

植物学报 ›› 2021, Vol. 56 ›› Issue (5): 520-532.DOI: 10.11983/CBB21119 cstr: 32102.14.CBB21119
收稿日期:2021-07-21
接受日期:2021-09-16
出版日期:2021-09-01
发布日期:2021-09-16
通讯作者:
李学勇
作者简介:* E-mail: lixueyong@caas.cn基金资助:
Jiangyuan Shang, Yan Chun, Xueyong Li*( )
)
Received:2021-07-21
Accepted:2021-09-16
Online:2021-09-01
Published:2021-09-16
Contact:
Xueyong Li
摘要: 穗型是决定水稻(Oryza sativa)产量的关键因素之一。我们从粳稻品种圣稻808 (SD808)的EMS诱变突变体库中发现4份短穗突变体, 这些突变体的穗长、一级枝梗数、二级枝梗数和穗粒数发生不同程度的降低。基因定位和图位克隆表明, 这些突变体的表型受同一基因控制, 将该基因命名为PAL3 (PANICLE LENGTH3)。PAL3编码一个含12个跨膜结构域的多肽转运蛋白。pal3-1和pal3-2的点突变造成保守区域的氨基酸发生非同义突变; pal3-3的点突变造成第1外显子和内含子拼接错误; pal3-4的点突变造成蛋白翻译提前终止, 导致第12个跨膜域缺失。对PAL3进行单倍型分析, 共鉴定出9个单倍型(Hap1-Hap9), 其中Hap1-Hap3为主要单倍型。Hap1以粳稻为主, Hap2同时包含籼稻和粳稻, Hap3则以籼稻为主。Hap1起源于普通野生稻(O. rufipogon), Hap2和Hap3可能起源于一年生普通野生稻(O. nivara)。统计分析结果表明, Hap3的穗长显著高于Hap1和Hap2, 其具有提高穗长的潜力。该研究揭示了多肽转运蛋白对水稻穗型的重要调控作用, 为水稻穗型改良奠定了理论基础。
尚江源, 淳雁, 李学勇. 水稻穗长基因PAL3的克隆及自然变异分析. 植物学报, 2021, 56(5): 520-532.
Jiangyuan Shang, Yan Chun, Xueyong Li. Map-based Cloning and Natural Variation Analysis of the PAL3 Gene Controlling Panicle Length in Rice. Chinese Bulletin of Botany, 2021, 56(5): 520-532.
| Primer name | Forward primer (5′→3′) | Reverse primer (5′→3′) |
|---|---|---|
| R11-3 | AGAGAGACATCCGGAGACAA | TAAGACGAAAGGTCAAACGT |
| R11-5 | GTGCTAACGTTTCGTCTAAC | AATAGCCTTCGGTGGTCTCA |
| R11-10 | GTTCGTAATGTGGGCGTCTT | TGGGCACTCTTCTCACACTG |
| R11-12 | CAATCTTGCTCTACTAGCTAGTG | GTGGCAACTAACAGATTAGATG |
| M1 | GCAGTATATATTCGGCGGCG | GCCGTCGCCATATAGCTG |
| M2 | GAGCCTCTCCTACTGTGCTA | AGAGCCCTCAGTTCCTCAAT |
| M3 | GCTGACTACAGTAAGATCATGC | AGACAAACGGTCAAACATGT |
| M4 | AAGGATCCAAGCTAGCCTCC | CCTGACAGCAAGCGAGAGAT |
| M5 | CTTCAGCAAGTGAACTACGA | CCTAAACTAGCACGGATCATAGC |
| M6 | TGTGAGGTTTAGGTTCTCGGA | TGAATAGAGATGCGGTCCAAC |
| M7 | GGATTCGGCCACTGGTTGTT | GAATGTACTCGGATAAACCC |
表1 定位引物
Table 1 Primers used for mapping
| Primer name | Forward primer (5′→3′) | Reverse primer (5′→3′) |
|---|---|---|
| R11-3 | AGAGAGACATCCGGAGACAA | TAAGACGAAAGGTCAAACGT |
| R11-5 | GTGCTAACGTTTCGTCTAAC | AATAGCCTTCGGTGGTCTCA |
| R11-10 | GTTCGTAATGTGGGCGTCTT | TGGGCACTCTTCTCACACTG |
| R11-12 | CAATCTTGCTCTACTAGCTAGTG | GTGGCAACTAACAGATTAGATG |
| M1 | GCAGTATATATTCGGCGGCG | GCCGTCGCCATATAGCTG |
