

植物学报 ›› 2023, Vol. 58 ›› Issue (3): 385-393.DOI: 10.11983/CBB22077 cstr: 32102.14.CBB22077
吴楠1, 覃磊1, 崔看1, 李海鸥1, 刘忠松2, 夏石头1( )
)
收稿日期:2022-04-17
接受日期:2022-07-03
出版日期:2023-05-01
发布日期:2023-05-17
通讯作者:
*E-mail: xstone0505@hunau.edu.cn
基金资助:
Nan Wu1, Lei Qin1, Kan Cui1, Haiou Li1, Zhongsong Liu2, Shitou Xia1( )
)
Received:2022-04-17
Accepted:2022-07-03
Online:2023-05-01
Published:2023-05-17
Contact:
*E-mail: xstone0505@hunau.edu.cn
摘要: 拟南芥(Arabidopsis thaliana) EXA1缺失会导致exa1-2突变体植株PR基因表达上调, 对病原菌的抗性提高。该研究通过克隆甘蓝型油菜(Brassica napus)中的BnaEXA1, 并将其在拟南芥exa1-2突变体中异源超表达, 发现BnaEXA1超表达不仅可恢复突变体的表型, 而且显著降低突变体中PR1和PR2基因的表达量, 导致其对核盘菌(Sclerotinia sclerotiorum)和卵菌H.a. Noco2易感, 表明BnaEXA1调控植物的基础抗性。
吴楠, 覃磊, 崔看, 李海鸥, 刘忠松, 夏石头. 甘蓝型油菜EXA1的克隆及其对植物抗病的调控作用. 植物学报, 2023, 58(3): 385-393.
Nan Wu, Lei Qin, Kan Cui, Haiou Li, Zhongsong Liu, Shitou Xia. Cloning of Brassica napus EXA1 Gene and Its Regulation on Plant Disease Resistance. Chinese Bulletin of Botany, 2023, 58(3): 385-393.
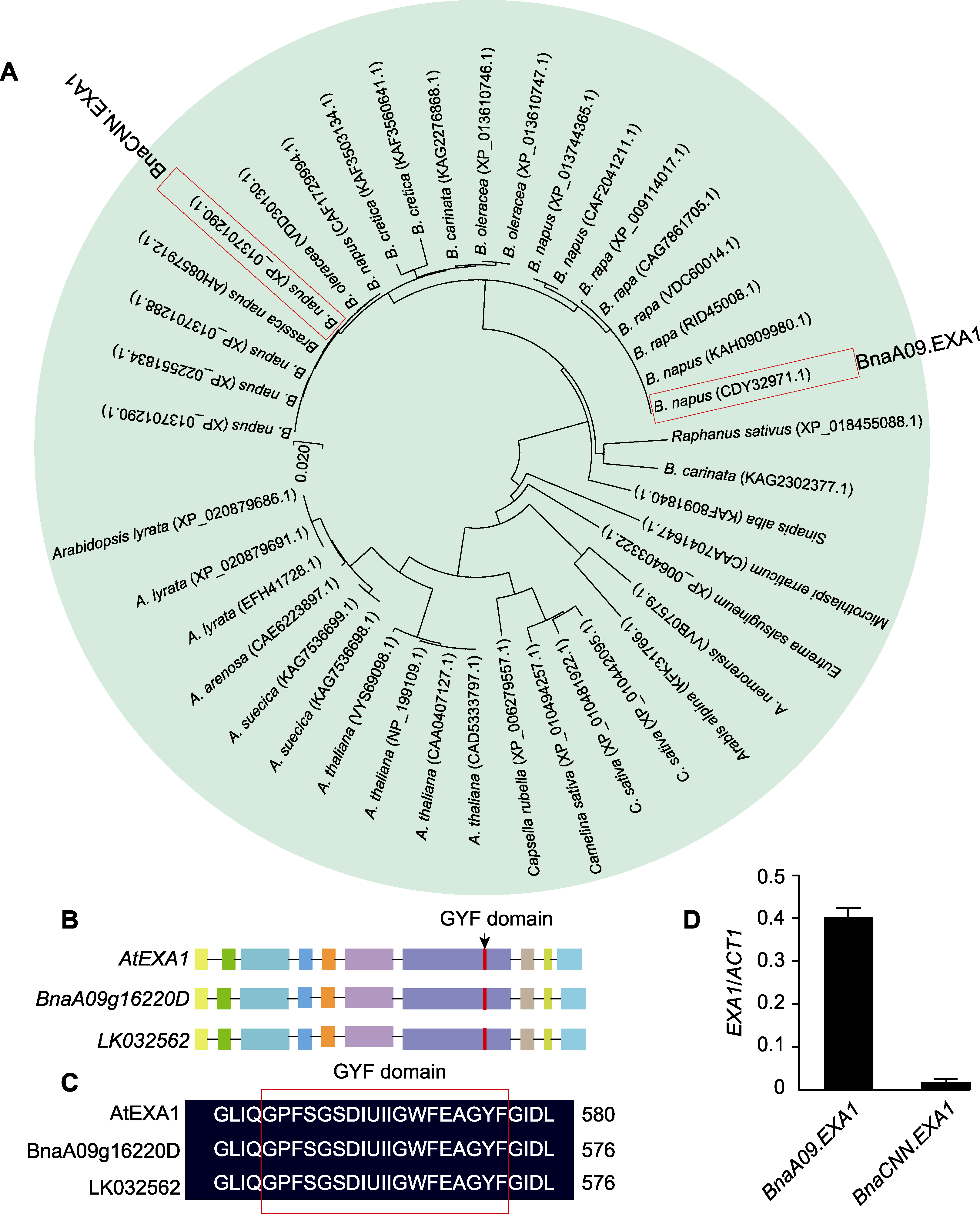
