

植物学报 ›› 2019, Vol. 54 ›› Issue (4): 474-485.DOI: 10.11983/CBB18200 cstr: 32102.14.CBB18200
王小龙,刘凤之,史祥宾,王孝娣,冀晓昊,王志强,王宝亮,郑晓翠,王海波( )
)
收稿日期:2018-09-19
接受日期:2019-01-15
出版日期:2019-07-01
发布日期:2020-01-08
通讯作者:
王海波
基金资助:
Xiaolong Wang,Fengzhi Liu,Xiangbin Shi,Xiaodi Wang,Xiaohao Ji,Zhiqiang Wang,Baoliang Wang,Xiaocui Zheng,Haibo Wang( )
)
Received:2018-09-19
Accepted:2019-01-15
Online:2019-07-01
Published:2020-01-08
Contact:
Haibo Wang
摘要: 9-顺式-环氧类胡萝卜素双加氧酶(NCED)是植物体内ABA生物合成的关键限速酶, 参与植物对干旱、外源ABA和高盐的响应过程, 降低环境胁迫对植株的危害。基于全基因组鉴定分析葡萄(Vitis vinifera) NCED基因家族成员, 探讨各成员的物种进化关系及各个基因成员在不同组织中的时空表达模式及对干旱、ABA和高盐(NaCl)胁迫的响应, 为进一步揭示该基因家族成员的生物学功能奠定基础。在葡萄基因组中共发现12个NCED基因。其推测的编码蛋白质长度在510 (VvNCED2)-625 aa (VvNCED10)之间。VvNCED蛋白的分子量最大值是70.53 kDa (VvNCED10), 最小值是57.85 kDa (VvNCED2)。在从祖先基因分化之后, 葡萄NCED基因发生了5次复制事件, 同时有2次丢失事件。NCED1/2、NCED3/4、NCED6/7和NCED9/10基因对被认为是通过片段复制产生。上述4对复制基因复制时间分布在3.08-120.0百万年前, 晚于单双子叶植物分化的时间。与对照相比, VvNCED1在ABA处理48小时后显著上调(72.1%), 而VvNCED2显著下调(84.0%)。VvNCED6只在干旱处理14、21和28天的根系中表达量高于对照, 分别为对照的2.49、1.05和1.09倍。VvNCED7只在干旱处理14天的根系中表达量高于对照, 为对照的1.07倍。在ABA处理72小时后, VvNCED3表达量较对照显著下调(59.5%), 而VvNCED4较对照显著上调(169.9%)。VvNCED3/VvNCED4分别在NaCl处理24和48小时出现显著性峰值, 较对照分别上调219.2%和114.4%。保守结构域不同组成和不同胁迫处理下差异表达模式是NCED蛋白发生功能分化的基础。推测NCED在进化过程中发生的功能分化有利于复制事件的发生。
王小龙,刘凤之,史祥宾,王孝娣,冀晓昊,王志强,王宝亮,郑晓翠,王海波. 葡萄NCED基因家族进化及表达分析. 植物学报, 2019, 54(4): 474-485.
Xiaolong Wang,Fengzhi Liu,Xiangbin Shi,Xiaodi Wang,Xiaohao Ji,Zhiqiang Wang,Baoliang Wang,Xiaocui Zheng,Haibo Wang. Evolution and Expression of NCED Family Genes in Vitis vinifera. Chinese Bulletin of Botany, 2019, 54(4): 474-485.
| Gene name | Forward primer (5'-3') | Reverse primer (5'-3') |
|---|---|---|
| VvNCED1 | GCTGGAGAAGCTGATAGTGAAG | GAAGATACCCAATGACCGGAAG |
| VvNCED2 | GGCACTTTCGGAGGTTGATAA | TGGATGAGCAGTGAAGGAATG |
| VvNCED3 | CGGTGGAGATGGTGAGAATAGA | CACTGCTGCGTACACGTATTT |
| VvNCED4 | CTCAGCAGTAGGTGATCCTTTG | CAGGCTCGTACATTCTCTTAGC |
| VvNCED6 | CTCGTGATTTGGGCTCTTTCT | GCTTGATGATGTGTGCTTTGG |
| VvNCED7 | CGCTCTTCTTCTTCCTCACTAC | GGCGTTCCCTCTTCTACTATTG |
| VvNCED9 | CCATGGACTTCCCGATGATAAA | ATCCCACAACTAGAGCTTGC |