| M2 | GAGCCTCTCCTACTGTGCTA | AGAGCCCTCAGTTCCTCAAT |
| M3 | GCTGACTACAGTAAGATCATGC | AGACAAACGGTCAAACATGT |
| M4 | AAGGATCCAAGCTAGCCTCC | CCTGACAGCAAGCGAGAGAT |
| M5 | CTTCAGCAAGTGAACTACGA | CCTAAACTAGCACGGATCATAGC |
| M6 | TGTGAGGTTTAGGTTCTCGGA | TGAATAGAGATGCGGTCCAAC |
| M7 | GGATTCGGCCACTGGTTGTT | GAATGTACTCGGATAAACCC |
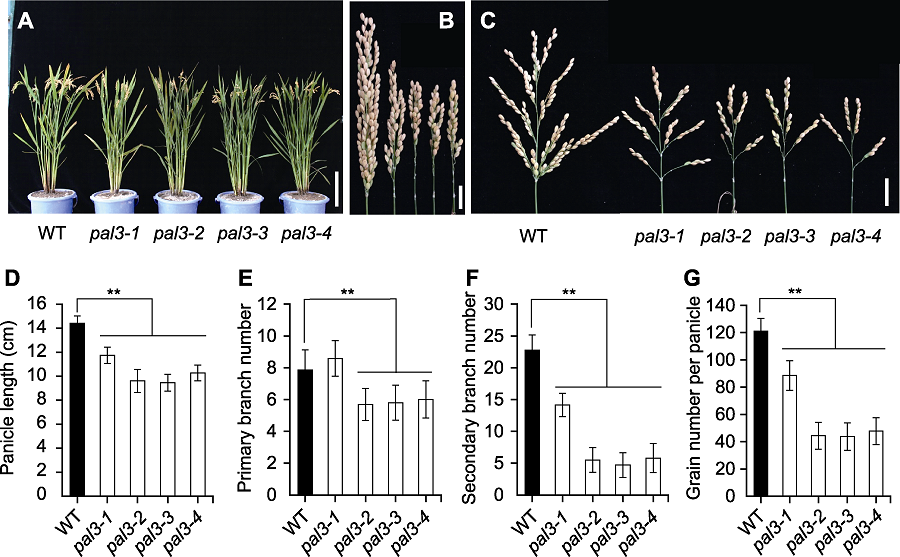
图1 水稻野生型与pal3突变体的表型比较 (A) 野生型与pal3突变体的成熟期株型(Bar=20 cm); (B) 闭合状态下野生型与pal3突变体的穗(Bar=2 cm); (C) 展开状态下野生型与pal3突变体的穗(右) (Bar=2 cm); (D)-(G) 野生型与pal3突变体的穗长(D)、一级枝梗数(E)、二级枝梗数(F)和穗粒数(G)的统计分析。WT: 野生型。图中数据为平均值±标准差(n=20)。**表示在P<0.01水平差异显著(经t检验)。
Figure 1 Comparison of the rice phenotype between wild type and the pal3 mutants (A) Gross morphology of wild type and the pal3 mutants at mature stage (Bar=20 cm); (B) Closed panicle of wild type and the pal3 mutants (Bar=2 cm); (C) Spread panicles of wild type and the pal3 mutants (right) (Bar=2 cm); (D)-(G) Statistic analysis of panicle length (D), primary branch number (E), secondary branch number (F) and grain number per panicle (G) of wild type and the pal3 mutants. WT: Wild type. Data in figure are means±SD (n=20). ** indicate the significant differences at P<0.01 level by Students’t test.
| Hybrid combi- nations | Phenotype of F1 | F2 population | χ23:1 | ||
|---|---|---|---|---|---|
| Wild-type plant number | Mutant-phenotypic plant number | Total number | |||
| pal3-1 × WT | WT | 190 | 60 | 250 | 0.13 |
| pal3-2 × WT | WT | 196 | 68 | 264 | 0.08 |
| pal3-3 × WT | WT | 180 | 63 | 243 | 0.11 |
| pal3-4 × WT | WT | 201 | 69 | 270 | 0.04 |
表2 pal3-1、pal3-2、pal3-3和pal3-4水稻突变体的遗传分析
Table 2 Genetic analysis of the pal3-1, pal3-2, pal3-3 and pal3-4 rice mutants
| Hybrid combi- nations | Phenotype of F1 | F2 population | χ23:1 | ||
|---|---|---|---|---|---|
| Wild-type plant number | Mutant-phenotypic plant number | Total number | |||
| pal3-1 × WT | WT | 190 | 60 | 250 | 0.13 |
| pal3-2 × WT | WT | 196 | 68 | 264 | 0.08 |
| pal3-3 × WT | WT | 180 | 63 | 243 | 0.11 |
| pal3-4 × WT | WT | 201 | 69 | 270 | 0.04 |
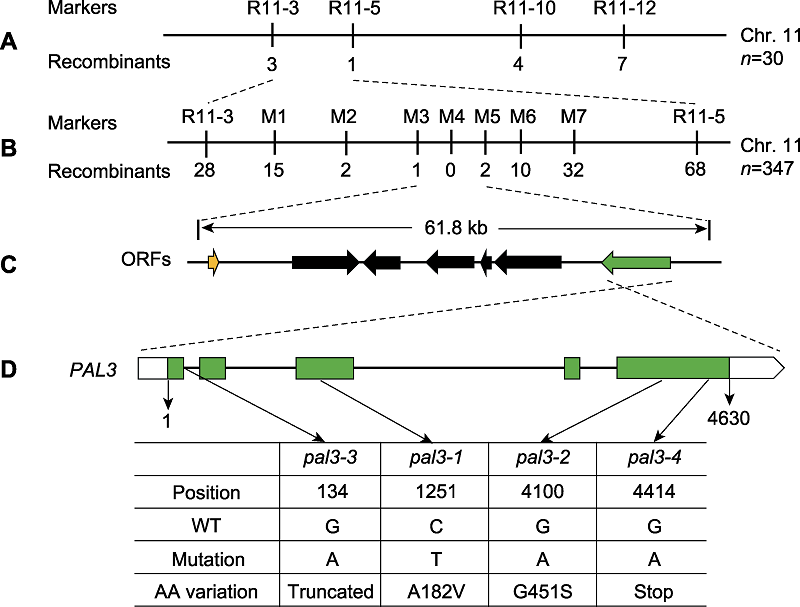
图2 PAL3基因的图位克隆 (A) PAL3基因定位于第11号染色体InDel标记R11-3和R11-5之间; (B), (C) PAL3的精细定位。进一步将PAL3基因定位于M3和M5标记之间, 该区间包含7个开放阅读框(ORFs)。标记下方的数字表示重组个体的数目; (D) PAL3基因的结构示意图及pal3突变体的各突变位点。WT: 野生型