图1 BnaEXA1的序列分析 (A) 十字花科EXA1进化树分析; (B) AtEXA1与BnaEXA1的基因组序列比对(矩形代表外显子, 黑色线条代表内含子); (C) BnaEXA1的结构示意图(红色框代表GYF结构域); (D) BnaEXA1在湘油15中的表达量分析
Figure 1 Sequence analysis of BnaEXA1 (A) Phylogenetic analysis of EXA1 in Brassicaceae; (B) Comparison of the derived genome sequences of AtEXA1 and BnaEXA1 (the rectangle represents the exon and the black line represents the intron); (C) Schematic structures of BnaEXA1 (red box indicates GYF domain); (D) Expression analysis of BnaEXA1 in XY-15
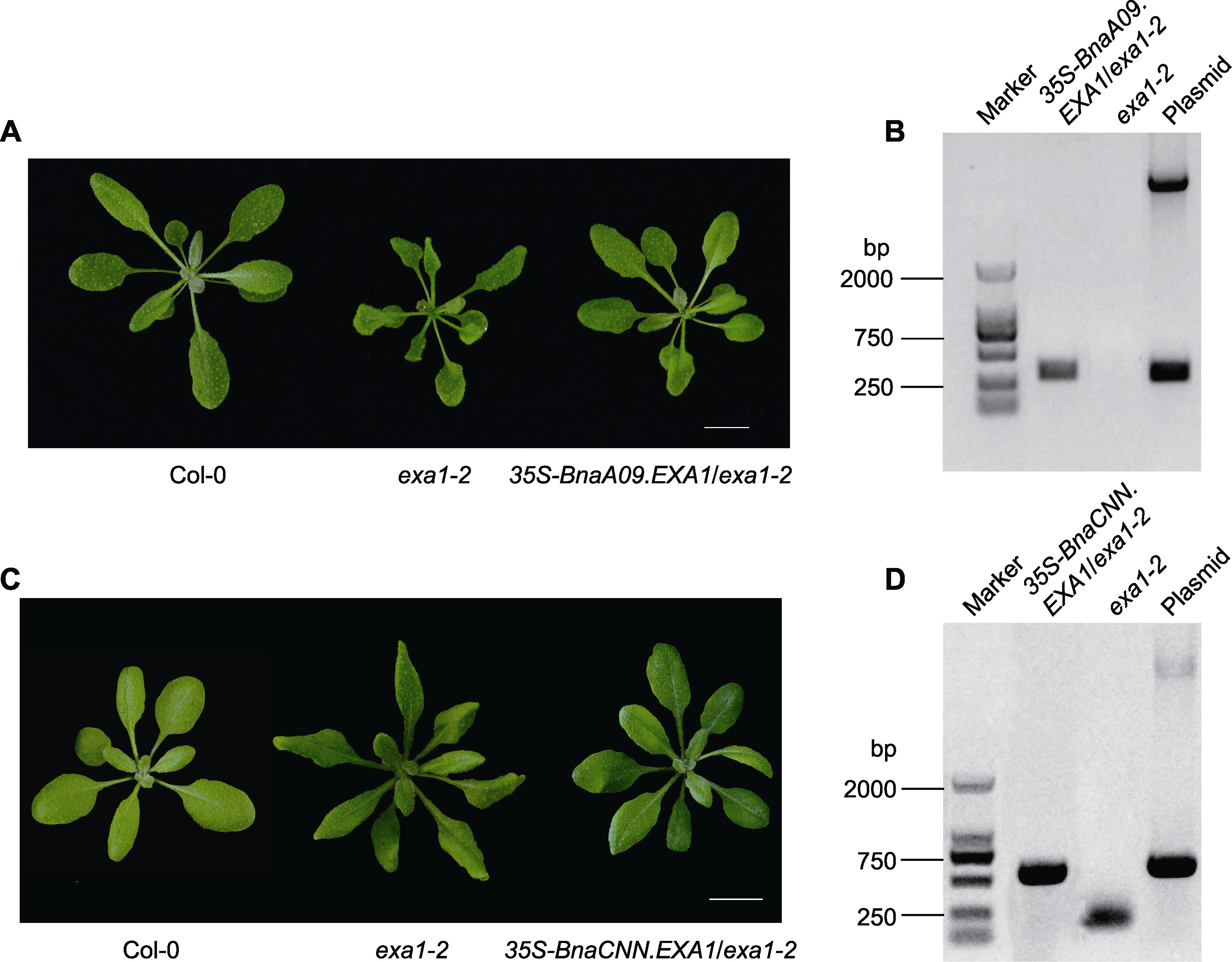
图2 BnaEXA1转基因拟南芥株系形态学表型及其观测 (A), (C) 4周龄Col-0、exa1-2及BnaEXA1转基因拟南芥株系形态学表型; (B), (D) BnaEXA1转基因拟南芥株系的PCR鉴定。Marker: DNA分子标记; 阴性对照: exa1-2突变体; 阳性对照: Plasmid (35S-BnaA09.EXA1和35S-BnaCNN.EXA1表达质粒)。Bars=1 cm
Figure 2 Morphological phenotype and detection of BnaEXA1 transgenic Arabidopsis thaliana (A), (C) Morphological phenotypes of 4-week-old Col-0, exa1-2 and BnaEXA1 transgenic Arabidopsis thaliana plants were used for analysis; (B), (D) Identification of BnaEXA1 transgenic Arabidopsis thaliana plants