

植物学报 ›› 2024, Vol. 59 ›› Issue (2): 188-203.DOI: 10.11983/CBB23107 cstr: 32102.14.CBB23107
赵晗茜, 宋佳怡, 杨洁, 赵永晶, 夏文念, 顾伟卓, 汪仲毅, 杨楠, 胡慧贞*( )
)
收稿日期:2023-08-07
接受日期:2024-01-30
出版日期:2024-03-10
发布日期:2024-03-10
通讯作者:
* 胡慧贞, 云南省高层次人才引进计划青年人才, 国家林业和草原局荷花及水生植物产业研发创新联盟秘书长。2017年于华中农业大学获农学博士学位, 长期致力于荷花、金鱼草和油菜等植物切花和株型形成机理及细胞壁抗性研究。发表高质量SCI论文近20篇, 累计影响因子超过100; 主持国家级和省部级项目6项。E-mail: 基金资助:
Hanqian Zhao, Jiayi Song, Jie Yang, Yongjing Zhao, Wennian Xia, Weizhuo Gu, Zhongyi Wang, Nan Yang, Huizhen Hu*( )
)
Received:2023-08-07
Accepted:2024-01-30
Online:2024-03-10
Published:2024-03-10
Contact:
* E-mail: 摘要: 木葡聚糖内转糖苷酶/水解酶(XTH)属于糖苷水解酶16家族(GH16), 是一类介导木葡聚糖(XyG)-纤维素骨架构建和重组的酶。为探明XTH家族基因在金鱼草(Antirrhinum majus)中的潜在生物学功能, 通过生物信息学分析, 结合转录组测序(RNA-seq)和实时荧光定量聚合酶链式反应(qRT-PCR)探究了XTH家族基因分别在金鱼草瓣化和非瓣化雄蕊以及抗感核盘菌材料中的表达水平。结果表明, 鉴定出的33个AmXTH蛋白主要保守基序为ExDxE, 分为3个亚组。AmXTH基因启动子的顺式作用元件多为生长发育、抗病及抗逆类。经RNA-seq和qRT-PCR验证, 最终挖掘出4个正向介导抗病的关键候选基因(AmXTH3、14、18和33), 1个负向介导抗病的关键候选基因(AmXTH23), 12个正向介导雄蕊瓣化的关键候选基因(AmXTH1、7、9、11、21、22、23、24、26、28、29和33)以及2个负向介导雄蕊瓣化的关键候选基因(AmXTH15和31); 其中AmXTH23 和AmXTH33可能同时在金鱼草抗核盘菌和雄蕊瓣化中发挥作用。该研究初步挖掘出参与金鱼草抗核盘菌及雄蕊瓣化的AmXTH候选基因, 为进一步揭示其生物学功能奠定了基础。
赵晗茜, 宋佳怡, 杨洁, 赵永晶, 夏文念, 顾伟卓, 汪仲毅, 杨楠, 胡慧贞. 金鱼草XTH家族基因鉴定及抗核盘菌和雄蕊瓣化相关基因筛选. 植物学报, 2024, 59(2): 188-203.
Hanqian Zhao, Jiayi Song, Jie Yang, Yongjing Zhao, Wennian Xia, Weizhuo Gu, Zhongyi Wang, Nan Yang, Huizhen Hu. Identification of XTH Family Genes in Antirrhinum majus and Screening of Genes Involoved in Sclerotinia sclerotiorum Resistance and Stamen Petalization. Chinese Bulletin of Botany, 2024, 59(2): 188-203.
| Gene name | Forward primer (5′-3′) | Reverse primer (5′-3′) |
|---|---|---|
| AmUBI | ATCCACCCTTCACCTTGTG | TTGTCAATGGTATCCGAGC |
| AmXTH1 | AAGGACAGAGGCAATGGGTG | AACCTGAGAACTCGGCAGTG |
| AmXTH2 | TATGGAACGGAACGGGTTGG | TGCCAATTGTTTGGTGCGTT |
| AmXTH3 | CGCCAAGATCTTCCGAGGAG | CAGCGACGAGTTTGAGTTGC |
| AmXTH4 | AAAAGACCAAGCAATGGGCG | CTGCATCGTCGCATGTTGTC |
| AmXTH5 | TATGGTTCGATCCCACTGCG | CGAACGGTGCCTTTTTCCAG |
| AmXTH6 | GTGTTTGTGAACGGGCAAGG | TGCCCAACCATCTCCATTCC |
| AmXTH7 | TGGTTTGATCCAACGGCTGA | TCCGTTGCCCAACTACTTCC |
| AmXTH8 | TGCAGCTCTCAATCTGCCAA | GTAAAACGCGACGACGATGC |
| AmXTH9 | GGCCGAATGCTTTCGCTATC | AGTTCCGGCTGAGTTTCCTG |
| AmXTH10 | TCGCATTCCCGACTAACCAG | TACACGAATTCGAGAGCCCG |
| AmXTH11 | ACGAAAGCTCCGTTTGTTGC | TCTCCCATTGGCGTGAAGTC |
| AmXTH12 | GGCCAAATGCTGTCGCTATC | CAGTTCCGGCTGAGTTTCCT |
| AmXTH13 | TACATCGGGCTCTGGTTTCG | ATCGTGTGCGTCTCCTTGAG |
| AmXTH14 | CCCAAAAGCCAGCCAATGAG | GGCTTCGCAAAAGTGCAAGT |
| AmXTH15 | CCACCGAGGAATCACTCCAG | TCTAGATTACGTCGTGCCGC |