| VvNCED10 | CAGGGAGGTGTTGAAGAAGATG | CCCTTTGAGGCAGTGTGATT |
| VvActin | TACAATTCCATCATGAAGTGTGATG | TTAGAAGCACTTCCTGTGAACAATG |
表1 RT-PCR分析所用引物序列
Table 1 The primer sequences used for quantitative RT-PCR
| Gene name | Forward primer (5'-3') | Reverse primer (5'-3') |
|---|---|---|
| VvNCED1 | GCTGGAGAAGCTGATAGTGAAG | GAAGATACCCAATGACCGGAAG |
| VvNCED2 | GGCACTTTCGGAGGTTGATAA | TGGATGAGCAGTGAAGGAATG |
| VvNCED3 | CGGTGGAGATGGTGAGAATAGA | CACTGCTGCGTACACGTATTT |
| VvNCED4 | CTCAGCAGTAGGTGATCCTTTG | CAGGCTCGTACATTCTCTTAGC |
| VvNCED6 | CTCGTGATTTGGGCTCTTTCT | GCTTGATGATGTGTGCTTTGG |
| VvNCED7 | CGCTCTTCTTCTTCCTCACTAC | GGCGTTCCCTCTTCTACTATTG |
| VvNCED9 | CCATGGACTTCCCGATGATAAA | ATCCCACAACTAGAGCTTGC |
| VvNCED10 | CAGGGAGGTGTTGAAGAAGATG | CCCTTTGAGGCAGTGTGATT |
| VvActin | TACAATTCCATCATGAAGTGTGATG | TTAGAAGCACTTCCTGTGAACAATG |
| Gene name | Accession No. | Chromosome location (start, end) | Length of amino acids (aa) | Molecular weight (kDa) | Theoretical pI | GRAVY |
|---|---|---|---|---|---|---|
| VvNCED1 | VIT_213s0064g00840.1 | Chr.13 (22672994, 22681910) | 546 | 61.63 | 6.13 | -0.271 |
| VvNCED2 | VIT_213s0064g00810.1 | Chr.13 (22587965, 22596719) | 510 | 57.85 | 6.23 | -0.250 |
| VvNCED3 | VIT_202s0087g00910.1 | Chr.2 (18560696, 18562591) | 599 | 65.90 | 6.88 | -0.236 |
| VvNCED4 | VIT_202s0087g00930.1 | Chr.2 (18588853, 18590786) | 589 | 65.61 | 6.63 | -0.187 |
| VvNCED5 | VIT_216s0039g01370.1 | Chr.16 (789473, 791221) | 558 | 62.09 | 5.57 | -0.197 |
| VvNCED6 | VIT_219s0093g00550.1 | Chr.19 (17645348, 17647649) | 609 | 67.13 | 6.38 | -0.317 |
| VvNCED7 | VIT_210s0003g03750.1 | Chr.10 (6374432, 6376728) | 605 | 67.34 | 6.36 | -0.365 |
| VvNCED8 | VIT_205s0051g00670.1 | Chr.5 (11589343, 11591102) | 575 | 63.14 | 8.24 | -0.202 |
| VvNCED9 | VIT_204s0008g03510.1 | Chr.4 (2883265, 2886523) | 567 | 63.72 | 5.73 | -0.335 |
| VvNCED10 | VIT_204s0008g03480.1 | Chr.4 (2873553, 2878309) | 625 | 70.53 | 6.43 | -0.305 |
| VvNCED11 | VIT_204s0008g03380.1 | Chr.4 (2784465, 2788790) | 563 | 62.34 | 7.27 | -0.339 |
| VvNCED12 | VIT_215s0021g02190.1 | Chr.15 (13131078, 13135539) | 610 | 68.46 | 7.31 | -0.313 |