Figure 2 Map-based cloning of the PAL3 gene (A) The PAL3 gene was mapped to a region between InDel markers R11-3 and R11-5 on chromosome 11; (B), (C) Fine-mapping of PAL3. The PAL3 gene was further delimited to the region between markers M3 and M5, which contains seven open reading frames (ORFs). The number of recombinants is indicated beneath the marker positions; (D) Schematic structure of the PAL3 gene and the mutation sites of pal3 mutants. WT: Wild type
| Gene ID | Annotation |
|---|---|
| LOC_Os11g12680 | Expressed protein |
| LOC_Os11g12690 | Retrotransposon protein, putative, unclassified, expressed |
| LOC_Os11g12700 | Retrotransposon protein, putative, unclassified, expressed |
| LOC_Os11g12710 | Retrotransposon protein, putative, unclassified, expressed |
| LOC_Os11g12720 | Retrotransposon, putative, centromere-specific |
| LOC_Os11g12730 | Transposon protein, putative, CACTA, En/Spm sub-class, expressed |
| LOC_Os11g12740 | Peptide transporter PTR2, putative, expressed |
表3 精细定位区间候选基因注释
Table 3 Annotations of candidate genes in the fine-mapping region
| Gene ID | Annotation |
|---|---|
| LOC_Os11g12680 | Expressed protein |
| LOC_Os11g12690 | Retrotransposon protein, putative, unclassified, expressed |
| LOC_Os11g12700 | Retrotransposon protein, putative, unclassified, expressed |
| LOC_Os11g12710 | Retrotransposon protein, putative, unclassified, expressed |
| LOC_Os11g12720 | Retrotransposon, putative, centromere-specific |
| LOC_Os11g12730 | Transposon protein, putative, CACTA, En/Spm sub-class, expressed |
| LOC_Os11g12740 | Peptide transporter PTR2, putative, expressed |
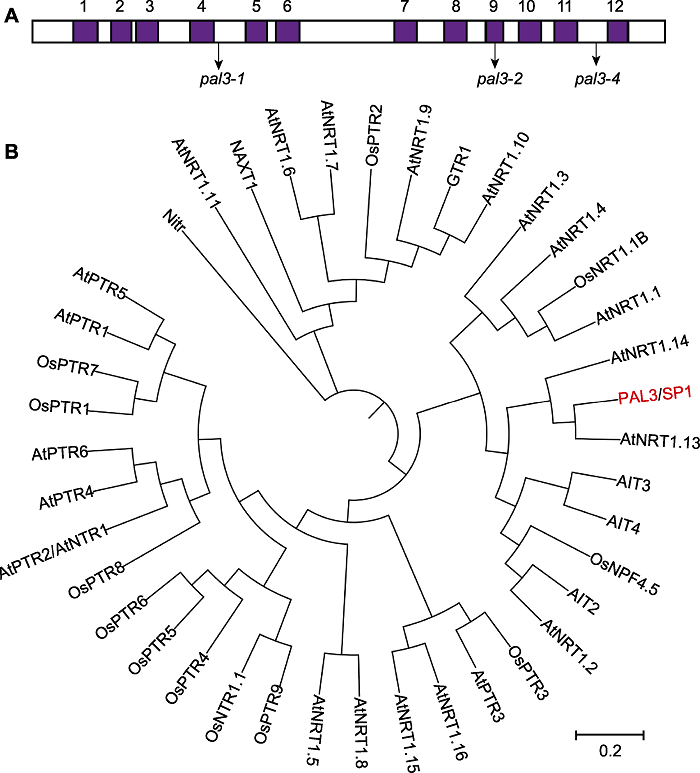
图3 PAL3跨膜域及系统发育分析 (A) PAL3的12个跨膜域及pal3-1、pal3-2和pal3-4突变体的突变位点; (B) PAL3及其同源蛋白的系统发育分析。系统发育树采用MEGA X软件, bootstrap重复1 000次, 最大似然距离法构建。
Figure 3 Transmembrane regions and phylogenetic analysis of PAL3 (A) 12 transmembrane domains of PAL3 and the mutation sites of the pal3-1, pal3-2 and pal3-4 mutants; (B) Phylogenetic analysis of PAL3 and its homologs. The phylogenetic analysis was carried out by MEGA X with 1 000 bootstrap replicates and was constructed using the distance method with maximum likelihood.
