.
DOI URL |
| 33 |
Sun WN, Bernard C, Van De Cotte B, Van Montagu M, Verbruggen N (2010). At-HSP17.6A, encoding a small heat-shock protein in Arabidopsis, can enhance osmotolerance upon overexpression. Plant J 27, 407-415.
DOI URL |
| 34 |
The French-Italian Public Consortium for Grapevine Genome Characterization (2007). The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449, 463-467.
DOI URL |
| 35 |
van Montfort RLM, Basha E, Friedrich KL, Slingsby C, Vierling E (2001). Crystal structure and assembly of a eukaryotic small heat shock protein. Nat Struct Biol 8, 1025-1030.
PMID |
| 36 |
Wang L, Zhang SL, Zhang XM, Hu XY, Guo CL, Wang XP, Song JY (2018). Evolutionary and expression analysis of Vitis vinifera OFP gene family. Plant Syst Evol 304, 995-1008.
DOI URL |
| 37 |
Wang WX, Vinocur B, Shoseyov O, Altman A (2004). Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci 9, 244-252.
DOI URL |
| 38 |
Waters ER, Vierling E (2020). Plant small heat shock proteins—evolutionary and functional diversity. New Phytol 227, 24-37.
DOI URL |
| 39 |
Zhai MZ, Sun YD, Jia CX, Peng SB, Liu ZX, Yang GY (2016). Over-expression of JrsHSP17.3 gene from Juglans regia confer the tolerance to abnormal temperature and NaCl stresses. J Plant Biol 59, 549-558.
DOI URL |
| 40 |
Zhang K, Han YT, Zhao FL, Hu Y, Gao YR, Ma YF, Zheng Y, Wang YJ, Wen YQ (2015). Genome-wide identification and expression analysis of the CDPK gene family in grape, Vitis spp. BMC Plant Biol 15, 164.
DOI PMID |
| 41 |
Zhang LS, Chen F, Zhang XT, Li Z, Zhao YY, Lohaus R, Chang XJ, Dong W, Ho SYW, Liu X, Song AX, Chen JH, Guo WL, Wang ZJ, Zhuang YY, Wang HF, Chen XQ, Hu J, Liu YH, Qin Y, Wang K, Dong SS, Liu Y, Zhang SZ, Yu XX, Wu Q, Wang LS, Yan XQ, Jiao YN, Kong HZ, Zhou XF, Yu CW, Chen YC, Li F, Wang JH, Chen W, Chen XL, Jia QD, Zhang C, Jiang YF, Zhang WB, Liu GH, Fu JY, Chen F, Ma H, van de Peer Y, Tang HB (2020a). The water lily genome and the early evolution of flowering plants. Nature 577, 79-84.
DOI URL |
| 42 |
Zhang LS, Wu SD, Chang XJ, Wang XJ, Zhao YP, Xia YP, Trigiano RN, Jiao YN, Chen F (2020b). The ancient wave of polyploidization events in flowering plants and their facilitated adaptation to environmental stress. Plant Cell Environ 43, 2847-2856.
DOI URL |
| 43 |
Zhang N, Shi JW, Zhao HY, Jiang J (2018). Activation of small heat shock protein (SlHSP17.7) gene by cell wall invertase inhibitor (SlCIF1) gene involved in sugar metabolism in tomato. Gene 679, 90-99.
DOI PMID |
| 44 |
Zhao P, Wang DD, Wang RQ, Kong NN, Zhang C, Yang CH, Wu WT, Ma HL, Chen Q (2018). Genome-wide analysis of the potato Hsp20 gene family: identification, genomic organization and expression profiles in response to heat stress. BMC Genomics 19, 61.
DOI URL |
| 45 | Zhu W, Lu MH, Gong ZH, Chen RG (2011). Cloning and expression of a small heat shock protein gene CaHSP24 from pepper under abiotic stress. Afr J Biotechnol 10, 4968-4975. |
| 46 |
Zhu YX, Yan HW, Wang YY, Feng L, Chen Z, Xiang Y (2016). Genome duplication and evolution of heat shock transcription factor (HSF) gene family in four model angiosperms. J Plant Growth Regul 35, 903-920.