表2 葡萄候选NCED基因及其详细信息
Table 2 NCED genes identified in grapevine and their detailed information
| Gene name | Accession No. | Chromosome location (start, end) | Length of amino acids (aa) | Molecular weight (kDa) | Theoretical pI | GRAVY |
|---|---|---|---|---|---|---|
| VvNCED1 | VIT_213s0064g00840.1 | Chr.13 (22672994, 22681910) | 546 | 61.63 | 6.13 | -0.271 |
| VvNCED2 | VIT_213s0064g00810.1 | Chr.13 (22587965, 22596719) | 510 | 57.85 | 6.23 | -0.250 |
| VvNCED3 | VIT_202s0087g00910.1 | Chr.2 (18560696, 18562591) | 599 | 65.90 | 6.88 | -0.236 |
| VvNCED4 | VIT_202s0087g00930.1 | Chr.2 (18588853, 18590786) | 589 | 65.61 | 6.63 | -0.187 |
| VvNCED5 | VIT_216s0039g01370.1 | Chr.16 (789473, 791221) | 558 | 62.09 | 5.57 | -0.197 |
| VvNCED6 | VIT_219s0093g00550.1 | Chr.19 (17645348, 17647649) | 609 | 67.13 | 6.38 | -0.317 |
| VvNCED7 | VIT_210s0003g03750.1 | Chr.10 (6374432, 6376728) | 605 | 67.34 | 6.36 | -0.365 |
| VvNCED8 | VIT_205s0051g00670.1 | Chr.5 (11589343, 11591102) | 575 | 63.14 | 8.24 | -0.202 |
| VvNCED9 | VIT_204s0008g03510.1 | Chr.4 (2883265, 2886523) | 567 | 63.72 | 5.73 | -0.335 |
| VvNCED10 | VIT_204s0008g03480.1 | Chr.4 (2873553, 2878309) | 625 | 70.53 | 6.43 | -0.305 |
| VvNCED11 | VIT_204s0008g03380.1 | Chr.4 (2784465, 2788790) | 563 | 62.34 | 7.27 | -0.339 |
| VvNCED12 | VIT_215s0021g02190.1 | Chr.15 (13131078, 13135539) | 610 | 68.46 | 7.31 | -0.313 |
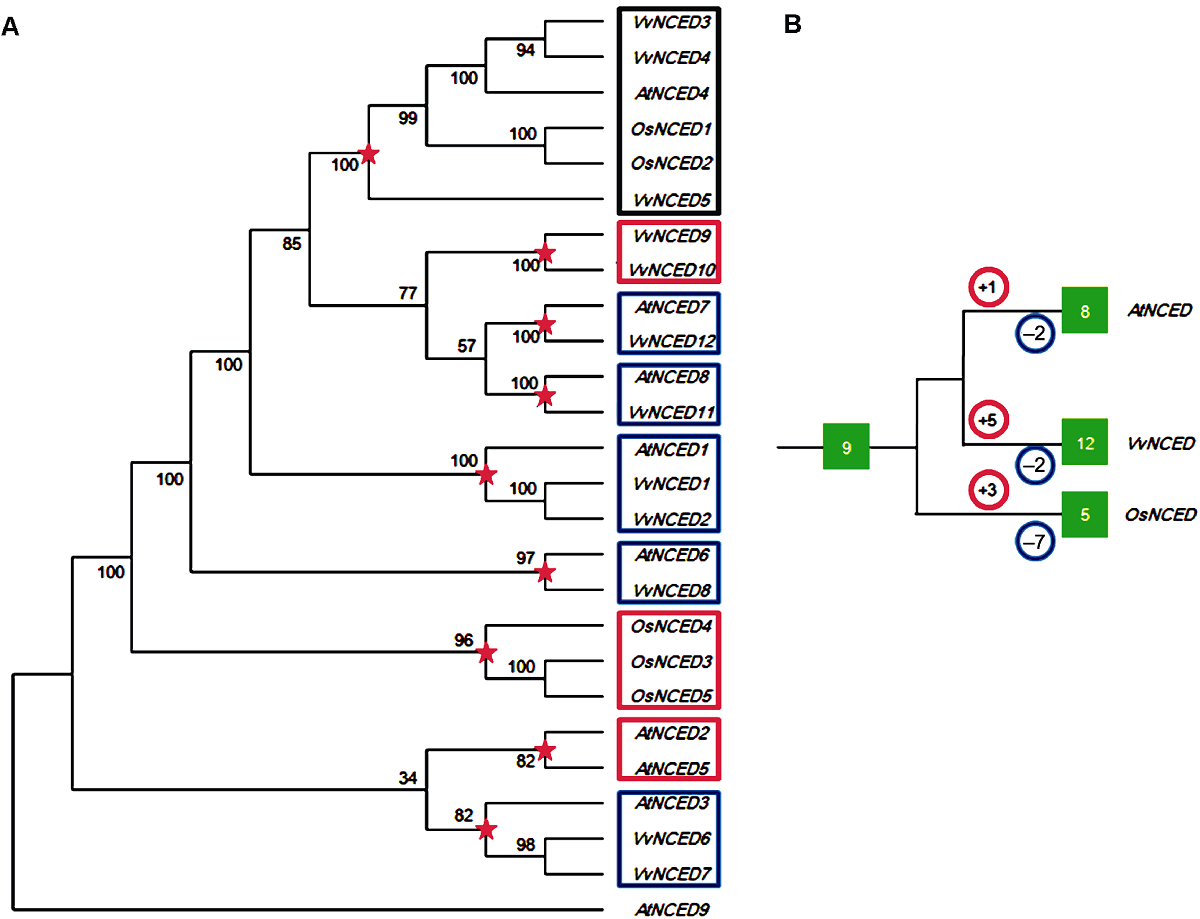
图1 葡萄、拟南芥和水稻NCED基因系统发育树(A)和扩增模式(B) 图中红色五角星代表物种分化前最近的共同祖先分化节点。红色和蓝色圆圈内的数字分别代表该物种在分化节点后发生的复制和丢失事件次数。绿色实心方框内的数字代表NCED基因的个数。
Figure 1 Joint phylogenetic tree (A) and amplification model (B) of NCED gene from grape, Arabidopsis and rice The red five-pointed stars in the figure represent the most recent common ancestral differentiation node before species differentiation. The numbers in the red and blue circles represent the number of duplication and loss events that occur after the species differentiated nodes, respectively. The numbers in the green solid box represent the number of NCED genes.