by PCR; Marker: DNA marker; Negative control: exa1-2 mutant; Positive control: Plasmid (35S-BnaA09.EXA1 and 35S-BnaCNN.EXA1 expression vectors). Bars=1 cm
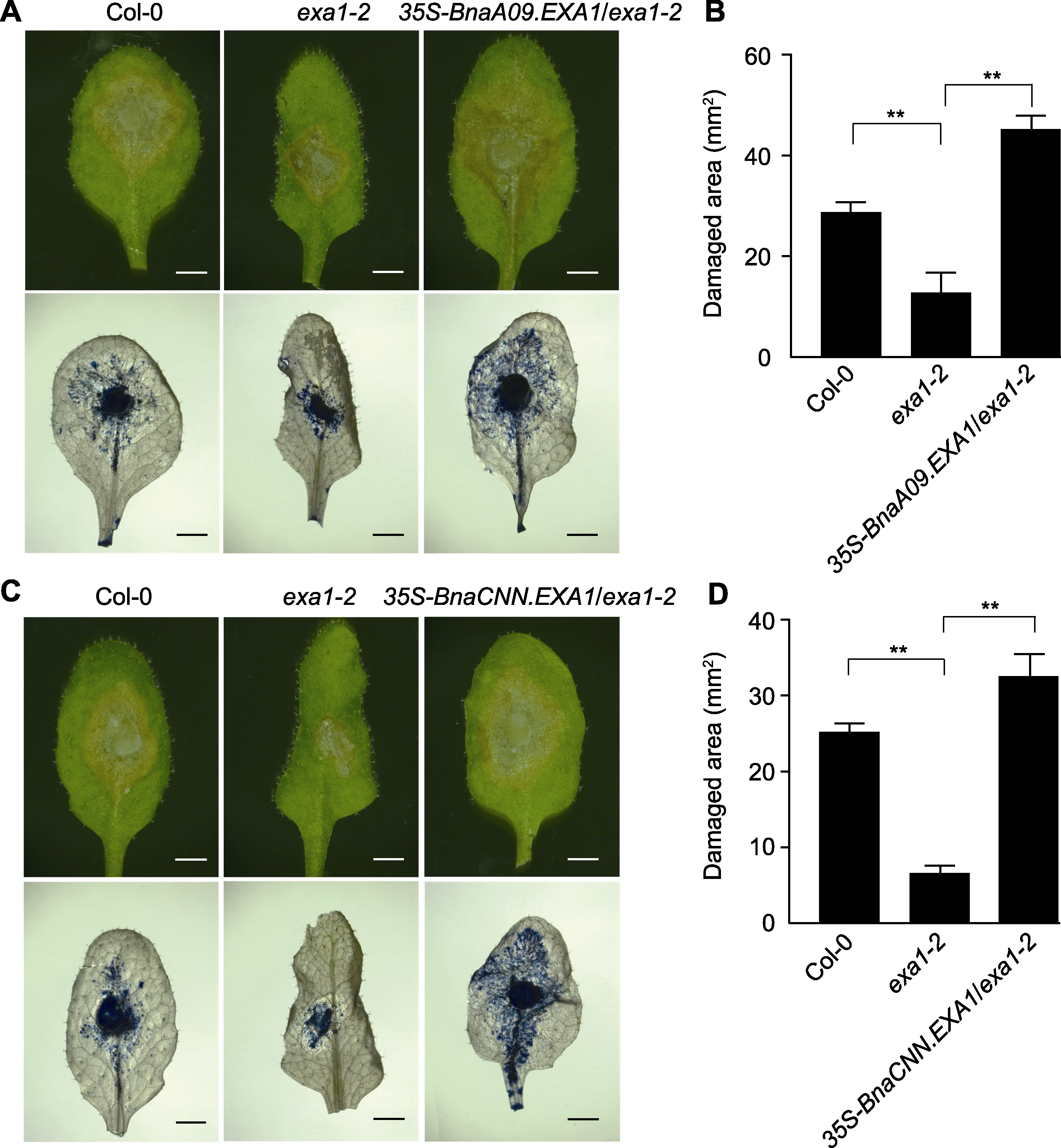
图3 核盘菌侵染后BnaEXA1转基因拟南芥株系的抗病表型 (A), (C) 核盘菌侵染后Col-0、exa1-2及BnaEXA1转基因拟南芥株系的抗病表型及台盼蓝染色(bars=1 cm); (B), (D) 核盘菌侵染后Col-0、exa1-2及BnaEXA1转基因拟南芥株系的损伤面积。**P<0.01
Figure 3 Disease resistance phenotype of BnaEXA1 transgenic Arabidopsis thaliana lineages after Sclerotinia sclerotiorum in-fection (A), (C) Disease resistance phenotype and trypan blue staining of Col-0, exa1-2 and BnaEXA1 transgenic A. thaliana plants infected by S. sclerotiorum (bars=1 cm); (B), (D) Damage area of Col-0, exa1-2 and BnaEXA1 transgenic A. thaliana lineages infected by S. sclerotiorum. ** P<0.01
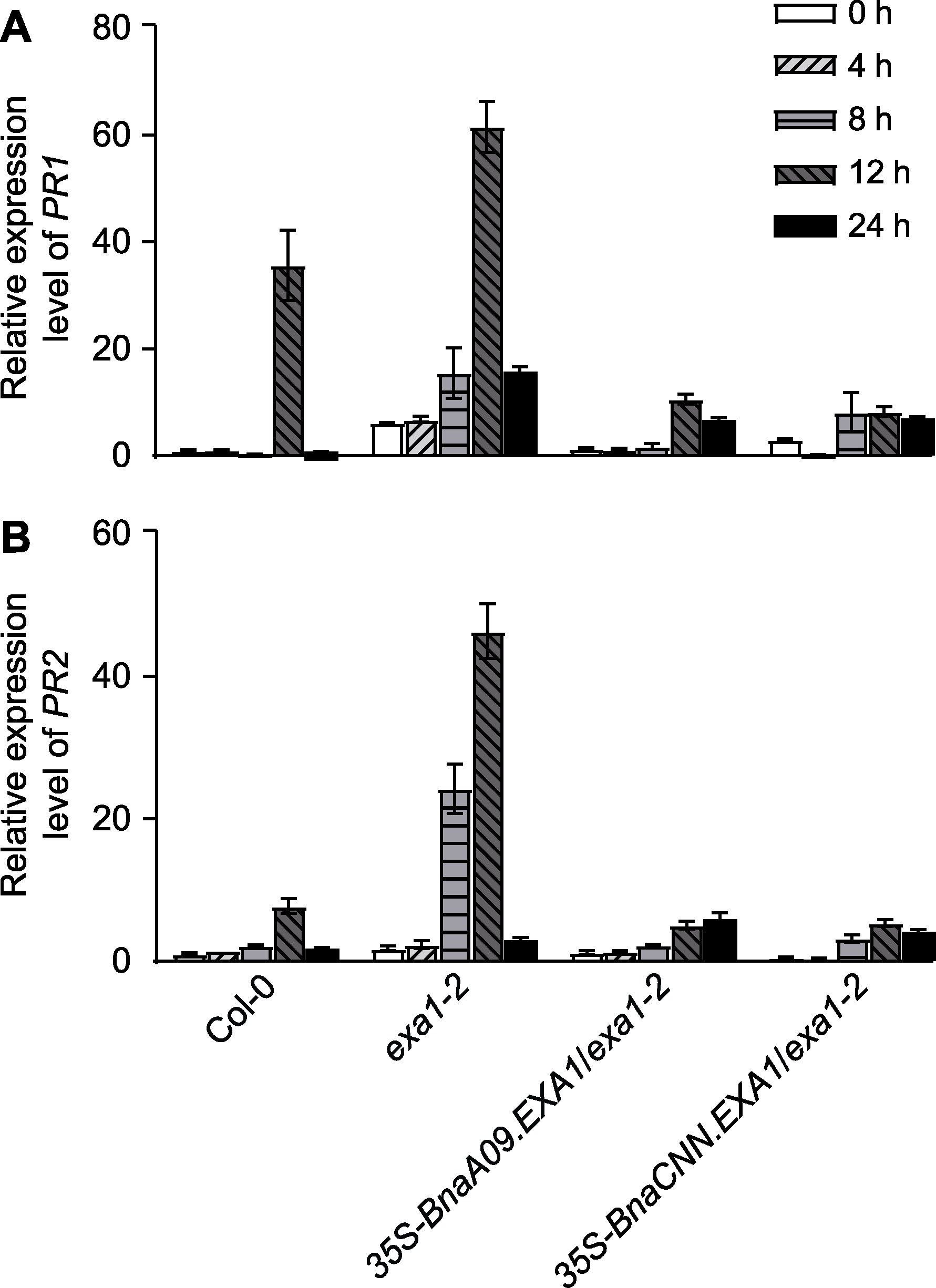
图4 核盘菌侵染诱导的PR基因表达 核盘菌侵染0、4、8、12和24小时Col-0、exa1-2及BnaEXA1转基因拟南芥株系的PR1 (A)和PR2 (B)基因的表达量