DOI URL |
| 47 |
Zhuo XK, Zheng TX, Zhang ZY, Zhang YC, Jiang LB, Ahmad S, Sun LD, Wang J, Cheng TR, Zhang QX (2018). Genome-wide analysis of the NAC transcription factor gene family reveals differential expression patterns and cold-stress responses in the woody plant Prunus mume. Genes (Basel) 9, 494.
DOI URL |
| [1] | 王子韵, 吕燕文, 肖钰, 吴超, 胡新生. 植物基因表达调控与进化机制研究进展[J]. 植物学报, 2025, 60(4): 1-0. |
| [2] | 夏琳凤, 李瑞, 王海政, 冯大领, 王春阳. 轮藻门植物基因组学研究进展[J]. 植物学报, 2025, 60(2): 271-282. |
| [3] | 赵来鹏, 王柏柯, 杨涛, 李宁, 杨海涛, 王娟, 闫会转. SlHVA22l基因调节番茄耐旱性[J]. 植物学报, 2024, 59(4): 558-573. |
| [4] | 段政勇, 丁敏, 王宇卓, 丁艺冰, 陈凌, 王瑞云, 乔治军. 糜子SBP基因家族全基因组鉴定及表达分析[J]. 植物学报, 2024, 59(2): 231-244. |
| [5] | 吴楠, 覃磊, 崔看, 李海鸥, 刘忠松, 夏石头. 甘蓝型油菜EXA1的克隆及其对植物抗病的调控作用[J]. 植物学报, 2023, 58(3): 385-393. |
| [6] | 褚振州, 古丽巴哈尔·依斯拉木, 屈泽众, 田新民. 同域分布的3种木蓼属植物叶绿体基因组比较[J]. 植物学报, 2023, 58(3): 417-432. |
| [7] | 王菲菲, 周振祥, 洪益, 谷洋洋, 吕超, 郭宝健, 朱娟, 许如根. 大麦NF-YC基因鉴定及在盐胁迫下的表达分析[J]. 植物学报, 2023, 58(1): 140-149. |
| [8] | 李永光, 任辉, 张英杰, 李瑞宁, 艾昊, 黄先忠. 十字花科植物PEBP基因家族的分子进化[J]. 生物多样性, 2022, 30(6): 21545-. |
| [9] | 何雨龙, 王佳歌, 赵珊珊, 高锦, 常英英, 赵喜亭, 聂碧华, 杨清香, 张江利, 李明军. 马铃薯Y病毒RPA-CRISPR/Cas12a检测技术体系的建立与应用[J]. 植物学报, 2022, 57(3): 308-319. |
| [10] | 谢露露, 崔青青, 董春娟, 尚庆茂. 植物嫁接愈合分子机制研究进展[J]. 植物学报, 2020, 55(5): 634-643. |
| [11] | 范业赓,丘立杭,黄杏,周慧文,甘崇琨,李杨瑞,杨荣仲,吴建明,陈荣发. 甘蔗节间伸长过程赤霉素生物合成关键基因的表达及相关植物激素动态变化[J]. 植物学报, 2019, 54(4): 486-496. |
| [12] | 王小龙,刘凤之,史祥宾,王孝娣,冀晓昊,王志强,王宝亮,郑晓翠,王海波. 葡萄NCED基因家族进化及表达分析[J]. 植物学报, 2019, 54(4): 474-485. |
| [13] | 化文平,陈尘,智媛,刘莉,王喆之,李翠芹. SmGGPPS2对丹参酮合成的影响[J]. 植物学报, 2019, 54(2): 217-226. |
| [14] | 刘铭, 刘霞, 孙然, 李玉灵, 杜克久. 多氯联苯促进毛白杨不定根分化的效应[J]. 植物学报, 2018, 53(6): 764-772. |
| [15] | 孙万梅, 王晓珠, 韩二琴, 韩丽, 孙丽萍, 彭再慧, 王邦俊. 亲免素在植物体内的功能研究进展[J]. 植物学报, 2017, 52(6): 808-819. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||