| AmXTH16 | TGCCGATGATTGGGCTACAC | TCAGGGGAAAGTTCACAGGC |
| AmXTH17 | CAGGCTCCCTTTACCGCTTA | CGGGGGTGAGATCCCAATAC |
| AmXTH18 | AGAACCACGACGAACTGGAC | AGGATGCTGTAACGATGCGA |
| AmXTH19 | ACCTTCCCCTGGCTACTACC | TTCCTGAGGTTCTGTCGAGC |
| AmXTH20 | TTCCTTGGCAATGTACCGGC | GCTGATCTGCAAGCTCATGG |
| AmXTH21 | CCTCTGGAATCCTCAACGCA | CACTAGTCCGCCTCTTGTCG |
| AmXTH22 | CGCTGAGTCAGTAGGTGTGG | TTTTCACTAGCCCGCCTCTC |
| AmXTH23 | AAGTTCTGCGACACTCAGGG | TAGTGTAGCTGCTGCGAACC |
| AmXTH24 | GTGGTTCGACCCGTCTAAGG | CTCCCCATTGCATCAACCCT |
| AmXTH25 | GATGGATCAGAGTGGGCGAC | CCCACTGCAACCCAGTAGTC |
| AmXTH26 | TGGGGCTATTTGAGCACTCG | GCCGAATGTGATTGTCAGCC |
| AmXTH27 | TGGAACCCAATGGCTTGGAC | GGGTTGAGTTGCAAACTGGG |
| AmXTH28 | CGACCCCTCCGAAGATTTCC | GCTATCCGCGTTCCAAATGC |
| AmXTH29 | CGATTCTCACATTCGGCAGC | AGCTCGTCGCGTACTGTATC |
| AmXTH30 | GAGCGGTTGTTCCGCATTTG | GCGTGAGAGCATGGTTCCTT |
| AmXTH31 | TTTCGACATCACGTGGGGAG | TCCCAGGGACAAGCTTCAAC |
| AmXTH32 | TCTACCGAGACAAATGGCCG | CAAGCGCATCTGGAAACGAG |
| AmXTH33 | ATGAAGATATTTATTGTGCA | AGATACCAATGGGAATCTTC |
表1 qRT-PCR引物序列
Table 1 The sequences of qRT-PCR primers
| Gene name | Forward primer (5′-3′) | Reverse primer (5′-3′) |
|---|---|---|
| AmUBI | ATCCACCCTTCACCTTGTG | TTGTCAATGGTATCCGAGC |
| AmXTH1 | AAGGACAGAGGCAATGGGTG | AACCTGAGAACTCGGCAGTG |
| AmXTH2 | TATGGAACGGAACGGGTTGG | TGCCAATTGTTTGGTGCGTT |
| AmXTH3 | CGCCAAGATCTTCCGAGGAG | CAGCGACGAGTTTGAGTTGC |
| AmXTH4 | AAAAGACCAAGCAATGGGCG | CTGCATCGTCGCATGTTGTC |
| AmXTH5 | TATGGTTCGATCCCACTGCG | CGAACGGTGCCTTTTTCCAG |
| AmXTH6 | GTGTTTGTGAACGGGCAAGG | TGCCCAACCATCTCCATTCC |
| AmXTH7 | TGGTTTGATCCAACGGCTGA | TCCGTTGCCCAACTACTTCC |
| AmXTH8 | TGCAGCTCTCAATCTGCCAA | GTAAAACGCGACGACGATGC |
| AmXTH9 | GGCCGAATGCTTTCGCTATC | AGTTCCGGCTGAGTTTCCTG |
| AmXTH10 | TCGCATTCCCGACTAACCAG | TACACGAATTCGAGAGCCCG |
| AmXTH11 | ACGAAAGCTCCGTTTGTTGC | TCTCCCATTGGCGTGAAGTC |
| AmXTH12 | GGCCAAATGCTGTCGCTATC | CAGTTCCGGCTGAGTTTCCT |
| AmXTH13 | TACATCGGGCTCTGGTTTCG | ATCGTGTGCGTCTCCTTGAG |
| AmXTH14 | CCCAAAAGCCAGCCAATGAG | GGCTTCGCAAAAGTGCAAGT |
| AmXTH15 | CCACCGAGGAATCACTCCAG | TCTAGATTACGTCGTGCCGC |
| AmXTH16 | TGCCGATGATTGGGCTACAC | TCAGGGGAAAGTTCACAGGC |
| AmXTH17 | CAGGCTCCCTTTACCGCTTA | CGGGGGTGAGATCCCAATAC |
| AmXTH18 | AGAACCACGACGAACTGGAC | AGGATGCTGTAACGATGCGA |
| AmXTH19 | ACCTTCCCCTGGCTACTACC | TTCCTGAGGTTCTGTCGAGC |
| AmXTH20 | TTCCTTGGCAATGTACCGGC | GCTGATCTGCAAGCTCATGG |
| AmXTH21 | CCTCTGGAATCCTCAACGCA | CACTAGTCCGCCTCTTGTCG |
| AmXTH22 | CGCTGAGTCAGTAGGTGTGG | TTTTCACTAGCCCGCCTCTC |
| AmXTH23 | AAGTTCTGCGACACTCAGGG | TAGTGTAGCTGCTGCGAACC |