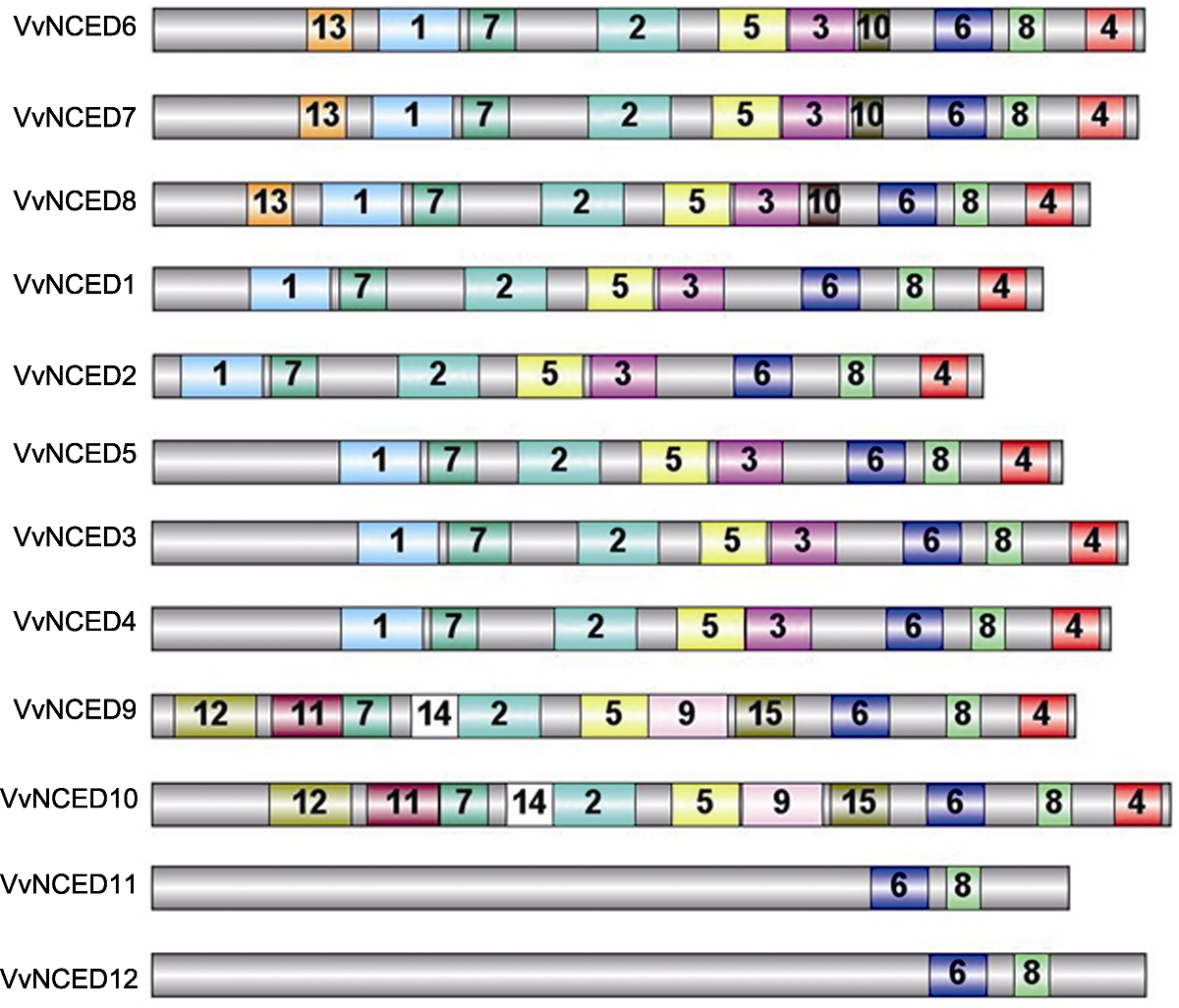
图3 葡萄NCED蛋白保守结构域示意图 灰色条形框代表NCED蛋白全长, 其它彩色框中的数字是NCED蛋白保守结构域的随机编号。相同的数字代表相同的结构域。
Figure 3 Schematic structure of conserved motifs identified in grapevine NCED proteins The grey bars represent the full length of NCED proteins, and the numbers in the other colored boxes are random numbers for the conserved domains located at NCED protein. The same number represents the same conserved domain.
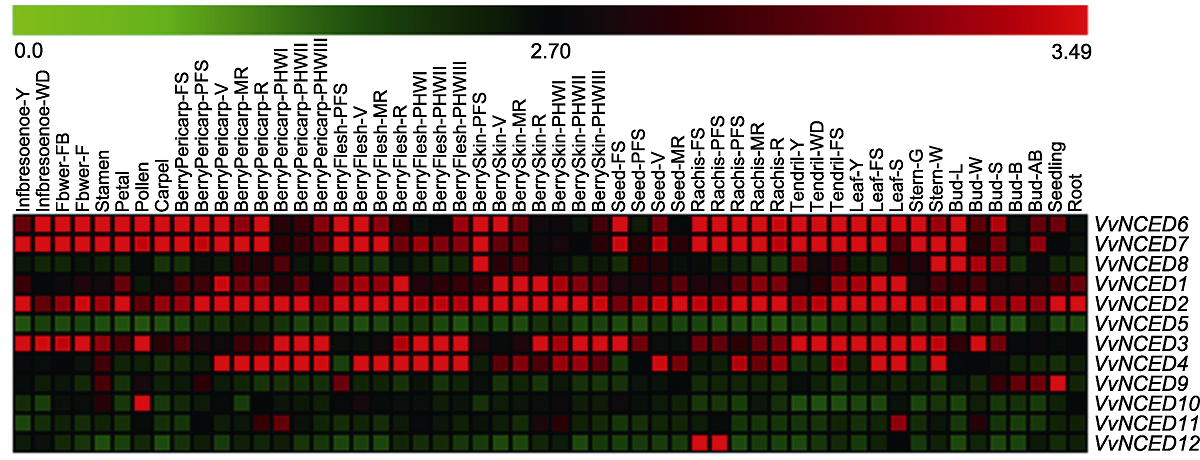
图4 NCED基因在葡萄不同发育阶段和组织中的表达模式 54种组织名称和基因名称分别位于热图的上方和右侧。热图上方比例尺表示基因表达量0.0-3.49。
Figure 4 Expression pattern of NCED genes at different developmental stages and in some specialized tissues of grapevine The 54 tissue names and gene names are located above and to the right of the heat map, respectively. The scale bar above the heat map indicates the geng expression level from 0.0 to 3.49.