Figure 4 PR gene expression induced by Sclerotinia scle- rotiorum PR1 (A) and PR2 (B) gene expression levels in Col-0, ex-a1-2 and BnaEXA1 transgenic Arabidopsis thaliana linea-ges after 0, 4, 8, 12, and 24 hours of infection by S. scle-rotiorum
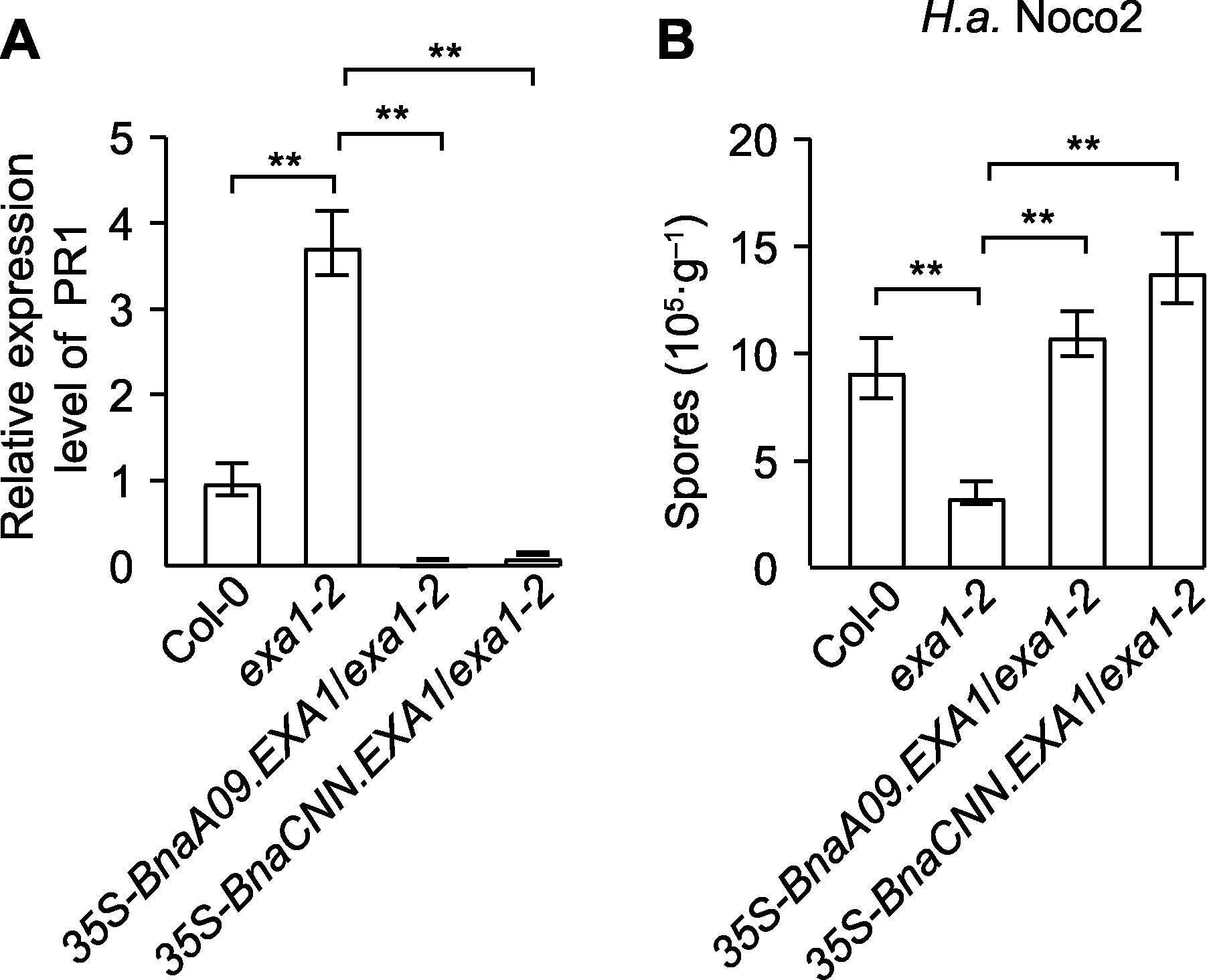
图5 BnaEXA1转基因拟南芥株系的抗病表型 (A) Col-0、exa1-2及BnaEXA1转基因拟南芥株系的PR1基因表达量; (B) Col-0、exa1-2及BnaEXA1转基因拟南芥株系对H.a. (Hyaloperonospora arabidopsidis) Noco2的抗性鉴定。** P<0.01
Figure 5 Disease resistance phenotype of BnaEXA1 trans- genic Arabidopsis thaliana lineages (A) Quantitative RT-PCR analysis of PR1 expressions in Col-0, exa1-2 and BnaEXA1 transgenic A. thaliana plant-s; (B) Identification of resistance of Col-0, exa1-2 and BnaEXA1 transgenic A. thaliana lineages to H.a. (Hyalo-peronospora arabidopsidis) Noco2. ** P<0.01
| [1] | 费维新, 李强生, 吴新杰, 侯树敏, 陈凤祥, 王文相, 胡宝成 (2002). 利用栽培措施控制油菜菌核病的研究. 中国油料作物学报 24(3), 47-49. |
| [2] | 王爱荣 (2006). 核盘菌与拟南芥互作的分子机制研究. 博士论文. 福州: 福建农林大学. pp. 1-106. |
| [3] | Amselem J, Cuomo CA, van Kan JAL, Viaud M, Benito EP, Couloux A, Coutinho PM, de Vries RP, Dyer PS, Fillinger S, Fournier E, Gout L, Hahn M, Kohn L, Lapalu N, Plummer