| AmXTH24 | GTGGTTCGACCCGTCTAAGG | CTCCCCATTGCATCAACCCT |
| AmXTH25 | GATGGATCAGAGTGGGCGAC | CCCACTGCAACCCAGTAGTC |
| AmXTH26 | TGGGGCTATTTGAGCACTCG | GCCGAATGTGATTGTCAGCC |
| AmXTH27 | TGGAACCCAATGGCTTGGAC | GGGTTGAGTTGCAAACTGGG |
| AmXTH28 | CGACCCCTCCGAAGATTTCC | GCTATCCGCGTTCCAAATGC |
| AmXTH29 | CGATTCTCACATTCGGCAGC | AGCTCGTCGCGTACTGTATC |
| AmXTH30 | GAGCGGTTGTTCCGCATTTG | GCGTGAGAGCATGGTTCCTT |
| AmXTH31 | TTTCGACATCACGTGGGGAG | TCCCAGGGACAAGCTTCAAC |
| AmXTH32 | TCTACCGAGACAAATGGCCG | CAAGCGCATCTGGAAACGAG |
| AmXTH33 | ATGAAGATATTTATTGTGCA | AGATACCAATGGGAATCTTC |
| Gene name | Gene ID | aa | Mw (Da) | PI | Instability index | Aliphatic index | GRAVY | TMHs | SP | Subcellular localization |
|---|---|---|---|---|---|---|---|---|---|---|
| AmXTH1 | Am01g06240.T01 | 332 | 38077.94 | 6.37 | 43.80 | 71.30 | -0.386 | 1 | 0 | Cell wall |
| AmXTH2 | Am01g11700.T01 | 260 | 29353.74 | 4.69 | 35.11 | 69.35 | -0.338 | 0 | 1 | Cell wall |
| AmXTH3 | Am01g14580.T01 | 285 | 32755.11 | 9.27 | 38.06 | 72.53 | -0.351 | 1 | 1 | Cell wall, cytoplasmic |
| AmXTH4 | Am01g53840.T01 | 290 | 33175.18 | 5.71 | 37.36 | 64.83 | -0.420 | 0 | 1 | Cell wall |
| AmXTH5 | Am02g07210.T01 | 299 | 34771.82 | 5.28 | 34.83 | 58.70 | -0.548 | 1 | 1 | Cell wall |
| AmXTH6 | Am02g15330.T01 | 221 | 25980.24 | 8.95 | 43.60 | 61.76 | -0.759 | 0 | 0 | Cell wall |
| AmXTH7 | Am02g15340.T01 | 292 | 33675.55 | 6.13 | 24.95 | 64.38 | -0.576 | 0 | 1 | Cell wall |
| AmXTH8 | Am02g39070.T01 | 340 | 39403.82 | 8.40 | 34.62 | 74.56 | -0.463 | 1 | 1 | Cell wall |
| AmXTH9 | Am03g04070.T01 | 287 | 32230.44 | 8.56 | 34.96 | 74.46 | -0.207 | 1 | 1 | Cell wall |
| AmXTH10 | Am03g04080.T01 | 287 | 32205.26 | 9.17 | 33.76 | 68.71 | -0.290 | 0 | 1 | Cell wall |
| AmXTH11 | Am03g04460.T01 | 285 | 32026.84 | 8.21 | 31.88 | 64.04 | -0.437 | 0 | 1 | Cell wall |
| AmXTH12 | Am03g04470.T01 | 278 | 31310.26 | 8.31 | 36.83 | 71.98 | -0.216 | 0 | 1 | Cell wall |
| AmXTH13 | Am03g04480.T01 | 284 | 32223.01 | 4.82 | 36.01 | 69.75 | -0.270 | 0 | 1 | Cell wall |
| AmXTH14 | Am03g08020.T01 | 288 | 32721.81 | 6.52 | 27.02 | 65.31 | -0.416 | 0 | 1 | Cell wall, cytoplasmic |
| AmXTH15 | Am03g08030.T01 | 320 | 36873.73 | 8.63 | 30.08 | 70.41 | -0.503 | 1 | 0 | Cell wall |
| AmXTH16 | Am04g01960.T01 | 303 | 35029.58 | 6.18 | 32.05 | 63.99 | -0.456 | 1 | 1 | Cell wall |
| AmXTH17 | Am04g29790.T01 | 276 | 32119.17 | 5.00 | 47.65 | 66.81 | -0.341 | 0 | 1 | Cell wall |
| AmXTH18 | Am05g24170.T01 | 350 | 40742.43 | 9.15 | 47.11 | 73.17 | -0.491 | 0 | 0 | Cell wall |