图5 葡萄NCED基因在干旱条件下的表达模式 干旱处理名称(DL1-DL5分别表示干旱处理0、7、14、21和28天叶组织; DR1-DR5分别表示干旱处理0、7、14、21和28天根组织)和基因名称分别位于热图的上方与右侧。热图上方比例尺表示基因表达量0.0-77.06。相同颜色的实心圆点代表复制基因对。
Figure 5 Expression pattern of grapevine NCED genes under drought treatments The drought treatments (DL1-DL5 indicate drought treatment for 0, 7, 14, 21 and 28 days of leaf tissue, respectively; DR1-DR5 indicate drought treatment for 0, 7, 14, 21 and 28 days of root tissue, respectively) and gene names are located above and to the right of the heat map, respectively. The scale bar above the heat map indicates the gene expression level from 0.0 to 77.06. Solid dots of the same color represent duplicated gene pairs.
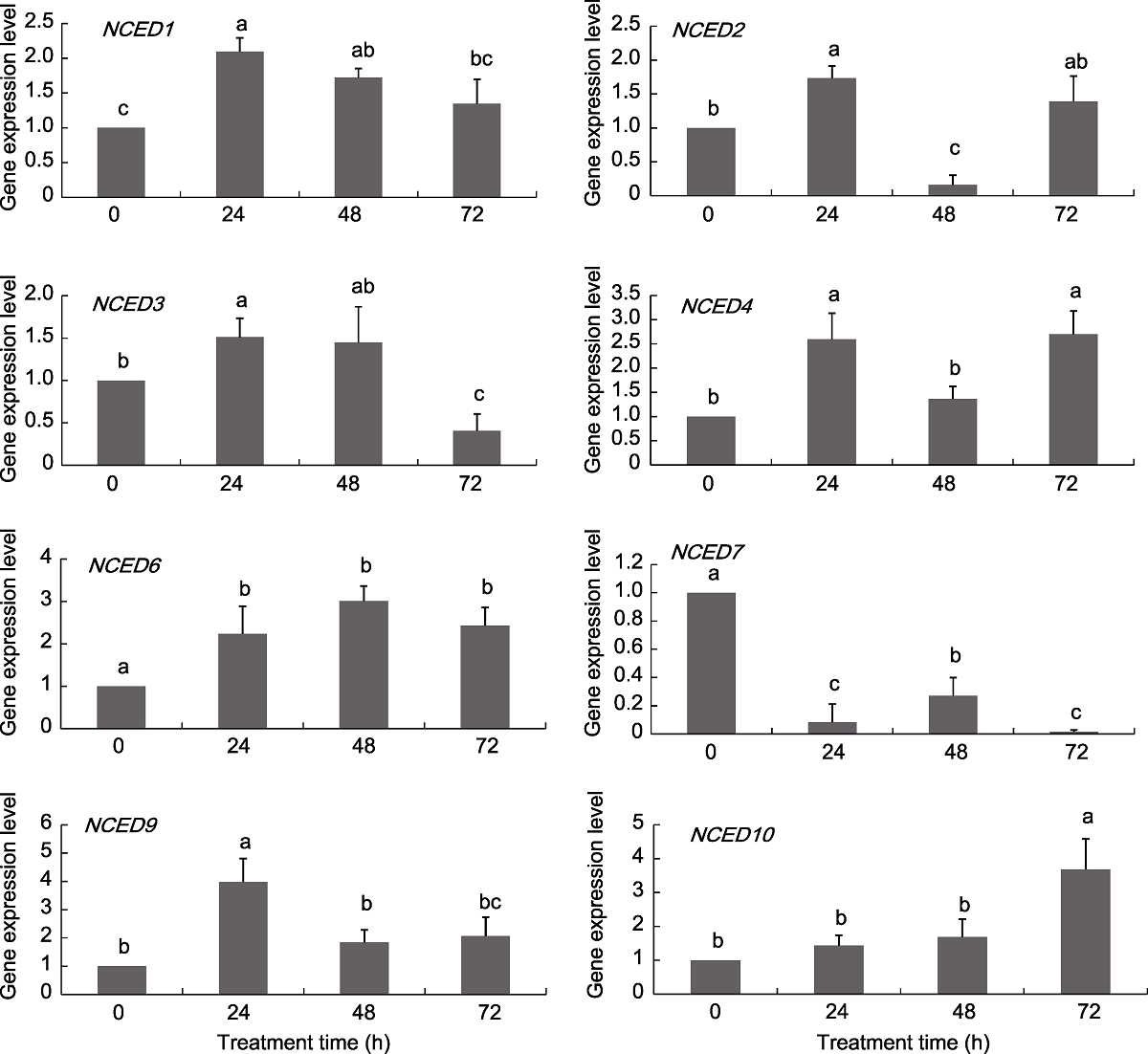
图6 葡萄NCED基因在ABA处理下的表达模式 不同小写字母表示不同处理时间点之间差异显著(P<0.05)。
Figure 6 Expression pattern of grapevine NCED genes under ABA treatments Different lowercase letters above the histogram indicate significant differences among the different time points of the treatment (P<0.05).