KM, Pradier JM, Quévillon E, Sharon A, Simon A, ten Have A, Tudzynski B, Tudzynski P, Wincker P, Andrew M, Anthouard V, Beever RE, Beffa R, Benoit I, Bouzid O, Brault B, Chen ZH, Choquer M, Collémare J, Cotton P, Danchin EG, Da Silva C, Gautier A, Giraud C, Giraud T, Gonzalez C, Grossetete S, Güldener U, Henrissat B, Howlett BJ, Kodira C, Kretschmer M, Lappartient A, Leroch M, Levis C, Mauceli E, Neuvéglise C, Oeser B, Pearson M, Poulain J, Poussereau N, Quesneville H, Rascle C, Schumacher J, Ségurens B, Sexton A, Silva E, Sirven C, Soanes DM, Talbot NJ, Templeton M, Yandava C, Yarden O, Zeng QD, Rollins JA, Lebrun MH, Dickman M(2011). Genomic analysis of the necrotrophic fungal pathogens Sclerotinia sclerotiorum and Botrytis cinerea. PLoS Genet 7, e1002230. |
| [4] |
Ash MR, Faelber K, Kosslick D, Albert GI, Roske Y, Kofler M, Schuemann M, Krause E, Freund C (2010). Conserved ß-hairpin recognition by the GYF domains of Smy2 and GIGYF2 in mRNA surveillance and vesicular transport complexes. Structure 18, 944-954.