| AmXTH19 | Am05g28140.T01 | 293 | 32825.66 | 7.08 | 40.82 | 65.60 | -0.503 | 0 | 1 | Cell wall |
| AmXTH20 | Am05g34130.T01 | 294 | 34194.77 | 8.56 | 37.68 | 72.96 | -0.397 | 1 | 1 | Cell wall |
| AmXTH21 | Am06g03100.T02 | 293 | 32816.60 | 5.12 | 40.48 | 65.97 | -0.393 | 0 | 0 | Cell wall, cytoplasmic |
| AmXTH22 | Am06g03110.T02 | 294 | 32795.53 | 5.12 | 36.12 | 67.41 | -0.404 | 1 | 1 | Cell wall, cytoplasmic |
| AmXTH23 | Am06g09100.T01 | 296 | 34193.50 | 8.22 | 48.24 | 68.21 | -0.451 | 0 | 1 | Cell wall, cytoplasmic |
| AmXTH24 | Am06g11340.T01 | 304 | 35193.12 | 5.93 | 38.12 | 55.43 | -0.663 | 0 | 1 | Cell wall |
| AmXTH25 | Am06g16890.T01 | 315 | 35592.11 | 7.14 | 51.07 | 73.59 | -0.295 | 0 | 1 | Cell wall |
| AmXTH26 | Am06g38850.T01 | 288 | 32908.11 | 5.84 | 43.84 | 71.46 | -0.362 | 1 | 1 | Cell wall |
| AmXTH27 | Am07g22440.T01 | 282 | 32198.49 | 8.70 | 39.91 | 74.65 | -0.420 | 0 | 1 | Cell wall |
| AmXTH28 | Am07g32570.T01 | 290 | 33298.41 | 8.13 | 54.79 | 62.90 | -0.449 | 0 | 1 | Cell wall |
| AmXTH29 | Am08g01110.T01 | 285 | 32630.86 | 6.38 | 35.18 | 69.12 | -0.371 | 0 | 1 | Cell wall |
| AmXTH30 | Am08g26720.T01 | 298 | 34067.63 | 9.48 | 56.25 | 66.48 | -0.351 | 1 | 1 | Cell wall |
| AmXTH31 | Am08g29760.T01 | 280 | 31431.47 | 5.93 | 39.93 | 68.64 | -0.286 | 1 | 1 | Cell wall, cytoplasmic |
| AmXTH32 | Am08g29770.T01 | 647 | 73526.80 | 6.98 | 38.61 | 88.42 | -0.058 | 1 | 0 | Cell wall, cytoplasmic |
| AmXTH33 | Am08g29790.T02 | 271 | 30595.34 | 6.41 | 31.72 | 63.32 | -0.379 | 0 | 1 | Cell wall, cytoplasmic |
表2 金鱼草XTH基因家族成员理化性质及亚细胞定位
Table 2 Physical and chemical characteristics, and subcellular localization of XTH gene family in Antirrhinum majus
| Gene name | Gene ID | aa | Mw (Da) | PI | Instability index | Aliphatic index | GRAVY | TMHs | SP | Subcellular localization |
|---|---|---|---|---|---|---|---|---|---|---|
| AmXTH1 | Am01g06240.T01 | 332 | 38077.94 | 6.37 | 43.80 | 71.30 | -0.386 | 1 | 0 | Cell wall |
| AmXTH2 | Am01g11700.T01 | 260 | 29353.74 | 4.69 | 35.11 | 69.35 | -0.338 | 0 | 1 | Cell wall |
| AmXTH3 | Am01g14580.T01 | 285 | 32755.11 | 9.27 | 38.06 | 72.53 | -0.351 | 1 | 1 | Cell wall, cytoplasmic |
| AmXTH4 | Am01g53840.T01 | 290 | 33175.18 | 5.71 | 37.36 | 64.83 | -0.420 | 0 | 1 | Cell wall |
| AmXTH5 | Am02g07210.T01 | 299 | 34771.82 | 5.28 | 34.83 | 58.70 | -0.548 | 1 | 1 | Cell wall |
| AmXTH6 | Am02g15330.T01 | 221 | 25980.24 | 8.95 | 43.60 | 61.76 | -0.759 | 0 | 0 | Cell wall |
| AmXTH7 | Am02g15340.T01 | 292 | 33675.55 | 6.13 | 24.95 | 64.38 | -0.576 | 0 | 1 | Cell wall |