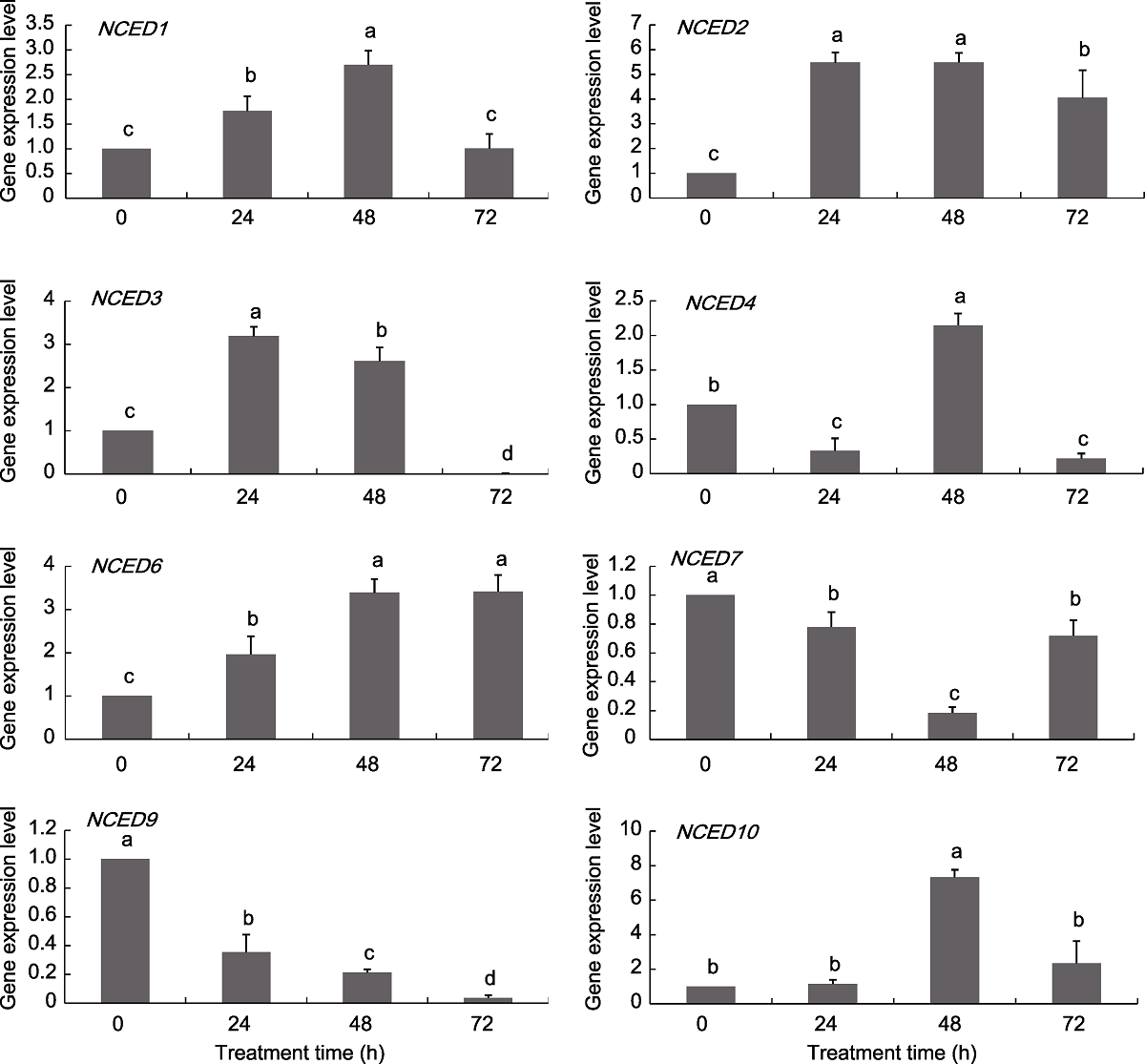
图7 葡萄NCED基因在NaCl处理下的表达模式 不同小写字母表示不同处理时间点之间差异显著(P<0.05)。
Figure 7 Expression pattern of grapevine NCED genes under NaCl treatments Different lowercase letters above the histogram indicate significant differences among the different time points of the treatment (P<0.05).