DOI URL |
| [5] |
Barbetti MJ, Banga SK, Fu TD, Li YC, Singh D, Liu SY, Ge XT, Banga SS (2014). Comparative genotype reactions to Sclerotinia sclerotiorum within breeding populations of Brassica napus and B. juncea from India and China. Euphytica 197, 47-59.
DOI URL |
| [6] |
Dai FM, Xu T, Wolf GA, He ZH (2006). Physiological and molecular features of the pathosystem Arabidopsis thaliana L.-Sclerotinia sclerotiorum Libert. J Integr Plant Biol 48, 44-52.
DOI URL |
| [7] |
Giovannone B, Tsiaras WG, de la Monte S, Klysik J, Lautier C, Karashchuk G, Goldwurm S, Smith RJ (2009). GIGYF2 gene disruption in mice results in neurodegeneration and altered insulin-like growth factor signaling. Hum Mol Genet 18, 4629-4639.
DOI PMID |
| [8] |
Guo XM, Stotz HU (2007). Defense against Sclerotinia sclerotiorum in Arabidopsis is dependent on jasmonic acid, salicylic acid, and ethylene signaling. Mol Plant Microbe Interact 20, 1384-1395.
DOI URL |
| [9] |
Hale VA, Guiney EL, Goldberg LY, Haduong JH, Kwartler CS, Scangos KW, Goutte C (2012). Notch signaling is antagonized by SAO-1, a novel GYF-domain protein that interacts with the E3 ubiquitin ligase SEL-10 in Caenorhabditis elegans. Genetics 190, 1043-1057.
DOI URL |
| [10] |
Hashimoto M, Neriya Y, Keima T, Iwabuchi N, Koinuma H, Hagiwara-Komoda Y, Ishikawa K, Himeno M, Maejima K, Yamaji Y, Namba S (2016). EXA1, a GYF domain protein, is responsible for loss-of-susceptibility to Plantago asiatica mosaic virus in Arabidopsis thaliana. Plant J 88, 120-131.
DOI URL |
| [11] |
Kofler M, Motzny K, Freund C (2005). GYF domain proteomics reveals interaction sites in known and novel target proteins. Mol Cell Proteomics 4, 1797-1811.
DOI PMID |
| [12] |
Letunic I, Doerks T, Bork P (2015). SMART: recent updates, new developments and status in 2015. Nucleic Acids Res 43, D257-D260.
DOI URL |
| [13] |
Li S, Li X, Zhou YJ (2018). Ribosomal protein L18 is an essential factor that promote rice stripe virus accumulation in small brown planthopper. Virus Res 247, 15-20.