| AmXTH8 | Am02g39070.T01 | 340 | 39403.82 | 8.40 | 34.62 | 74.56 | -0.463 | 1 | 1 | Cell wall |
| AmXTH9 | Am03g04070.T01 | 287 | 32230.44 | 8.56 | 34.96 | 74.46 | -0.207 | 1 | 1 | Cell wall |
| AmXTH10 | Am03g04080.T01 | 287 | 32205.26 | 9.17 | 33.76 | 68.71 | -0.290 | 0 | 1 | Cell wall |
| AmXTH11 | Am03g04460.T01 | 285 | 32026.84 | 8.21 | 31.88 | 64.04 | -0.437 | 0 | 1 | Cell wall |
| AmXTH12 | Am03g04470.T01 | 278 | 31310.26 | 8.31 | 36.83 | 71.98 | -0.216 | 0 | 1 | Cell wall |
| AmXTH13 | Am03g04480.T01 | 284 | 32223.01 | 4.82 | 36.01 | 69.75 | -0.270 | 0 | 1 | Cell wall |
| AmXTH14 | Am03g08020.T01 | 288 | 32721.81 | 6.52 | 27.02 | 65.31 | -0.416 | 0 | 1 | Cell wall, cytoplasmic |
| AmXTH15 | Am03g08030.T01 | 320 | 36873.73 | 8.63 | 30.08 | 70.41 | -0.503 | 1 | 0 | Cell wall |
| AmXTH16 | Am04g01960.T01 | 303 | 35029.58 | 6.18 | 32.05 | 63.99 | -0.456 | 1 | 1 | Cell wall |
| AmXTH17 | Am04g29790.T01 | 276 | 32119.17 | 5.00 | 47.65 | 66.81 | -0.341 | 0 | 1 | Cell wall |
| AmXTH18 | Am05g24170.T01 | 350 | 40742.43 | 9.15 | 47.11 | 73.17 | -0.491 | 0 | 0 | Cell wall |
| AmXTH19 | Am05g28140.T01 | 293 | 32825.66 | 7.08 | 40.82 | 65.60 | -0.503 | 0 | 1 | Cell wall |
| AmXTH20 | Am05g34130.T01 | 294 | 34194.77 | 8.56 | 37.68 | 72.96 | -0.397 | 1 | 1 | Cell wall |
| AmXTH21 | Am06g03100.T02 | 293 | 32816.60 | 5.12 | 40.48 | 65.97 | -0.393 | 0 | 0 | Cell wall, cytoplasmic |
| AmXTH22 | Am06g03110.T02 | 294 | 32795.53 | 5.12 | 36.12 | 67.41 | -0.404 | 1 | 1 | Cell wall, cytoplasmic |
| AmXTH23 | Am06g09100.T01 | 296 | 34193.50 | 8.22 | 48.24 | 68.21 | -0.451 | 0 | 1 | Cell wall, cytoplasmic |
| AmXTH24 | Am06g11340.T01 | 304 | 35193.12 | 5.93 | 38.12 | 55.43 | -0.663 | 0 | 1 | Cell wall |
| AmXTH25 | Am06g16890.T01 | 315 | 35592.11 | 7.14 | 51.07 | 73.59 | -0.295 | 0 | 1 | Cell wall |
| AmXTH26 | Am06g38850.T01 | 288 | 32908.11 | 5.84 | 43.84 | 71.46 | -0.362 | 1 | 1 | Cell wall |
| AmXTH27 | Am07g22440.T01 | 282 | 32198.49 | 8.70 | 39.91 | 74.65 | -0.420 | 0 | 1 | Cell wall |
| AmXTH28 | Am07g32570.T01 | 290 | 33298.41 | 8.13 | 54.79 | 62.90 | -0.449 | 0 | 1 | Cell wall |
| AmXTH29 | Am08g01110.T01 | 285 | 32630.86 | 6.38 | 35.18 | 69.12 | -0.371 | 0 | 1 | Cell wall |
| AmXTH30 | Am08g26720.T01 | 298 | 34067.63 | 9.48 | 56.25 | 66.48 | -0.351 | 1 | 1 | Cell wall |
| AmXTH31 | Am08g29760.T01 | 280 | 31431.47 | 5.93 | 39.93 | 68.64 | -0.286 | 1 | 1 | Cell wall, cytoplasmic |
| AmXTH32 | Am08g29770.T01 | 647 | 73526.80 | 6.98 | 38.61 | 88.42 | -0.058 | 1 | 0 | Cell wall, cytoplasmic |