| [1] | 白戈, 杨大海, 姚恒, 谢贺 ( 2017). 烟草NtNCED基因的鉴定分析. 分子植物育种 15, 3907-3912. |
| [2] | 徐学中, 汪婷, 万旺, 李思慧, 朱国辉 ( 2018). 水稻ABA生物合成基因OsNCED3响应干旱胁迫. 作物学报 44, 24-31. |
| [3] | Adams KL, Cronn R, Percifield R, Wendel JF ( 2003). Genes duplicated by polyploidy show unequal contributions to the transcriptome and organ-specific reciprocal silencing. Proc Natl Acad Sci USA 100, 4649-4654. |
| [4] | Ahrazem O, Rubio-Moraga A, Trapero A, Gómez-Gómez L ( 2012). Developmental and stress regulation of gene expression for a 9-cis-epoxycarotenoid dioxygenase, Cst NCED, isolated from Crocus sativus stigmas. J Exp Bot 63, 681-694. |
| [5] | Chaw SM, Chang CC, Chen HL, Li WH ( 2004). Dating the monocot-dicot divergence and the origin of core eudicots using whole chloroplast genomes. J Mol Evol 58, 424-441. |
| [6] | Cohen-Gihon I, Sharan R, Nussinov R ( 2011). Processes of fungal proteome evolution and gain of function: gene duplication and domain rearrangement. Phys Biol 8, 035009. |
| [7] | Fasoli M, Dal Santo S, Zenoni S, Tornielli GB, Farina L, Zamboni A, Porceddu A, Venturini L, Bicego M, Murino V, Ferrarini A, Delledonne M, Pezzotti M ( 2012). The grapevine expression atlas reveals a deep transcriptome shift driving the entire plant into a maturation program. Plant Cell 24, 3489-3505. |
| [8] | Gaut BS, Morton BR, McCaig BC, Clegg MT ( 1996). Substitution rate comparisons between grasses and palms: synonymous rate differences at the nuclear gene Adh parallel rate differences at the plastid gene rbcL. Proc Natl Acad Sci USA 93, 10274-10279. |
| [9] | Gu ZL, Nicolae D, Lu HHS, Li WH ( 2002). Rapid divergence in expression between duplicate genes inferred from microarray data. Trends Genet 18, 609-613. |
| [10] | Gu ZL, Rifkin SA, White KP, Li WH ( 2004). Duplicate genes increase gene expression diversity within and between species. Nat Genet 36, 577-579. |
| [11] | Guo CL, Guo RR, Xu XZ, Gao M, Li XQ, Song JY, Zheng Y, Wang XP ( 2014). Evolution and expression analysis of the grape (Vitis vinifera L.) WRKY gene family. J Exp Bot 65, 1513-1528. |
| [12] | Jaillon O, Aury JM, Noel B, Policriti A, Clepet C, Casagrande A, Choisne N, Aubourg S, Vitulo N, Jubin C, Vezzi A, Legeai F, Hugueney P, Dasilva C, Horner D, Mica E, Jublot D, Poulain J, Bruyère C, Billault A, Segurens B, Gouyvenoux M, Ugarte E, Cattonaro F, Anthouard V, Vico V, Del Fabbro C, Alaux M, Di Gaspero G, Dumas V, Felice N, Paillard S, Juman I, Moroldo M, Scalabrin S, Canaguier A, Le Clainche I, Malacrida G, Durand E, Pesole G, Laucou V, Chatelet P, Merdinoglu D, Delledonne M, Pezzotti M, Lecharny A, Scarpelli C, Artiguenave F, Pè ME, Valle G, Morgante M, Caboche M, Adam-Blondon AF, Weissenbach J, Quétier F, Wincker P ( 2007). The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449, 463-467. |
| [13] | Malacarne G, Perazzolli M, Cestaro A, Sterck L, Fontana P, Van de Peer Y, Viola R, Velasco R, Salamini F ( 2012). Deconstruction of the (Paleo) polyploid grapevine genome based on the analysis of transposition events involving NBS resistance genes. PLoS One 7, e29762. |
| [14] | McAdam SAM, Brodribb TJ ( 2015). The evolution of mechanisms driving the stomatal response to vapor pressure deficit. Plant Physiol 167, 833-843. |