DOI PMID |
| [14] | Matsui H, Nomura Y, Egusa M, Hamada T, Hyon GS, Kaminaka H, Watanabe Y, Ueda T, Trujillo M, Shirasu K, Nakagami H (2017). The GYF domain protein PSIG1 dampens the induction of cell death during plant-pathogen interactions. PLoS Genet 13, e1007037. |
| [15] | Rowe HC, Walley JW, Corwin J, Chan EKF, Dehesh K, Kliebenstein DJ (2010). Deficiencies in jasmonate-mediated plant defense reveal quantitative variation in Botrytis cinerea pathogenesis. PLoS Pathog 6, e1000861. |
| [16] |
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28, 2731-2739.
DOI PMID |
| [17] | Wu ZS, Huang S, Zhang XB, Wu D, Xia ST, Li X (2017). Regulation of plant immune receptor accumulation through translational repression by a glycine-tyrosine-phenylalanine (GYF) domain protein. eLife 6, e23684. |
| [1] | 杨柳卿, 王劲, 燕敬利, 陈芹芹, 程浩坤, 李春, 赵培玉, 杨博, 江元清. 甘蓝型油菜转录因子BnaABF2的表征分析及互作蛋白鉴定[J]. 植物学报, 2025, 60(1): 49-61. |
| [2] | 李青洋, 刘翠, 何李, 彭姗, 马嘉吟, 胡子祎, 刘宏波. 甘蓝型油菜BnaA02.CPSF6基因的克隆及功能分析(长英文摘要)[J]. 植物学报, 2025, 60(1): 62-73. |
| [3] | 赵晗茜, 宋佳怡, 杨洁, 赵永晶, 夏文念, 顾伟卓, 汪仲毅, 杨楠, 胡慧贞. 金鱼草XTH家族基因鉴定及抗核盘菌和雄蕊瓣化相关基因筛选[J]. 植物学报, 2024, 59(2): 188-203. |
| [4] | 张盈川, 吴晓明玉, 陶保龙, 陈丽, 鲁海琴, 赵伦, 文静, 易斌, 涂金星, 傅廷栋, 沈金雄. Bna-miR43介导甘蓝型油菜响应干旱胁迫[J]. 植物学报, 2023, 58(5): 701-711. |
| [5] | 王钢, 王二涛. “卫青不败由天幸”——WeiTsing的广谱抗根肿病机理被揭示[J]. 植物学报, 2023, 58(3): 356-358. |
| [6] | 贺闽, 尹俊杰, 冯志明, 朱孝波, 赵剑华, 左示敏, 陈学伟. 水稻稻瘟病和纹枯病抗性鉴定方法[J]. 植物学报, 2020, 55(5): 577-587. |
| [7] | 张娜,刘秀霞,陈学森,吴树敬. 基于转录组分析鉴定苹果茉莉素响应基因[J]. 植物学报, 2019, 54(6): 733-743. |
| [8] | 宋敏,张瑶,王丽莹,彭向永. 甘蓝型油菜ZF-HD基因家族的鉴定与系统进化分析[J]. 植物学报, 2019, 54(6): 699-710. |
| [9] | 刘凯歌, 齐双慧, 段绍伟, 李东, 金倡宇, 高晨浩, 刘绚霞, 陈明训. 甘蓝型油菜BnTTG1-1基因的功能分析[J]. 植物学报, 2017, 52(6): 713-722. |
| [10] | 高虎虎, 张云霄, 胡胜武, 郭媛. 甘蓝型油菜MADS-box基因家族的鉴定与系统进化分析[J]. 植物学报, 2017, 52(6): 699-712. |
| [11] | 牛毅, 高远, 李隔萍, 任安芝, 高玉葆. 内生真菌对羽茅抗病性的影响[J]. 植物生态学报, 2016, 40(9): 925-932. |
| [12] | 贾乐东, 李施蒙, 许代香, 曲存民, 李加纳, 王瑞. 甘蓝型油菜BnMYB80基因的生物信息学分析[J]. 植物学报, 2016, 51(5): 620-630. |
| [13] | 刘振, 刘霞, 刘建中. 亚硝基化在植物细胞死亡及防御反应中的作用[J]. 植物学报, 2016, 51(1): 130-143. |
| [14] | 李春宏, 付三雄, 戚存扣. 应用基因芯片分析甘蓝型油菜柱头特异表达基因[J]. 植物学报, 2014, 49(3): 246-253. |
| [15] | 付三雄, 李成磊, 尼玛卓玛, 唐林, 戚存扣. 气象因子对油菜种子中油分积累的影响[J]. 植物学报, 2014, 49(1): 41-48. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||