| AmXTH33 | Am08g29790.T02 | 271 | 30595.34 | 6.41 | 31.72 | 63.32 | -0.379 | 0 | 1 | Cell wall, cytoplasmic |
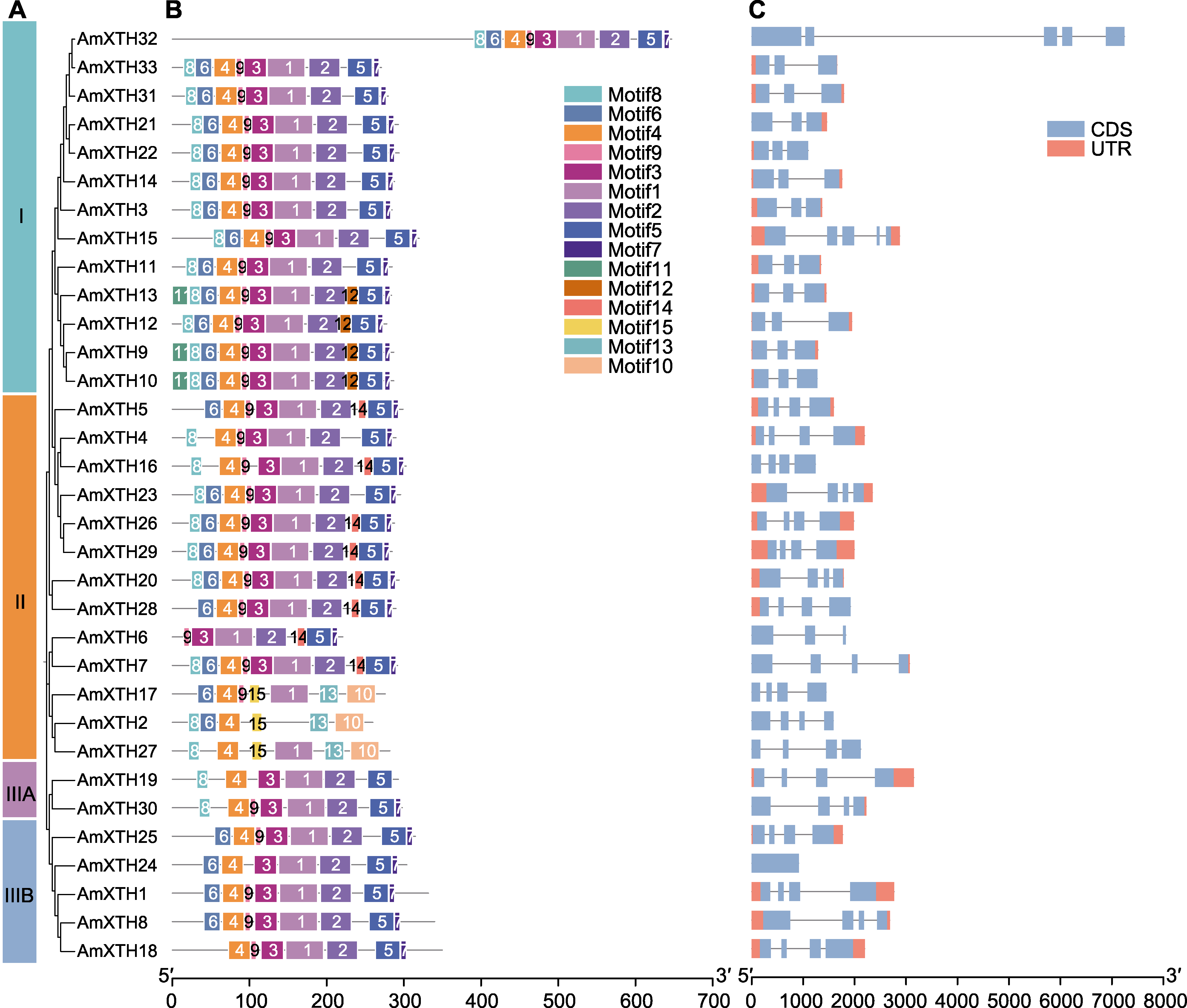
图2 金鱼草XTH家族系统进化关系(A)、保守基序(B)和基因结构(C) CDS: 编码序列; UTR: 非翻译区
Figure 2 Phylogenetic relationship (A), conserved motif (B) and gene structures (C) of XTH family in Antirrhinum majus CDS: Coding sequence; UTR: Untranslated region
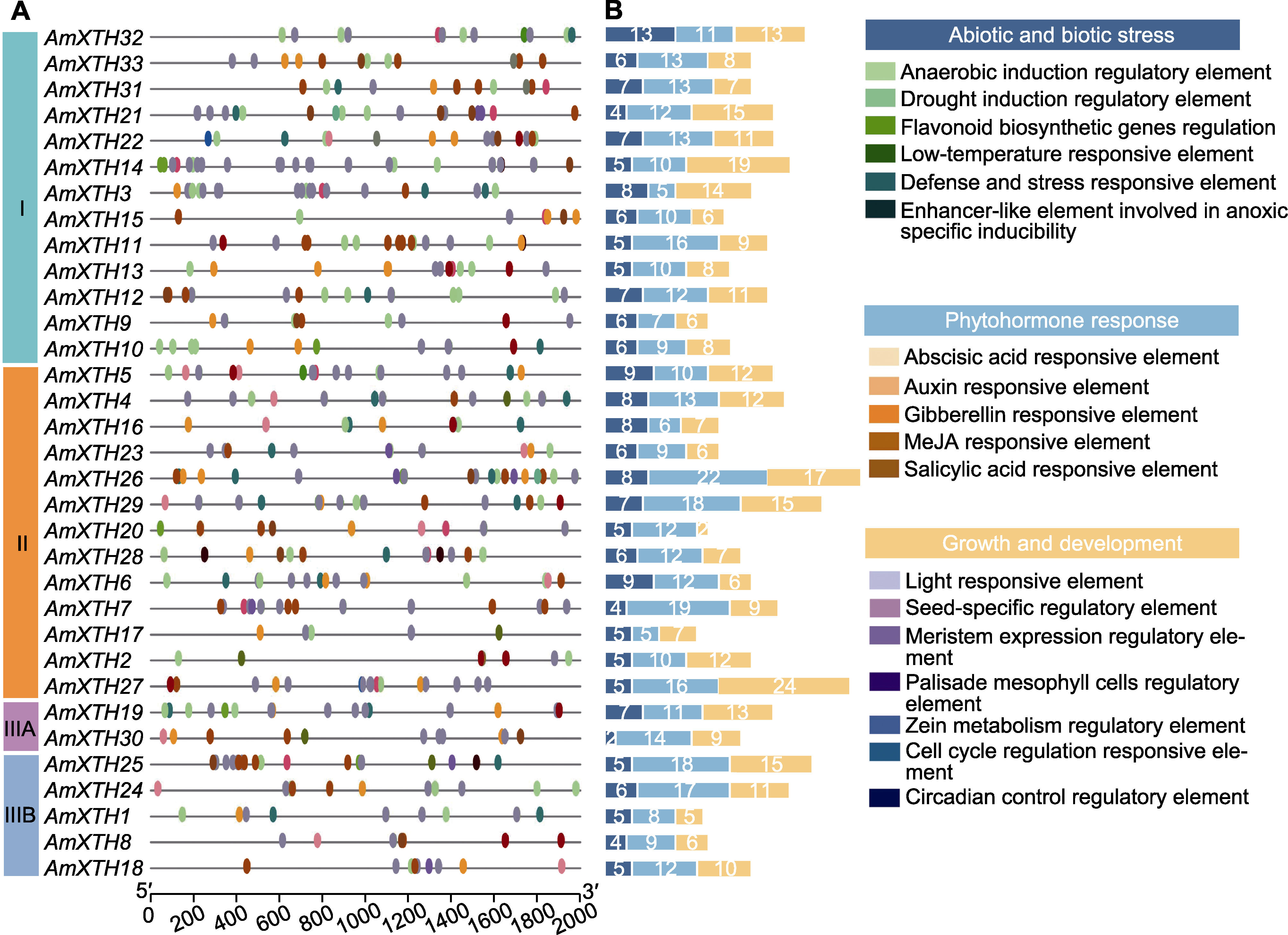
图4 金鱼草XTH家族基因顺式作用元件分析 (A) 响应元件; (B) 响应类型及响应元件数量
Figure 4 Analysis of cis-acting elements of the XTH family gene in Antirrhinum majus (A) Response elements; (B) Response type and number of response elements
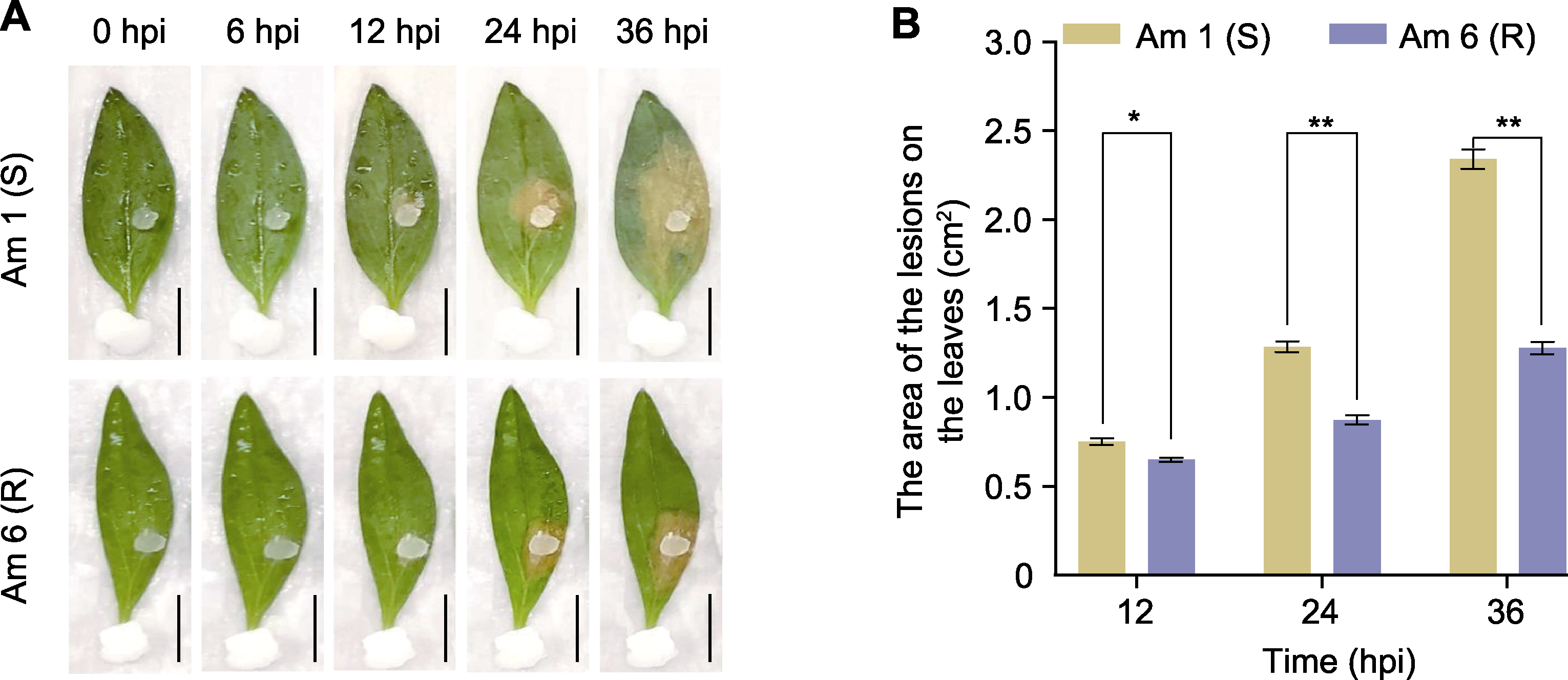
图5 金鱼草叶片离体接种核盘菌表型(A)及菌斑面积分析(B) * P<0.05; ** P<0.01; hpi: 接菌后小时数。Bars=1 cm
Figure 5 Phenotype (A) and the area of the lesions on Antirrhinum majus leaves (B) in vitro inoculation of Sclerotinia sclerotiorum * P<0.05; ** P<0.01; hpi: Hours post-inoculation. Bars=1 cm
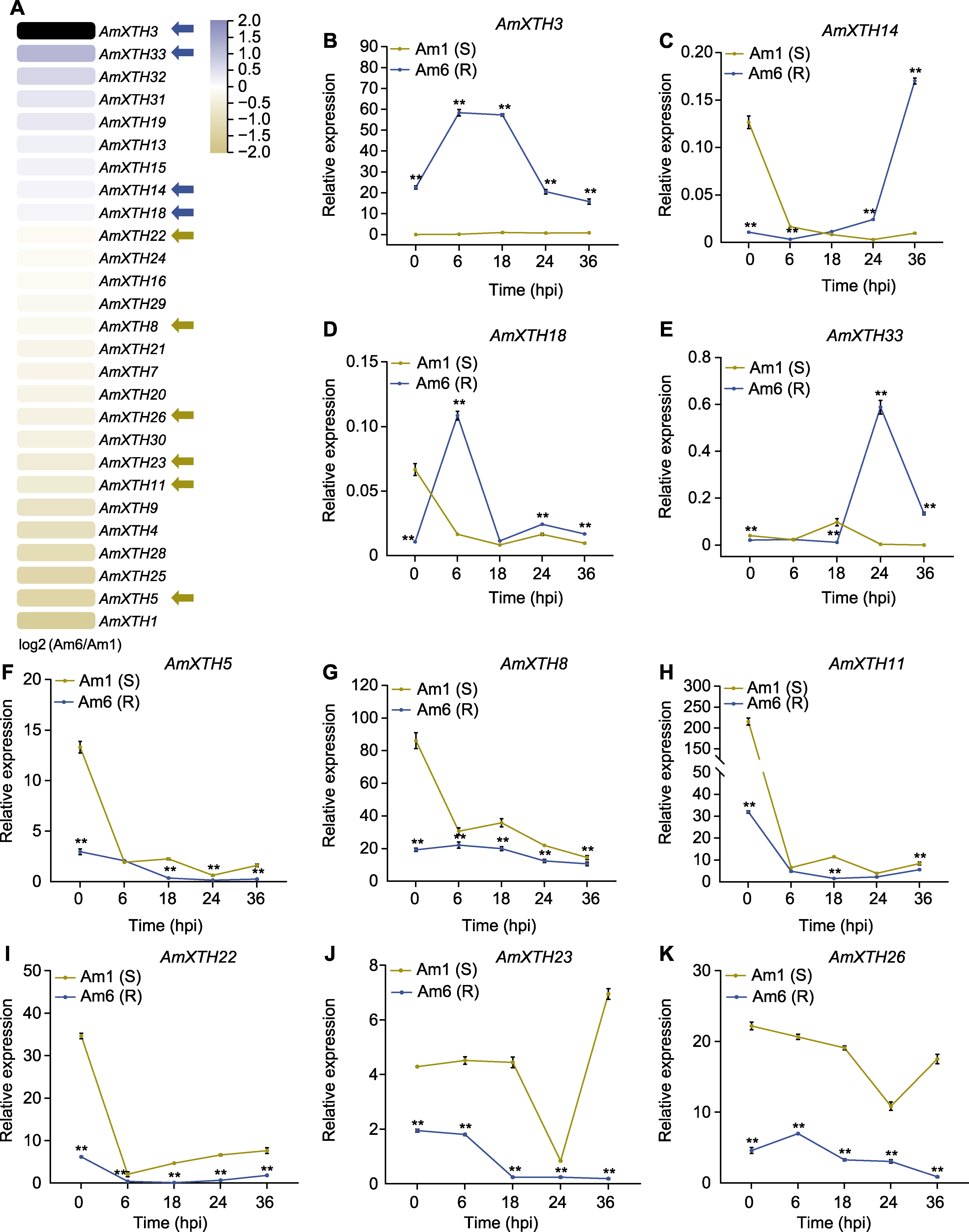
图6 RNA-seq分析差异表达AmXTH基因(A)和qRT-PCR验证(B)-(K) 蓝色箭头指示在抗病材料中表达量显著高于感病材料的基因, 黄色箭头指示在抗病材料中表达量显著低于感病材料的基因。* P<0.05; ** P<0.01; hpi: 接菌后小时数
Figure 6 Differentially expressed AmXTH genes in RNA-seq analysis (A) and qRT-PCR verification (B)-(K) Blue arrows indicate genes expression significant higher in resistant material than in susceptible material, and yellow arrows indicate genes expression significant lower in resistant material than in susceptible material. * P<0.05; ** P<0.01; hpi: Hours post-inoculation
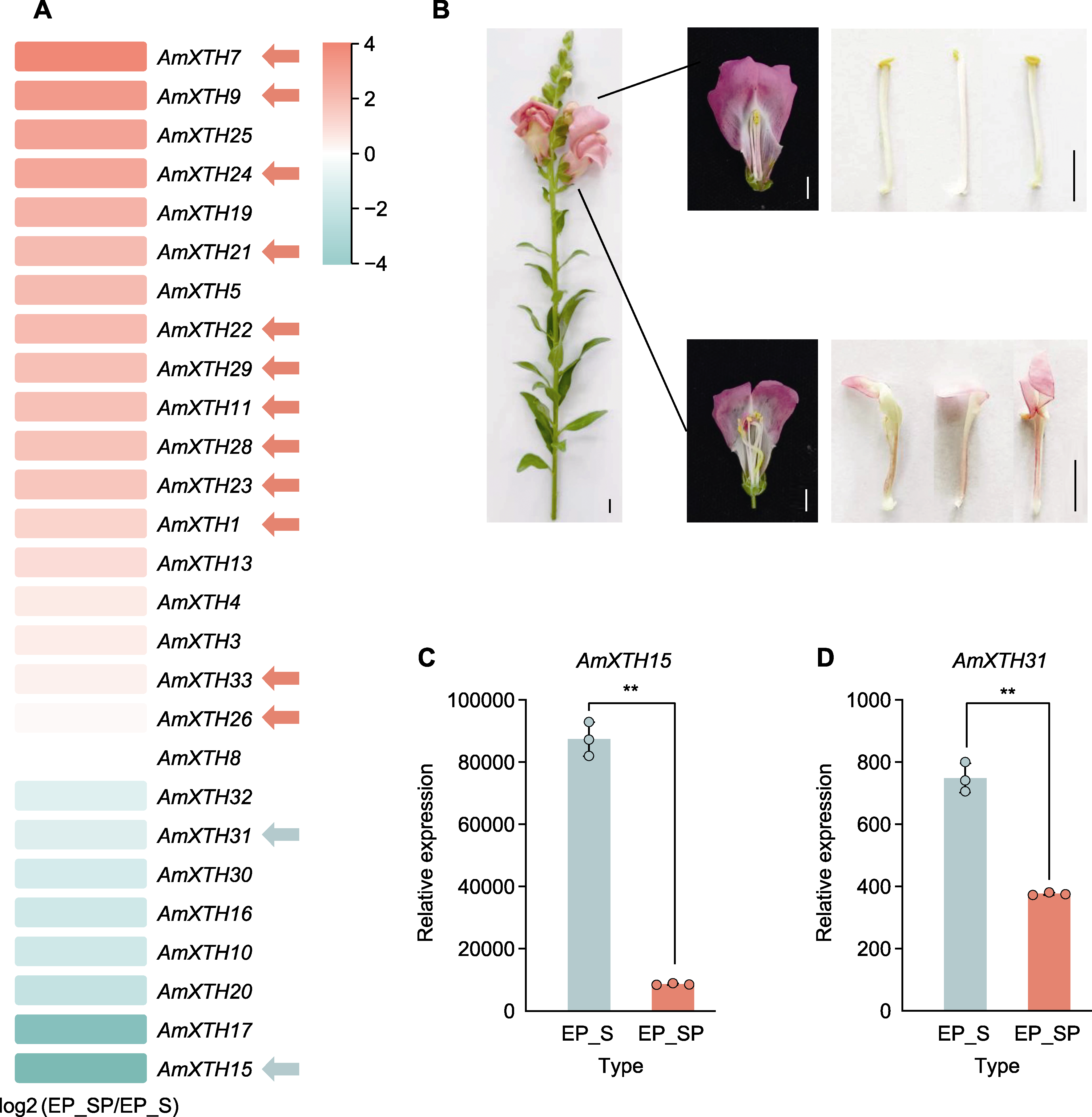
图7 RNA-seq分析差异表达AmXTH基因(A)、雄蕊瓣化表型观察(B)和qRT-PCR验证介导雄蕊瓣化的AmXTH基因(C), (D) 红色箭头指示在瓣化雄蕊中表达量显著高于正常雄蕊的基因, 绿色箭头指示在瓣化雄蕊中表达量显著低于正常雄蕊的基因。** P<0.01。Bars=1 cm
Figure 7 Differentially expressed AmXTH genes in RNA-seq analysis (A), phenotype observation of stamen petalization (B) and qRT-PCR verification of AmXTH genes involved in stamen petalization (C), (D) Red arrows indicate genes expression significant higher in petalized stamen than in normal stamen, and green arrows indicate genes expression significant lower in petalized stamen than in normal stamen. ** P<0.01。Bars=1 cm
| [1] | Cardarelli M, Cecchetti V (2014). Auxin polar transport in stamen formation and development: how many actors? Front Plant Sci 5, 333. |
| [2] |
Claverie J, Balacey S, Lemaître-Guillier C, Brulé D, Chiltz A, Granet L, Noirot E, Daire X, Darblade B, Héloir MC, Poinssot B (2018). The cell wall-derived xyloglucan is a new DAMP triggering plant immunity in Vitis vinifera and Arabidopsis thaliana. Front Plant Sci 9, 1725.