| Species | Locus ID | Gene name |
|---|---|---|
| Rice | LOC_Os11g12740 | PAL3/SP1 |
| LOC_Os07g01070 | OsPTR1 | |
| LOC_Os12g44100 | OsPTR2 | |
| LOC_Os10g33210 | OsPTR3 | |
| LOC_Os07g41250 | OsPTR4 | |
| LOC_Os04g50940 | OsPTR5 | |
| LOC_Os04g50950 | OsPTR6 | |
| LOC_Os01g04950 | OsPTR7 | |
| LOC_Os03g51050 | OsPTR8 | |
| LOC_Os06g49250 | OsPTR9 | |
| LOC_Os03g13274 | OsNTR1.1 | |
| LOC_Os10g40600 | OsNRT1.1B | |
| LOC_Os01g54515 | OsNPF4.5 | |
| Arabidopsis | At1g12110 | AtNRT1.1 |
| At5g62680 | AtNRT1.10 | |
| At1g52190 | AtNRT1.11 | |
| At1g33440 | AtNRT1.13 | |
| At1g59740 | AtNRT1.14 | |
| At1g72120 | AtNRT1.15 | |
| At1g72125 | AtNRT1.16 | |
| At1g69850 | AtNRT1.2 | |
| At3g21670 | AtNRT1.3 | |
| At2g26690 | AtNRT1.4 | |
| At1g32450 | AtNRT1.5 | |
| At1g27080 | AtNRT1.6 | |
| At1g69870 | AtNRT1.7 | |
| At4g21680 | AtNRT1.8 | |
| At1g18880 | AtNRT1.9 | |
| At3g54140 | AtPTR1 | |
| At2g02040 | AtPTR2/AtNTR1 | |
| At5g46050 | AtPTR3 | |
| At2g02020 | AtPTR4 | |
| At5g01180 | AtPTR5 | |
| At1g62200 | AtPTR6 | |
| At1g27040 | AIT2 | |
| At3g25260 | AIT3 | |
| At3g25280 | AIT4 | |
| At3g47960 | GTR1 | |
| At3g45650 | NAXT1 | |
| At1g68570 | Nitr |
表4 PAL3及其同源家族基因的基因号和名称
Table 4 Locus ID and gene name of PAL3 and its homologs
| Species | Locus ID | Gene name |
|---|---|---|
| Rice | LOC_Os11g12740 | PAL3/SP1 |
| LOC_Os07g01070 | OsPTR1 | |
| LOC_Os12g44100 | OsPTR2 | |
| LOC_Os10g33210 | OsPTR3 | |
| LOC_Os07g41250 | OsPTR4 | |
| LOC_Os04g50940 | OsPTR5 | |
| LOC_Os04g50950 | OsPTR6 | |
| LOC_Os01g04950 | OsPTR7 | |
| LOC_Os03g51050 | OsPTR8 | |
| LOC_Os06g49250 | OsPTR9 | |
| LOC_Os03g13274 | OsNTR1.1 | |
| LOC_Os10g40600 | OsNRT1.1B | |
| LOC_Os01g54515 | OsNPF4.5 | |
| Arabidopsis | At1g12110 | AtNRT1.1 |
| At5g62680 | AtNRT1.10 | |
| At1g52190 | AtNRT1.11 | |
| At1g33440 | AtNRT1.13 | |
| At1g59740 | AtNRT1.14 | |
| At1g72120 | AtNRT1.15 | |
| At1g72125 | AtNRT1.16 | |
| At1g69850 | AtNRT1.2 | |
| At3g21670 | AtNRT1.3 | |
| At2g26690 | AtNRT1.4 | |
| At1g32450 | AtNRT1.5 | |
| At1g27080 | AtNRT1.6 | |
| At1g69870 | AtNRT1.7 | |
| At4g21680 | AtNRT1.8 | |
| At1g18880 | AtNRT1.9 | |
| At3g54140 | AtPTR1 | |
| At2g02040 | AtPTR2/AtNTR1 | |
| At5g46050 | AtPTR3 | |
| At2g02020 | AtPTR4 | |
| At5g01180 | AtPTR5 | |
| At1g62200 | AtPTR6 | |
| At1g27040 | AIT2 | |
| At3g25260 | AIT3 | |
| At3g25280 | AIT4 | |
| At3g47960 | GTR1 | |
| At3g45650 | NAXT1 | |
| At1g68570 | Nitr |
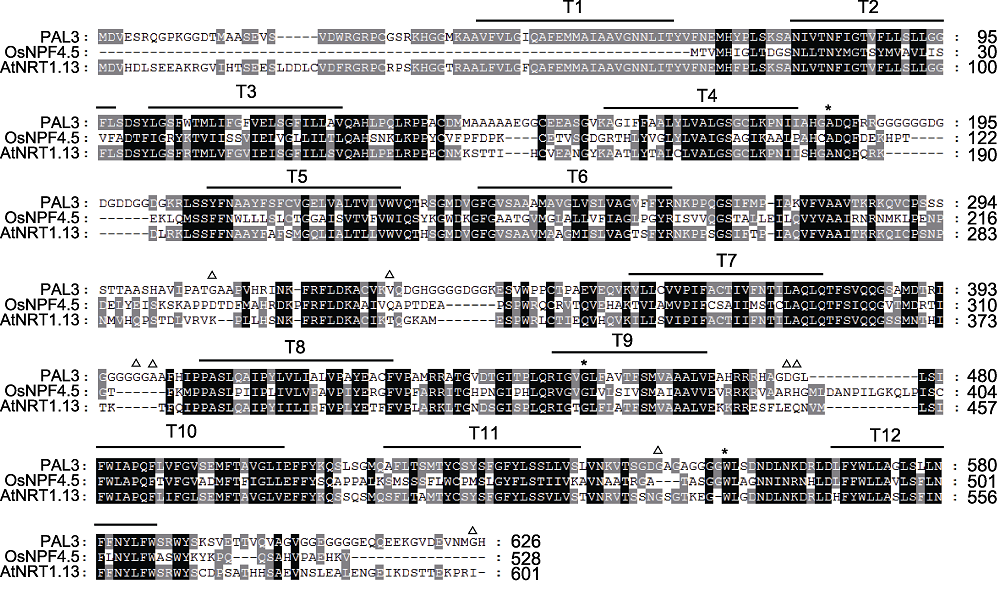
图4 PAL3及其同源蛋白序列比对 黑色背景表示所有氨基酸序列都高度同源, 灰色部分表示部分氨基酸序列同源, 白色部分表示氨基酸序列完全不同源。*依次为pal3-1、pal3-2和pal3-4突变体的突变位点; △为Hap1和Hap3的差异位点。
Figure 4 Sequences alignment of PAL3 and its homologs The black background represent that all amino acid sequences are homologous. The gray part represent that some of the amino acid sequences are homologous. The white part represent that the amino acid sequences are completely different. * indicate the mutation sites of the pal3-1, pal3-2 and pal3-4 mutants in turn; △: The variation sites between Hap1 and Hap3.
