

植物学报 ›› 2024, Vol. 59 ›› Issue (2): 217-230.DOI: 10.11983/CBB23161 cstr: 32102.14.CBB23161
朱超宇1, 胡程翔1, 朱哲楠1, 张芷宁1, 汪理海1, 陈钧1, 李三峰2, 连锦瑾1, 唐璐瑶1, 钟芊芊1, 殷文晶1, 王跃星2,*( ), 饶玉春1,*(
), 饶玉春1,*( )
)
收稿日期:2023-12-04
接受日期:2024-01-30
出版日期:2024-03-10
发布日期:2024-03-10
通讯作者:
* E-mail: 基金资助:
Chaoyu Zhu1, Chengxiang Hu1, Zhenan Zhu1, Zhining Zhang1, Lihai Wang1, Jun Chen1, Sanfeng Li2, Jinjin Lian1, Luyao Tang1, Qianqian Zhong1, Wenjing Yin1, Yuexing Wang2,*( ), Yuchun Rao1,*(
), Yuchun Rao1,*( )
)
Received:2023-12-04
Accepted:2024-01-30
Online:2024-03-10
Published:2024-03-10
Contact:
* E-mail: 摘要: 水稻(Oryza sativa)穗部性状与产量直接相关, 其相关基因的挖掘与功能解析对于保障国家粮食安全意义重大。以籼稻华占(HZ)和粳稻热研2号(Nekken2)及构建的120个重组自交系(RILs)为实验材料, 测定了穗长、每穗粒数、结实率、柱头外露率及一次枝梗数等穗部性状。结合高密度分子遗传图谱进行QTL定位, 结果共检测到31个QTLs, 分别位于第1、2、3、4、5、6、10和11号染色体上, 其中2个位点的LOD值分别高达5.45与5.28。通过分析筛选QTL区间内可能影响穗部性状的相关基因, 并利用qRT-PCR进行基因表达检测, 发现LOC_Os05g05490、LOC_Os05g06150、LOC_Os03g11700、LOC_Os03g12430、LOC_Os05g28720、LOC_Os05g30890、LOC_Os05g31740和LOC_Os02g17880在双亲间的表达水平差异显著。其中, 前5个基因编码三角状五肽重复蛋白, 而后3个基因编码糖基转移酶。研究挖掘到31个与穗部性状相关的QTLs, 为进一步定位和克隆相关基因, 从而选育高产水稻新品种奠定理论基础。
朱超宇, 胡程翔, 朱哲楠, 张芷宁, 汪理海, 陈钧, 李三峰, 连锦瑾, 唐璐瑶, 钟芊芊, 殷文晶, 王跃星, 饶玉春. 水稻穗部性状QTL定位及候选基因分析. 植物学报, 2024, 59(2): 217-230.
Chaoyu Zhu, Chengxiang Hu, Zhenan Zhu, Zhining Zhang, Lihai Wang, Jun Chen, Sanfeng Li, Jinjin Lian, Luyao Tang, Qianqian Zhong, Wenjing Yin, Yuexing Wang, Yuchun Rao. Mapping of QTLs Associated with Rice Panicle Traits and Candidate Gene Analysis. Chinese Bulletin of Botany, 2024, 59(2): 217-230.