DOI PMID |
| [3] | de Azevedo Souza C, Li SD, Lin AZ, Boutrot F, Grossmann G, Zipfel C, Somerville SC (2017). Cellulose-derived oligomers act as damage-associated molecular patterns and trigger defense-like responses. Plant Physiol 173, 2383-2398. |
| [4] |
Divol F, Vilaine F, Thibivilliers S, Kusiak C, Sauge MH, Dinant S (2007). Involvement of the xyloglucan endotransglycosylase/hydrolases encoded by celery XTH1 and Arabidopsis XTH33 in the phloem response to aphids. Plant Cell Environ 30, 187-201.
DOI PMID |
| [5] | Fan TG (2014). Action of Excessive Petal Formation Caused by Rose AGAMOUS Gene Related to Floral Organ Development Under Low Temperature. Master’s thesis. Baoding: Hebei Agricultural University. pp. 9-48. (in Chinese) |
| 范天刚 (2014). 月季花器官发育基因AGAMOUS对低温导致花朵过度重瓣化的作用研究. 硕士论文. 保定: 河北农业大学. pp. 9-48. | |
| [6] | Guo XM, Stotz HU (2007). Defense against Sclerotinia sclerotiorum in Arabidopsis is dependent on jasmonic acid, salicylic acid, and ethylene signaling. Mol Plant Mirobe Interact 20, 1384-1395. |
| [7] |
Han Y, Ban QY, Hou YL, Meng K, Suo JT, Rao JP (2016). Isolation and characterization of two persimmon xyloglucan endotransglycosylase/hydrolase (XTH) genes that have divergent functions in cell wall modification and fruit postharvest softening. Front Plant Sci 7, 624.
DOI PMID |
| [8] |
Harada T, Torii Y, Morita S, Onodera R, Hara Y, Yokoyama R, Nishitani K, Satoh S (2011). Cloning, characterization, and expression of xyloglucan endotransglucosylase/hydrolase and expansion genes associated with petal growth and development during carnation flower opening. J Exp Bot 62, 815-823.
DOI PMID |
| [9] |
Hu HZ, Tang YW, Wu J, Chen FZ, Yang YD, Pan XC, Dong X, Jin XD, Liu S, Du XZ (2021). Brassica napus mediator subunit16 induces BnMED25- and BnWRKY33-activated defense signaling to confer Sclerotinia sclerotiorum resistance. Front Plant Sci 12, 663536.
DOI URL |
| [10] |
Hu HZ, Zhang R, Feng SQ, Wang YM, Wang YT, Fan CF, Li Y, Liu ZY, Schneider R, Xia T, Ding SY, Persson S, Peng LC (2018). Three AtCesA6-like members enhance biomass production by distinctively promoting cell growth in Arabidopsis. Plant Biotechnol J 16, 976-988.
DOI PMID |
| [11] | Huang X, Tian DK, Zhang WW, Zeng SJ, Mo HB (2014). Comparison of floral organ morphological development between single and double flowers in Nelumbo nucifera. Plant Diver Resour 36, 303-309. (in Chinese) |
| 黄秀, 田代科, 张微微, 曾宋君, 莫海波 (2014). 荷花“重瓣化”的花器官形态发育比较观察. 植物分类与资源学报 36, 303-309. | |
| [12] |
Hyodo H, Yamakawa S, Takeda Y, Tsuduki M, Yokota A, Nishitani K, Kohchi T (2003). Active gene expression of a xyloglucan endotransglucosylase/hydrolase gene, XTH9, in inflorescence apices is related to cell elongation in Arabidopsis thaliana. Plant Mol Biol 52, 473-482.
DOI PMID |
| [13] | Jia XL, He BX, Guo DD, Guo ML (2018). Research progress in the function of expansins and xyloglucan endotransglucosylase/hydrolase. Plant Physiol J 54, 1659-1668. (in Chinese) |
| 贾鑫磊, 何贝轩, 郭丹丹, 郭美丽 (2018). 膨胀素和木葡聚糖内转葡糖基酶/水解酶基因的功能研究进展. 植物生理学报 54, 1659-1668. | |
| [14] | Jiang JF (2022). Bioinformatics Analysis of Ginseng XTH Family Genes and Function Analysis of Two PgXTH Genes. Master’s thesis. Changchun: Jilin University. pp. 1-69. (in Chinese) |
| 江俊峰 (2022). 人参XTH家族基因生物信息学分析及两个PgXTH基因的功能分析. 硕士论文. 长春: 吉林大学. pp. 1-69. | |
| [15] |
Li MM, Zhang DF, Gao Q, Luo YF, Zhang H, Ma B, Chen CH, Whibley A, Zhang Y, Cao YH, Li Q, Guo H, Li JH, Song YZ, Zhang Y, Copsey L, Li Y, Li XX, Qi M, Wang JW, Chen Y, Wang D, Zhao JY, Liu GC, Wu B, Yu LL, Xu CY, Li J, Zhao SC, Zhang YJ, Hu SN, Liang CZ, Yin Y, Coen E, Xue YB (2019a). Genome structure and evolution of Antirrhinum majus L. Nat Plants 5, 174-183.
DOI |
| [16] |
Li Q, Hu AH, Dou WF, Qi JJ, Long Q, Zou XP, Lei TG, Yao LX, He YR, Chen SC (2019b). Systematic analysis and functional validation of citrus XTH genes reveal the role of Csxth04 in citrus bacterial canker resistance and tolerance. Front Plant Sci 10, 1109.
DOI URL |
| [17] |
Livak KJ, Schmittgen TD (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25, 402-408.
DOI PMID |
| [18] | Luo WP, Chen SP, Gong YX (2008). Breeding and pest control of Antirrhinum majus. Chin Flower Horticul (8), 23-25. (in Chinese) |
| 罗维平, 陈少萍, 龚衍熙 (2008). 金鱼草的繁殖与病虫害防治. 中国花卉园艺 (8), 23-25. | |
| [19] |
Malinowski R, Fry SC, Zuzga S, Wiśniewska A, Godlewski M, Noyszewski A, Barczak-Brzyżek A, Malepszy S, Filipecki M (2018). Developmental expression of the cucumber Cs-XTH1 and Cs-XTH3 genes, encoding xyloglucan endotransglucosylase/hydrolases, can be influenced by mechanical stimuli. Acta Physiol Plant 40, 130.
DOI |
| [20] |
Marciniak K, Przedniczek K (2019). Comprehensive insight into gibberellin- and jasmonate-mediated stamen development. Genes 10, 811.
DOI URL |
| [21] | Miedes E, Suslov D, Vandenbussche F, Kenobi K, Ivakov A, Van Der straeten D, Lorences EP, Mellerowicz EJ, Verbelen JP, Vissenberg K (2013). Xyloglucan endotransglucosylase/hydrolase (XTH) overexpression affects growth and cell wall mechanics in etiolated Arabidopsis hypocotyls. J Exp Bot 64, 2481-2497. |
| [22] |
Pitaksaringkarn W, Matsuoka K, Asahina M, Miura K, Sage-Ono K, Ono M, Yokoyama R, Nishitani K, Ishii T, Iwai H, Satoh S (2014). XTH20 and XTH19 regulated by ANAC071 under auxin flow are involved in cell proliferation in incised Arabidopsis inflorescence stems. Plant J 80, 604-614.