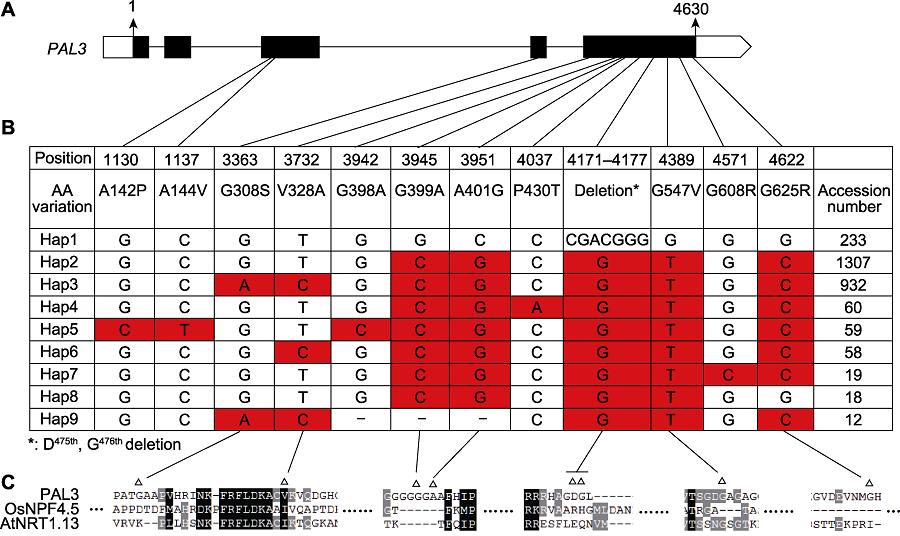
图5 PAL3基因的单倍型分析 (A) PAL3基因结构及各单倍型变异所在位置; (B) PAL3在栽培稻中的单倍型。单倍型数据通过水稻分子育种整合组学知识库(MBKbase) (Peng et al., 2020)分析而来, 共包含2 187个栽培稻品种, 分析时去除了内含子变异、杂合变异和同义变异(-: 碱基对缺失); (C) Hap1和Hap3差异位点的保守性分析(Δ: Hap1和Hap3的差异位点)。
Figure 5 Haplotype analysis of PAL3 (A) Structure of PAL3 gene and the location of haplotypes; (B) Haplotypes of PAL3 in the cultivated rice. Haplotype data were analyzed by Rice Molecular Breeding Knowledgebase (MBKbase) (Peng et al., 2020), including 2 187 cultivated rice varieties. Variations on intron, heterozygous variations and synonymous variations were removed during analysis (-: The deletion of base pair); (C) Sequence alignment around the variation sites between Hap1 and hap3 (Δ: The variation sites between Hap1 and Hap3).
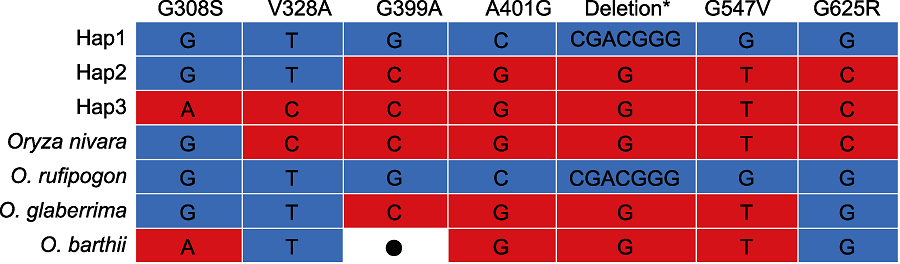
图6 PAL3在野生稻中的单倍型 野生稻单倍型数据来源于禾本科基因组数据库(Gramene)和水稻相关物种基因组数据库(Rice Relatives-GD) (Mao et al., 2019)。*: D475和G476缺失; ●: 无数据
Figure 6 Haplotype of PAL3 in the wild rice Haplotype data of wild rice were from Gramene and Rice Relatives-GD (Mao et al., 2019). *: D475 and G476 deletion; ●: No datum
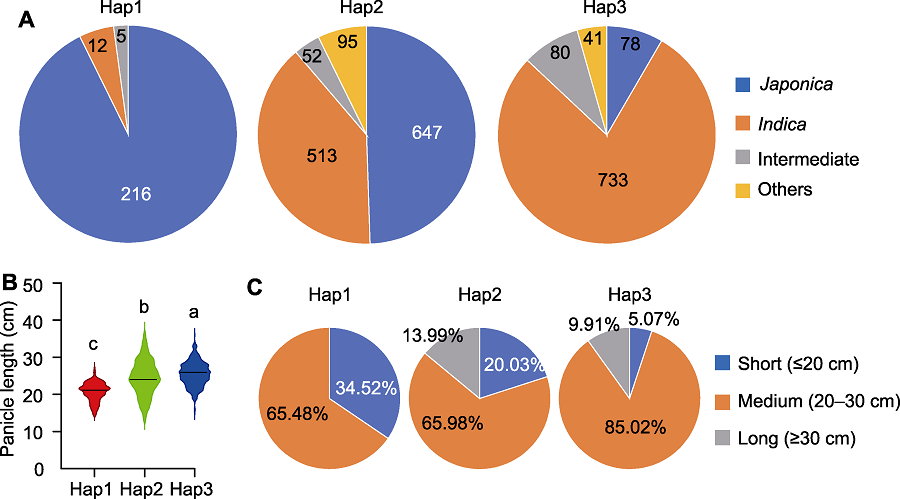
图7 PAL3基因Hap1、Hap2和Hap3的亚种组成及穗型分析 (A) Hap1、Hap2和Hap3的亚种构成; (B) Hap1、Hap2和Hap3的穗长比较; (C) Hap1、Hap2和Hap3的穗长统计。不同小写字母表示差异显著(P<0.05)。
Figure 7 Subspecies composition and panicle phenotype analysis of Hap1, Hap2 and Hap3 of the PAL3 gene (A) Subspecies composition of Hap1, Hap2 and Hap3; (B) Comparison of panicle length among Hap1, Hap2 and Hap3; (C) Statistic of panicle length of Hap1, Hap2 and Hap3. Different lowercase letters indicate significant differences (P<0.05).