| Traits | Primer name | Forward primer (5′-3′) | Reverse primer (5′-3′) |
|---|---|---|---|
| OsActin | TGGCATCTCAGCACATTCC | TGCACAATGGATGGGTCAGA | |
| Panicle length | LOC_Os03g08900 | CTCTTCTTCTGGGTGCTCAT | GATGCCGATGAGGTAGTAGG |
| LOC_Os03g10210 | AGTTGCTGAAGCTGAAGGAG | CGCGTACTCACTCATGTAGG | |
| LOC_Os03g10520 | GCTCCTTATCCGACTCAAGA | AACCAACTGCTCCACAATTT | |
| LOC_Os03g10620 | GACAACCTCGACGCCTAC | ACCTGCTGTATCTCCTCCAG | |
| LOC_Os03g11670 | TGTACAACATGCTCCTTTCG | CGAGGTGAGGATGAAGATGT | |
| LOC_Os03g12120 | TGCAGATTCTTCTGGACAAA | AACTGAAGATAGCGGGACTG | |
| Grain number per panicle | LOC_Os05g30890 | GAATCTCGGTGGCATCAAAT | TCATGCTGAGAGCCATCTTG |
| LOC_Os05g31040 | CTGACATTGCCACAACCATC | GTTCTCCTCCTGAAGCATCG | |
| LOC_Os05g31280 | GAACAACTCGTCGTCGTCGT | GACTGCTTGTTGCCGTAGGT | |
| LOC_Os05g31380 | CAGTTCCATCCGGTGCTATC | CATCGTATGCTGGCTGGAT | |
| LOC_Os05g31740 | GACTGGTACACCGACCTGCT | TAGGACGCCACTTTCCTCTG | |
| Seed-setting rate | LOC_Os02g50240 | CACCAACAAGAGGCACAATG | TCAACAATATCACGCCCAAA |
| LOC_Os02g50740 | GTTCCCTTCCTTCCCTTGAC | TGCTGCTCCTCCTGACTTCT | |
| LOC_Os02g52990 | CGATCGCCTACCTCTACCAC | CACGGGGAGAAGGAGTAGTG | |
| LOC_Os02g53680 | GAAATTGAGGATGCCCAGAA | CACAGGAAAGATCCCACGTT | |
| LOC_Os02g54910 | CCAGAAATCGAAGGAACCAA | CTCGCGGAAGTATCCATCAT | |
| LOC_Os02g56140 | GCTCCAGGTCCAGATGTTGT | CGGTTGCATTAGGAACACCT | |
| Stigma exsertion rate | LOC_Os05g02870 | CAAGCTCAAGGTGCTCGAC | TTGTTCTTGCTGCCAAAGTG |
| LOC_Os05g04160 | GTGCGGGTCTACAATCACCT | ACTATCGTCCCATGGCTCAC | |
| LOC_Os05g04770 | ATCCGTCTGTGTTTCGATCC | TATTCGTCCATCCGAAAAGC | |
| LOC_Os05g05280 | GCCACCAGCCTAATCATTGT | GCACAAGAACCGACAAGACA | |
| LOC_Os05g05490 | TGGAGTTCTTCCAAGGATGG | GCAACACATAAGCAGCCGTA | |
| LOC_Os05g06150 | GGAAATTGTTTGGGGTGATG | GCATCCCTTGCTTCAACAAT | |
| LOC_Os05g08350 | ATCCCCGGAGATTCGATTAC | AGTAAATTTCCACCGCAACG | |
| Number of primary branches | LOC_Os05g09740 | AACATCATCGGCTGGAAGAC | TGCCTGATCATATGGCTGAA |
| LOC_Os05g10210 | AACATCATCGGCTGGAAGAC | GCGTGGTCGTATGGCTTAGT | |
| LOC_Os05g10370 | CTGCTCCTCCTCCTCTTCCT | ATCACGTTGTGTGCCTCCAC | |
| LOC_Os05g10690 | CTTGACAAGCTCGGAAAAGG | TGCAGTTCAGATGCAGATCC | |
| LOC_Os05g10770 | TGGTGCTGATCTGGAGACTG | GTCTTCAACATGCCAGCAGA | |
| Number of secondary branches | LOC_Os03g11614 | CTCTTCGAGTTCTCCAGCTC | CCAGCTCCTTCATGCTTAGT |
| LOC_Os03g11700 | ACTGCTCCTATGCCGTATCG | AGGTACAAGCGGCTTCCTTT | |
| LOC_Os03g11790 | TGAATCGGCTCATGTCCATA | GGCAGCATCTTGAAGTGACA | |
| LOC_Os03g12430 | GATGACAGGAAGAGGGGTGA | TCATCATCCTCTGCACAAGC | |
| LOC_Os03g12940 | ACCCTGCCAGAGCTTGTAGA | TCTGCTGTGCTCACCTCATC | |