DOI URL |
| [23] | Sasidharan R, Pierik R (2010). Cell wall modification involving XTHs controls phytochrome-mediated petiole elongation in Arabidopsis thaliana. Plant Signal Behav 5, 1491-1492. |
| [24] | Shi YY, Li DY, Zhang HJ, Song FM (2011). Cell wall-mediated disease resistance and its molecular mechanism in plants. Plant Physiol J 47, 661-668. (in Chinese) |
| 师莹莹, 李大勇, 张慧娟, 宋凤鸣 (2011). 植物细胞壁介导的抗病性及其分子机制. 植物生理学报 47, 661-668. | |
| [25] | Sun R, Yang YL, Li YJ, Zhang H, Li XK (2023). Genome- wide identification and analysis of PLATZ transcription factor gene family in foxtail millet. Chin Bull Bot 58, 548-559. (in Chinese) |
| 孙蓉, 杨宇琭, 李亚军, 张会, 李旭凯 (2023). 谷子PLATZ转录因子基因家族的鉴定和分析. 植物学报 58, 548-559. | |
| [26] |
Takahashi D, Johnson KL, Hao PF, Tuong T, Erban A, Sampathkumar A, Bacic A, Livingston DP, Kopka J, Kuroha T, Yokoyama R, Nishitani K, Zuther E, Hincha DK (2021). Cell wall modification by the xyloglucan endotransglucosylase/hydrolase XTH19 influences freezing tolerance after cold and sub-zero acclimation. Plant Cell Environ 44, 915-930.
DOI URL |
| [27] | Tian YR, Fan TG, Zhang G, Li YH (2016). Expression and analysis of key genes of excessive double flowers in rose caused by low temperature. Chin J Tropical Crops 37, 1147-1154. (in Chinese) |
| 田亚然, 范天刚, 张钢, 李永红 (2016). 低温引起月季花朵过度重瓣化关键基因的表达及分析. 热带作物学报 37, 1147-1154. | |
| [28] | Wang RS (2019). Cloning and Functional Analysis of the AmDEFH28 Gene of Antirrhinum majus. Master’s thesis. Hefei: Anhui Agricultural University.pp:1-67. (in Chinese) |
| 王瑞生 (2019). 金鱼草AmDEFH28基因克隆和功能分析. 硕士论文. 合肥: 安徽农业大学. pp. 1-67. | |
| [29] |
Watanabe Y, Niki T, Norikoshi R, Nakano M, Ichimura K (2022). Soluble carbohydrate concentration and expression of expansin and xyloglucan endotransglucosylase/ hydrolase genes in epidermal and parenchyma cells during lily flower opening. J Plant Physiol 270, 153615.
DOI URL |
| [30] |
Witasari LD, Huang FC, Hoffmann T, Rozhon W, Fry SC, Schwab W (2019). Higher expression of the strawberry xyloglucan endotransglucosylase/hydrolase genes FvXTH9 and FvXTH6 accelerates fruit ripening. Plant J 100, 1237-1253.
DOI |
| [31] | Wu D, Liu AQ, Qu XY, Liang JY, Song M (2020). Genome- wide identification, and phylogenetic and expression profiling analyses of XTH gene families in Brassica rapa L. and Brassica oleracea L. BMC Genomics 21, 782. |
| [32] |
Wu J, Zhou YM, Wang YP (2018). Research progress on molecular mechanisms of Brassica napus-Sclerotinia sclerotiorum interaction. Chin J Oil Crop Sci 40, 721-729. (in Chinese)
DOI |
|
吴健, 周永明, 王幼平 (2018). 油菜与核盘菌互作分子机理研究进展. 中国油料作物学报 40, 721-729.
DOI |
|
| [33] | Wu N, Qin L, Cui K, Li HO, Liu ZS, Xia ST (2023). Cloning of Brassica napus EXA1 gene and its regulation on plant disease resistance. Chin Bull Bot 58, 385-393. (in Chinese) |
| 吴楠, 覃磊, 崔看, 李海鸥, 刘忠松, 夏石头 (2023). 甘蓝型油菜EXA1的克隆及其对植物抗病的调控作用. 植物学报 58, 385-393. | |
| [34] |
Xu PP, Fang S, Chen HY, Cai WM (2020). The brassinosteroid-responsive xyloglucan endotransglucosylase/hydrolase 19 (XTH19) and XTH23 genes are involved in lateral root development under salt stress in Arabidopsis. Plant J 104, 59-75.
DOI URL |
| [35] | Xu TS (2021). Cell Wall Components Changes in Red Skin Ginseng and Function Analysis of Two Non-typical XTH Family Genes. Master’s thesis. Changchun: Jilin University. pp. 1-59. (in Chinese) |
| 徐天舒 (2021). 红皮病人参细胞壁组分变化及两个非典型XTH家族基因的功能探究. 硕士论文. 长春: 吉林大学. pp. 1-59. | |
| [36] | Xu XW (2023). Identification and Functional Analysis of Broad-spectrum Resistance Protein Induced by P1 Protein of Sugarcane Streak Mosaic Virus. Master’s thesis. Yangzhou: Yangzhou University. pp. 6-79. (in Chinese) |
| 徐小伟 (2023). 甘蔗线条花叶病毒P1蛋白诱导的广谱抗病蛋白的鉴定及功能研究. 硕士论文. 扬州: 扬州大学. pp. 6-79. | |
| [37] | Xuan Y (2020). Excavation of the Key Genes of Xyloglucan Metabolic Pathway and Primary Funcational Analysis of MtXTH 3 in Medicago truncatula Under Environmental Stresses. Doctoral dissertation. Nanjing: Nanjing Agricultural University. pp. 1-151. (in Chinese) |
| 宣云 (2020). 响应环境胁迫的蒺藜苜蓿XG代谢途径关键基因的挖掘及MtXTH3功能初步研究. 博士论文. 南京: 南京农业大学. pp. 1-151. | |
| [38] | Yan J, Liu YQ, Hou SW (2018). Recent advances in disease resistance proteins in plant immunity. Chin Bull Bot 53, 250-263. (in Chinese) |
|
闫佳, 刘雅琼, 侯岁稳 (2018). 植物抗病蛋白研究进展. 植物学报 53, 250-263.
DOI |
|
| [39] |
Yang Y, Miao YF, Zhong SW, Fang Q, Wang YG, Dong B, Zhao HB (2022). Genome-wide identification and expression analysis of XTH gene family during floweropening stages in Osmanthus fragrans. Plants 11, 1015.
DOI URL |
| [40] |
Zhang R, Hu Z, Wang YT, Hu HZ, Li FC, Li M, Ragauskas A, Xia T, Han HY, Tang JF, Yu HZ, Xu BQ, Peng LC (2023). Single-molecular insights into the breakpoint of cellulose nanofibers assembly during saccharification. Nat Commun 14, 1100.
DOI PMID |
| [41] | Zhao XH, Wang QJ, Li C, Chen XD, Xiao W, Gao DS, Fu XL (2018). Genome-wide identification of ethylene responsive factor (ERF) family genes in peach and screening of genes related to germination. Chin Bull Bot 53, 612-624. (in Chinese) |
|
赵雪惠, 王庆杰, 李晨, 陈修德, 肖伟, 高东升, 付喜玲 (2018). 桃ERF转录因子家族生物信息学分析及芽萌发相关基因筛选. 植物学报 53, 612-624.
DOI |
| No related articles found! |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||