| [1] |
淳雁, 李学勇 (2017). 水稻穗型的遗传调控研究进展. 植物学报 52, 19-29.
DOI |
| [2] | 刘丹, 王嘉宇, 刘进, 马殿荣, 赵明辉, 陈温福 (2015). 水稻散状穗突变体sp的遗传分析及基因初定位. 植物学报 50, 198-205. |
| [3] |
刘玉良, 郑术芝 (2017). 水稻产量相关性状驯化研究进展. 植物学报 52, 113-121.
DOI |
| [4] | 田翠 (2010). 复粒稻小穗簇生突变体基因的遗传分析和初步定位. 硕士论文. 重庆: 重庆大学. pp. 19-23. |
| [5] | 吴光南, 张云桥 (1962). 稻穗发育过程及其控制途径的研究. 作物学报 1, 43-52. |
| [6] | 张淑红 (2002). 水稻中控制形态结构建成相关基因的功能研究. 博士论文. 上海: 复旦大学. pp. 70-78. |
| [7] | 郑雷英, 朱旭东, 钱前, 赵忠, 张建军, 胡筱荷, 林鸿宣, 罗达 (2003). 水稻穗部突变体CL的形态和定位分析. 科学通报 48, 264-267. |
| [8] | 周鹏 (2009). 水稻簇生穗突变体Cl-dz的形态特征及遗传定位. 硕士论文. 成都: 四川农业大学. pp. 28-30. |
| [9] |
Chiang CS, Stacey G, Tsay YF (2004). Mechanisms and functional properties of two peptide transporters, AtPTR2 and fPTR2. J Biol Chem 279, 30150-30157.
DOI URL |
| [10] |
Choi JY, Lye ZN, Groen SC, Dai XG, Rughani P, Zaaijer S, Harrington ED, Juul S, Purugganan MD (2020). Nanopore sequencing-based genome assembly and evolutionary genomics of circum-basmati rice. Genome Biol 21, 21.
DOI URL |
| [11] | Choi JY, Platts AE, Fuller DQ, Hsing YI, Wing RA, Purugganan MD (2017). The rice paradox: multiple origins but single domestication in Asian rice. Mol Biol Evol 34, 969-979. |
| [12] |
Grillo MA, Li CB, Fowlkes AM, Briggeman TM, Zhou AL, Schemske DW, Sang T (2009). Genetic architecture for the adaptive origin of annual wild rice, Oryza nivala. Evolution 63, 870-883.
DOI URL |
| [13] |
Gross BL, Zhao ZJ (2014). Archaeological and genetic insights into the origins of domesticated rice. Proc Natl Acad Sci USA 111, 6190-6197.
DOI URL |
| [14] |
Huang XH, Kurata N, Wei XH, Wang ZX, Wang AH, Zhao Q, Zhao Y, Liu KY, Lu HY, Li WJ, Guo YL, Lu YQ, Zhou CC, Fan DL, Weng QJ, Zhu CR, Huang T, Zhang L, Wang YC, Feng L, Furuumi H, Kubo T, Miyabayashi T, Yuan XP, Xu Q, Dong GJ, Zhan QL, Li CY, Fujiyama A, Toyoda A, Lu TT, Feng Q, Qian Q, Li JY, Han B (2012). A map of rice genome variation reveals the origin of cultivated rice. Nature 490, 497-501.
DOI URL |
| [15] |
Huang XZ, Qian Q, Liu ZB, Sun HY, He SY, Luo D, Xia GM, Chu CC, Li JY, Fu XD (2009). Natural variation at the DEP1 locus enhances grain yield in rice. Nat Genet 41, 494-497.
DOI URL |
| [16] |
Huang Y, Bai XF, Luo MF, Xing YZ (2019). Short Panicle 3 controls panicle architecture by upregulating APO2/RFL and increasing cytokinin content in rice. J Integr Plant Biol 61, 987-999.
DOI |
| [17] |
Ishii T, Numaguchi K, Miura K, Yoshida K, Thanh PT, Htun TM, Yamasaki M, Komeda N, Matsumoto T, Terauchi R, Ishikawa R, Ashikari M (2013). OsLG1 regulates a closed panicle trait in domesticated rice. Nat Genet 45, 462-465.