| [15] | McAdam SAM, Brodribb TJ, Banks JA, Hedrich R, Atallah NM, Cai C, Geringer MA, Lind C, Nichols DS, Stachowski K, Geiger D, Sussmilch FC ( 2016). Abscisic acid controlled sex before transpiration in vascular plants. Proc Natl Acad Sci USA 113, 12862-12867. |
| [16] | Raghavendra AS, Gonugunta VK, Christmann A, Grill E ( 2010). ABA perception and signaling. Trends Plant Sci 15, 395-401. |
| [17] | Roychoudhury A, Paul S, Basu S ( 2013). Cross-talk between abscisic acid-dependent and abscisic acid-independent pathways during abiotic stress. Plant Cell Rep 32, 985-1006. |
| [18] | Saeed AI, Bhagabati NK, Braisted JC, Liang W, Sharov V, Howe EA, Li JW, Thiagarajan M, White JA, Quackenbush J ( 2006). TM4 microarray software suite. Methods Enzymol 411, 134-193. |
| [19] | Sussmilch FC, Brodribb TJ, McAdam SAM ( 2017). What are the evolutionary origins of stomatal responses to abscisic acid in land plants? J Integr Plant Biol 59, 240-260. |
| [20] | Tan BC, Joseph LM, Deng WT, Liu LJ, Li QB, Cline K, McCarty DR ( 2010). Molecular characterization of the Arabidopsis 9-cis epoxycarotenoid dioxygenase gene family. Plant J 35, 44-56. |
| [21] | Wang RY, Yang Y, Wang HG, Chen L, Wang L, Lu P, Liu MX, Qiao ZJ ( 2018). Cloning of gene PmNCED1 and its response to PEG stress in common millet. J Nuclear Agric Sci 32, 244-256. |
| [22] | Yang SH, Zhang XH, Yue JX, Tian DC, Chen JQ ( 2008). Recent duplications dominate NBS-encoding gene expan- sion in two woody species. Mol Genet Genom 280, 187-198. |
| [23] | Ye X, Kang BG, Osburn LD, Li Y, Cheng ZM ( 2009). Identification of the flavin-dependent monooxygenase- encoding YUCCA gene family in Populus trichocarpa and their expression in vegetative tissues and in response to hormone and environmental stresses. Plant Cell Tissue Organ Cult 97, 271-283. |
| [24] | Zhang JZ ( 2003). Evolution by gene duplication: an update. Trends Ecol Evol 18, 292-298. |
| [1] | 王子韵, 吕燕文, 肖钰, 吴超, 胡新生. 植物基因表达调控与进化机制研究进展[J]. 植物学报, 2025, 60(4): 1-0. |
| [2] | 熊良林, 梁国鲁, 郭启高, 景丹龙. 基因可变剪接调控植物响应非生物胁迫研究进展[J]. 植物学报, 2025, 60(3): 435-448. |
| [3] | 杜庆国, 李文学. lncRNA调控玉米生长发育和非生物胁迫研究进展[J]. 植物学报, 2024, 59(6): 950-962. |
| [4] | 周文杰, 张文瀚, 贾玮, 许自成, 黄五星. 植物miRNA响应非生物胁迫研究进展[J]. 植物学报, 2024, 59(5): 810-833. |
| [5] | 赵来鹏, 王柏柯, 杨涛, 李宁, 杨海涛, 王娟, 闫会转. SlHVA22l基因调节番茄耐旱性[J]. 植物学报, 2024, 59(4): 558-573. |
| [6] | 段政勇, 丁敏, 王宇卓, 丁艺冰, 陈凌, 王瑞云, 乔治军. 糜子SBP基因家族全基因组鉴定及表达分析[J]. 植物学报, 2024, 59(2): 231-244. |
| [7] | 仲昭暄, 张冬瑞, 李璐, 苏颖, 王黛宁, 王泽冉, 刘洋, 常缨. 香鳞毛蕨dfr-miR160a和靶基因DfARF10的生物信息学及表达模式分析[J]. 植物学报, 2024, 59(1): 22-33. |
| [8] | 张悦婧, 桑鹤天, 王涵琦, 石珍珍, 李丽, 王馨, 孙坤, 张继, 冯汉青. 植物对非生物胁迫系统性反应中信号传递的研究进展[J]. 植物学报, 2024, 59(1): 122-133. |
| [9] | 许亚楠, 闫家榕, 孙鑫, 王晓梅, 刘玉凤, 孙周平, 齐明芳, 李天来, 王峰. 红光和远红光在调控植物生长发育及应答非生物胁迫中的作用[J]. 植物学报, 2023, 58(4): 622-637. |
| [10] | 张嘉, 李启东, 李翠, 王庆海, 侯新村, 赵春桥, 李树和, 郭强. 植物MATE转运蛋白研究进展[J]. 植物学报, 2023, 58(3): 461-474. |
| [11] | 任晓童, 张冉冉, 魏绍巍, 罗晓峰, 徐佳慧, 舒凯. 种子际微生物研究展望[J]. 植物学报, 2023, 58(3): 499-509. |
| [12] | 吴楠, 覃磊, 崔看, 李海鸥, 刘忠松, 夏石头. 甘蓝型油菜EXA1的克隆及其对植物抗病的调控作用[J]. 植物学报, 2023, 58(3): 385-393. |
| [13] | 王菲菲, 周振祥, 洪益, 谷洋洋, 吕超, 郭宝健, 朱娟, 许如根. 大麦NF-YC基因鉴定及在盐胁迫下的表达分析[J]. 植物学报, 2023, 58(1): 140-149. |
| [14] | 王浩, 王明, 梁婷, 姚玉新, 杜远鹏, 高振. 气温和根区温度对葡萄叶片光合荧光特性的影响[J]. 植物学报, 2022, 57(2): 209-216. |
| [15] | 吴霖升, 张永光, 章钊颖, 张小康, 吴云飞. 日光诱导叶绿素荧光遥感及其在陆地生态系统监测中的应用[J]. 植物生态学报, 2022, 46(10): 1167-1199. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||