| LOC_Os05g28720 | TGATCCAAGAGATGGGGAAG | GCATTCTGTCGAACACCTGA | |
| Thousand grain weight | LOC_Os02g17880 | TGTACGCCTACTCGAGCATA | ATCCTGAAGTTGTCCTGCAC |
| LOC_Os02g18820 | TCTCAGCCTTCAGAACATGA | TCTAGAACTGCCGACATCAA | |
| LOC_Os02g19820 | CATGAAAATGGTGATCCAGA | GCTGCGTCGACTTGTAGTAG | |
| LOC_Os02g29530 | ACTTCTCGCACCCACTTATC | ATTTTTCCAGAGGCTTTGTG | |
| LOC_Os02g30850 | GTGAAGCGCCTCTTCTGC | GAGGGAGCCGTTGATGTG | |
| LOC_Os02g32370 | GCAAACCCTGTACCACTTCT | AAGACGCCTTCTCGGTAAC | |
| LOC_Os05g11700 | CGGACTCGGTGAGCTATAAT | CAACAAATTTCCTCACACCA | |
| Grain length | LOC_Os03g44430 | CAAGGGTCGGAAGACAGTTC | TATCATCCGCATCCTCATCA |
| LOC_Os03g44500 | GTGTGTGATGGATGGCTTTG | GTTGGAGTCGGTGGTCTGTT | |
| LOC_Os03g45830 | GATGTGGAGGAGGAAGAGCA | GCCGTACCCGTACTGCTG | |
| LOC_Os03g46180 | AAGAGGAGGAGGAGGAGCAG | CTTCTTCAGCACGCTGTTCA | |
| LOC_Os03g46560 | GGCTTTGTCCAAAAATGGAA | CTGGCAGTCCAAAGACATCA | |
| Grain width | LOC_Os02g30910 | TCACCTTTGGCATCTTAGGT | AGGTACATGGCGAGGTAGAC |
| LOC_Os06g10950 | GTACCAATCCGCCATGTACC | GCGGTAGGAGATGGAGACAA | |
| LOC_Os06g10960 | ACCTCGACCACGACTACACC | GACCCCAGATGTGGTTGTTC | |
| LOC_Os06g11270 | GGTTACCAAGAACGGACGAA | CACCTGCGATCCTACCTTGT | |
| LOC_Os06g09270 | TACGTGCTGTCCTGGGATCT | AAGGTTTGTGCCCATCTCAG |
表1 qRT-PCR引物序列
Table 1 Primer sequences for qRT-PCR
| Traits | Primer name | Forward primer (5′-3′) | Reverse primer (5′-3′) |
|---|---|---|---|
| OsActin | TGGCATCTCAGCACATTCC | TGCACAATGGATGGGTCAGA | |
| Panicle length | LOC_Os03g08900 | CTCTTCTTCTGGGTGCTCAT | GATGCCGATGAGGTAGTAGG |
| LOC_Os03g10210 | AGTTGCTGAAGCTGAAGGAG | CGCGTACTCACTCATGTAGG | |
| LOC_Os03g10520 | GCTCCTTATCCGACTCAAGA | AACCAACTGCTCCACAATTT | |
| LOC_Os03g10620 | GACAACCTCGACGCCTAC | ACCTGCTGTATCTCCTCCAG | |
| LOC_Os03g11670 | TGTACAACATGCTCCTTTCG | CGAGGTGAGGATGAAGATGT | |
| LOC_Os03g12120 | TGCAGATTCTTCTGGACAAA | AACTGAAGATAGCGGGACTG | |
| Grain number per panicle | LOC_Os05g30890 | GAATCTCGGTGGCATCAAAT | TCATGCTGAGAGCCATCTTG |
| LOC_Os05g31040 | CTGACATTGCCACAACCATC | GTTCTCCTCCTGAAGCATCG | |
| LOC_Os05g31280 | GAACAACTCGTCGTCGTCGT | GACTGCTTGTTGCCGTAGGT | |
| LOC_Os05g31380 | CAGTTCCATCCGGTGCTATC | CATCGTATGCTGGCTGGAT | |
| LOC_Os05g31740 | GACTGGTACACCGACCTGCT | TAGGACGCCACTTTCCTCTG | |
| Seed-setting rate | LOC_Os02g50240 | CACCAACAAGAGGCACAATG | TCAACAATATCACGCCCAAA |
| LOC_Os02g50740 | GTTCCCTTCCTTCCCTTGAC | TGCTGCTCCTCCTGACTTCT | |
| LOC_Os02g52990 | CGATCGCCTACCTCTACCAC | CACGGGGAGAAGGAGTAGTG | |
| LOC_Os02g53680 | GAAATTGAGGATGCCCAGAA | CACAGGAAAGATCCCACGTT | |
| LOC_Os02g54910 | CCAGAAATCGAAGGAACCAA | CTCGCGGAAGTATCCATCAT | |