DOI URL |
| [18] |
Komatsu K, Maekawa M, Ujiie S, Satake Y, Furutani I, Okamoto H, Shimamoto K, Kyozuka J (2003). LAX and SPA: major regulators of shoot branching in rice. Proc Natl Acad Sci USA 100, 11765-11770.
DOI URL |
| [19] |
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018). MEGA X: Molecular Evolutionary Genetics Analysis ac-ross computing platforms. Mol Biol Evol 35, 1547-1549.
DOI URL |
| [20] |
Lee J, Park JJ, Kim SL, Yim J, An G (2007). Mutations in the rice liguleless gene result in a complete loss of the auricle, ligule, and laminar joint. Plant Mol Biol 65, 487-499.
DOI URL |
| [21] |
Léran S, Varala K, Boyer JC, Chiurazzi M, Crawford N, Daniel-Vedele F, David L, Dickstein R, Fernandez E, Forde B, Gassmann W, Geiger D, Gojon A, Gong JM, Halkier BA, Harris JM, Hedrich R, Limami AM, Rentsch D, Seo M, Tsay YF, Zhang MY, Coruzzi G, Lacombe B (2014). A unified nomenclature of NITRATE TRANSPORTER 1/PEPTIDE TRANSPORTER family members in plants. Trends Plant Sci 19, 5-9.
DOI URL |
| [22] |
Li F, Liu WB, Tang JY, Chen JF, Tong HN, Hu B, Li CL, Fang J, Chen MS, Chu CC (2010). Rice DENSE AND ERECT PANICLE 2 is essential for determining panicle outgrowth and elongation. Cell Res 20, 838-849.
DOI URL |
| [23] |
Li SB, Qian Q, Fu ZM, Zeng DL, Meng XB, Kyozuka J, Maekawa M, Zhu XD, Zhang J, Li JY, Wang YH (2009). Short panicle1 encodes a putative PTR family transporter and determines rice panicle size. Plant J 58, 592-605.
DOI URL |
| [24] |
Liu TM, Li LZ, Zhang YS, Xu CG, Li XH, Xing YZ (2011). Comparison of quantitative trait loci for rice yield, panicle length and spikelet density across three connected populations. J Genet 90, 377-382.
DOI URL |
| [25] | Mao LF, Chen MH, Chu QJ, Jia L, Sultana MH, Wu DY, Kong XD, Qiu J, Ye CY, Zhu QH, Chen X, Fan LJ (2019). RiceRelativesGD: a genomic database of rice relatives for rice research. Database 2019, baz110. |
| [26] |
Molina J, Sikora M, Garud N, Flowers JM, Rubinstein S, Reynolds A, Huang P, Jackson S, Schaal BA, Bustamante CD, Boyko AR, Purugganan MD (2011). Molecular evidence for a single evolutionary origin of domesticated rice. Proc Natl Acad Sci USA 108, 8351-8356.
DOI URL |
| [27] |
Nakagawa M, Shimamoto K, Kyozuka J (2002). Overexpression of RCN1 and RCN2, rice TERMINAL FLOWER 1/CENTRORADIALIS homologs, confers delay of phase transition and altered panicle morphology in rice. Plant J 29, 743-750.
PMID |
| [28] | Peng H, Wang K, Chen Z, Cao YH, Gao Q, Li Y, Li XX, Lu HW, Du HL, Lu M, Yang X, Liang CZ (2020). MBKbase for rice: an integrated omics knowledgebase for molecular breeding in rice. Nucleic Acids Res 48, D1085-D1092. |
| [29] |
Piao RH, Jiang WZ, Ham TH, Choi MS, Qiao YL, Chu SH, Park JH, Woo MO, Jin ZX, An G, Lee J, Koh HJ (2009). Map-based cloning of the ERECT PANICLE 3 gene in rice. Theor Appl Genet 119, 1497-1506.
DOI URL |
| [30] |
Qiao YL, Piao RH, Shi JX, Lee SI, Jiang WZ, Kim BK, Lee J, Han LZ, Ma WB, Koh HJ (2011). Fine mapping and candidate gene analysis of Dense and Erect Panicle 3, DEP3, which confers high grain yield in rice (Oryza sativa L.). Theor Appl Genet 122, 1439-1449.
DOI URL |
| [31] |
Tabuchi H, Zhang Y, Hattori S, Omae M, Shimizu-sato S, Oikawa T, Qian Q, Nishimura M, Kitano H, Xie H, Fang XH, Yoshida H, Kyozuka J, Chen F, Sato Y (2011). LAX PANICLE2 of rice encodes a novel nuclear protein and regulates the formation of axillary meristems. Plant Cell 23, 3276-3287.
DOI URL |
| [32] |
Vaughan DA, Morishima H, Kadowaki K (2003). Diversity in the Oryza genus. Curr Opin Plant Biol 6, 139-146.
PMID |
| [33] |
Wu YZ, Fu YC, Zhao SS, Gu P, Zhu ZF, Sun CQ, Tan LB (2016). CLUSTERED PRIMARY BRANCH 1, a new allele of DWARF11, controls panicle architecture and seed size in rice. Plant Biotechnol J 14, 377-386.
DOI URL |
| [34] |
Yan CJ, Zhou JH, Yan S, Chen F, Yeboah M, Tang SZ, Liang GH, Gu MH (2007). Identification and characterization of a major QTL responsible for erect panicle trait in japonica rice (Oryza sativa L.). Theor Appl Genet 115, 1093-1100.
DOI URL |
| [35] |
Yoshida A, Sasao M, Yasuno N, Takagi K, Daimon Y, Chen RH, Yamazaki R, Tokunaga H, Kitaguchi Y, Sato Y, Nagamura Y, Ushijima T, Kumamaru T, Iida S, Maekawa M, Kyozuka J (2013). TAWAWA1, a regulator of rice inflorescence architecture, functions through the suppression of meristem phase transition. Proc Natl Acad Sci USA 110, 767-772.
DOI URL |
| [36] |
Yu HY, Murchie EH, González-carranza ZH, Pyke KA, Roberts JA (2015). Decreased photosynthesis in the erect panicle 3 (ep3) mutant of rice is associated with reduced stomatal conductance and attenuated guard cell development. J Exp Bot 66, 1543-1552.
DOI URL |
| [37] |
Zhou Y, Zhu JY, Li ZY, Yi CD, Liu J, Zhang HG, Tang SZ, Gu MH, Liang GH (2009). Deletion in a quantitative trait gene qPE9-1 associated with panicle erectness improves plant architecture during rice domestication. Genetics 183, 315-324.
DOI URL |
| [38] |
Zhu ZF, Tan LB, Fu YC, Liu FX, Cai HW, Xie DX, Wu F, Wu JZ, Matsumoto T, Sun CQ (2013). Genetic control of inflorescence architecture during rice domestication. Nat Commun 4, 2200.
DOI URL |
| [1] | 叶灿, 姚林波, 金莹, 高蓉, 谭琪, 李旭映, 张艳军, 陈析丰, 马伯军, 章薇, 张可伟. 水稻水杨酸代谢突变体高通量筛选方法的建立与应用[J]. 植物学报, 2025, 60(4): 1-0. |
| [2] | 赵凌, 管菊, 梁文化, 张勇, 路凯, 赵春芳, 李余生, 张亚东. 基于高密度Bin图谱的水稻苗期耐热性QTL定位[J]. 植物学报, 2025, 60(3): 342-353. |
| [3] | 李新宇, 谷月, 徐非非, 包劲松. 水稻胚乳淀粉合成相关蛋白的翻译后修饰研究进展[J]. 植物学报, 2025, 60(2): 256-270. |
| [4] | 李建国, 张怡, 张文君. 水稻根系铁膜形成及对磷吸收的影响[J]. 植物学报, 2025, 60(1): 132-143. |
| [5] | 姚瑞枫, 谢道昕. 水稻独脚金内酯信号感知的激活和终止[J]. 植物学报, 2024, 59(6): 873-877. |
| [6] | 连锦瑾, 唐璐瑶, 张伊诺, 郑佳兴, 朱超宇, 叶语涵, 王跃星, 商文楠, 傅正浩, 徐昕璇, 吴日成, 路梅, 王长春, 饶玉春. 水稻抗氧化性状遗传位点挖掘及候选基因分析[J]. 植物学报, 2024, 59(5): 738-751. |
| [7] | 黄佳慧, 杨惠敏, 陈欣雨, 朱超宇, 江亚楠, 胡程翔, 连锦瑾, 芦涛, 路梅, 张维林, 饶玉春. 水稻突变体pe-1对弱光胁迫的响应机制[J]. 植物学报, 2024, 59(4): 574-584. |
| [8] | 周俭民. 收放自如的明星战车[J]. 植物学报, 2024, 59(3): 343-346. |
| [9] | 朱超宇, 胡程翔, 朱哲楠, 张芷宁, 汪理海, 陈钧, 李三峰, 连锦瑾, 唐璐瑶, 钟芊芊, 殷文晶, 王跃星, 饶玉春. 水稻穗部性状QTL定位及候选基因分析[J]. 植物学报, 2024, 59(2): 217-230. |
| [10] | 夏婧, 饶玉春, 曹丹芸, 王逸, 柳林昕, 徐雅婷, 牟望舒, 薛大伟. 水稻中乙烯生物合成关键酶OsACS和OsACO调控机制研究进展[J]. 植物学报, 2024, 59(2): 291-301. |
| [11] | 朱宝, 赵江哲, 张可伟, 黄鹏. 水稻细胞分裂素氧化酶9参与调控水稻叶夹角发育[J]. 植物学报, 2024, 59(1): 10-21. |
| [12] | 方妍力, 田传玉, 苏如意, 刘亚培, 王春连, 陈析丰, 郭威, 纪志远. 水稻抗细菌性条斑病基因挖掘与初定位[J]. 植物学报, 2024, 59(1): 1-9. |
| [13] | 贾绮玮, 钟芊芊, 顾育嘉, 陆天麒, 李玮, 杨帅, 朱超宇, 胡程翔, 李三峰, 王跃星, 饶玉春. 水稻茎秆细胞壁相关组分含量QTL定位及候选基因分析[J]. 植物学报, 2023, 58(6): 882-892. |
| [14] | 戴若惠, 钱心妤, 孙静蕾, 芦涛, 贾绮玮, 陆天麒, 路梅, 饶玉春. 水稻叶色调控机制及相关基因研究进展[J]. 植物学报, 2023, 58(5): 799-812. |
| [15] | 田传玉, 方妍力, 沈晴, 王宏杰, 陈析丰, 郭威, 赵开军, 王春连, 纪志远. 2019-2021年我国南方稻区白叶枯病菌的毒力与遗传多样性调查研究[J]. 植物学报, 2023, 58(5): 743-749. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||