| LOC_Os02g56140 | GCTCCAGGTCCAGATGTTGT | CGGTTGCATTAGGAACACCT | |
| Stigma exsertion rate | LOC_Os05g02870 | CAAGCTCAAGGTGCTCGAC | TTGTTCTTGCTGCCAAAGTG |
| LOC_Os05g04160 | GTGCGGGTCTACAATCACCT | ACTATCGTCCCATGGCTCAC | |
| LOC_Os05g04770 | ATCCGTCTGTGTTTCGATCC | TATTCGTCCATCCGAAAAGC | |
| LOC_Os05g05280 | GCCACCAGCCTAATCATTGT | GCACAAGAACCGACAAGACA | |
| LOC_Os05g05490 | TGGAGTTCTTCCAAGGATGG | GCAACACATAAGCAGCCGTA | |
| LOC_Os05g06150 | GGAAATTGTTTGGGGTGATG | GCATCCCTTGCTTCAACAAT | |
| LOC_Os05g08350 | ATCCCCGGAGATTCGATTAC | AGTAAATTTCCACCGCAACG | |
| Number of primary branches | LOC_Os05g09740 | AACATCATCGGCTGGAAGAC | TGCCTGATCATATGGCTGAA |
| LOC_Os05g10210 | AACATCATCGGCTGGAAGAC | GCGTGGTCGTATGGCTTAGT | |
| LOC_Os05g10370 | CTGCTCCTCCTCCTCTTCCT | ATCACGTTGTGTGCCTCCAC | |
| LOC_Os05g10690 | CTTGACAAGCTCGGAAAAGG | TGCAGTTCAGATGCAGATCC | |
| LOC_Os05g10770 | TGGTGCTGATCTGGAGACTG | GTCTTCAACATGCCAGCAGA | |
| Number of secondary branches | LOC_Os03g11614 | CTCTTCGAGTTCTCCAGCTC | CCAGCTCCTTCATGCTTAGT |
| LOC_Os03g11700 | ACTGCTCCTATGCCGTATCG | AGGTACAAGCGGCTTCCTTT | |
| LOC_Os03g11790 | TGAATCGGCTCATGTCCATA | GGCAGCATCTTGAAGTGACA | |
| LOC_Os03g12430 | GATGACAGGAAGAGGGGTGA | TCATCATCCTCTGCACAAGC | |
| LOC_Os03g12940 | ACCCTGCCAGAGCTTGTAGA | TCTGCTGTGCTCACCTCATC | |
| LOC_Os05g28720 | TGATCCAAGAGATGGGGAAG | GCATTCTGTCGAACACCTGA | |
| Thousand grain weight | LOC_Os02g17880 | TGTACGCCTACTCGAGCATA | ATCCTGAAGTTGTCCTGCAC |
| LOC_Os02g18820 | TCTCAGCCTTCAGAACATGA | TCTAGAACTGCCGACATCAA | |
| LOC_Os02g19820 | CATGAAAATGGTGATCCAGA | GCTGCGTCGACTTGTAGTAG | |
| LOC_Os02g29530 | ACTTCTCGCACCCACTTATC | ATTTTTCCAGAGGCTTTGTG | |
| LOC_Os02g30850 | GTGAAGCGCCTCTTCTGC | GAGGGAGCCGTTGATGTG | |
| LOC_Os02g32370 | GCAAACCCTGTACCACTTCT | AAGACGCCTTCTCGGTAAC | |
| LOC_Os05g11700 | CGGACTCGGTGAGCTATAAT | CAACAAATTTCCTCACACCA | |
| Grain length | LOC_Os03g44430 | CAAGGGTCGGAAGACAGTTC | TATCATCCGCATCCTCATCA |
| LOC_Os03g44500 | GTGTGTGATGGATGGCTTTG | GTTGGAGTCGGTGGTCTGTT | |
| LOC_Os03g45830 | GATGTGGAGGAGGAAGAGCA | GCCGTACCCGTACTGCTG | |
| LOC_Os03g46180 | AAGAGGAGGAGGAGGAGCAG | CTTCTTCAGCACGCTGTTCA | |
| LOC_Os03g46560 | GGCTTTGTCCAAAAATGGAA | CTGGCAGTCCAAAGACATCA | |
| Grain width | LOC_Os02g30910 | TCACCTTTGGCATCTTAGGT | AGGTACATGGCGAGGTAGAC |
| LOC_Os06g10950 | GTACCAATCCGCCATGTACC | GCGGTAGGAGATGGAGACAA | |
| LOC_Os06g10960 | ACCTCGACCACGACTACACC | GACCCCAGATGTGGTTGTTC | |
| LOC_Os06g11270 | GGTTACCAAGAACGGACGAA | CACCTGCGATCCTACCTTGT | |
| LOC_Os06g09270 | TACGTGCTGTCCTGGGATCT | AAGGTTTGTGCCCATCTCAG |
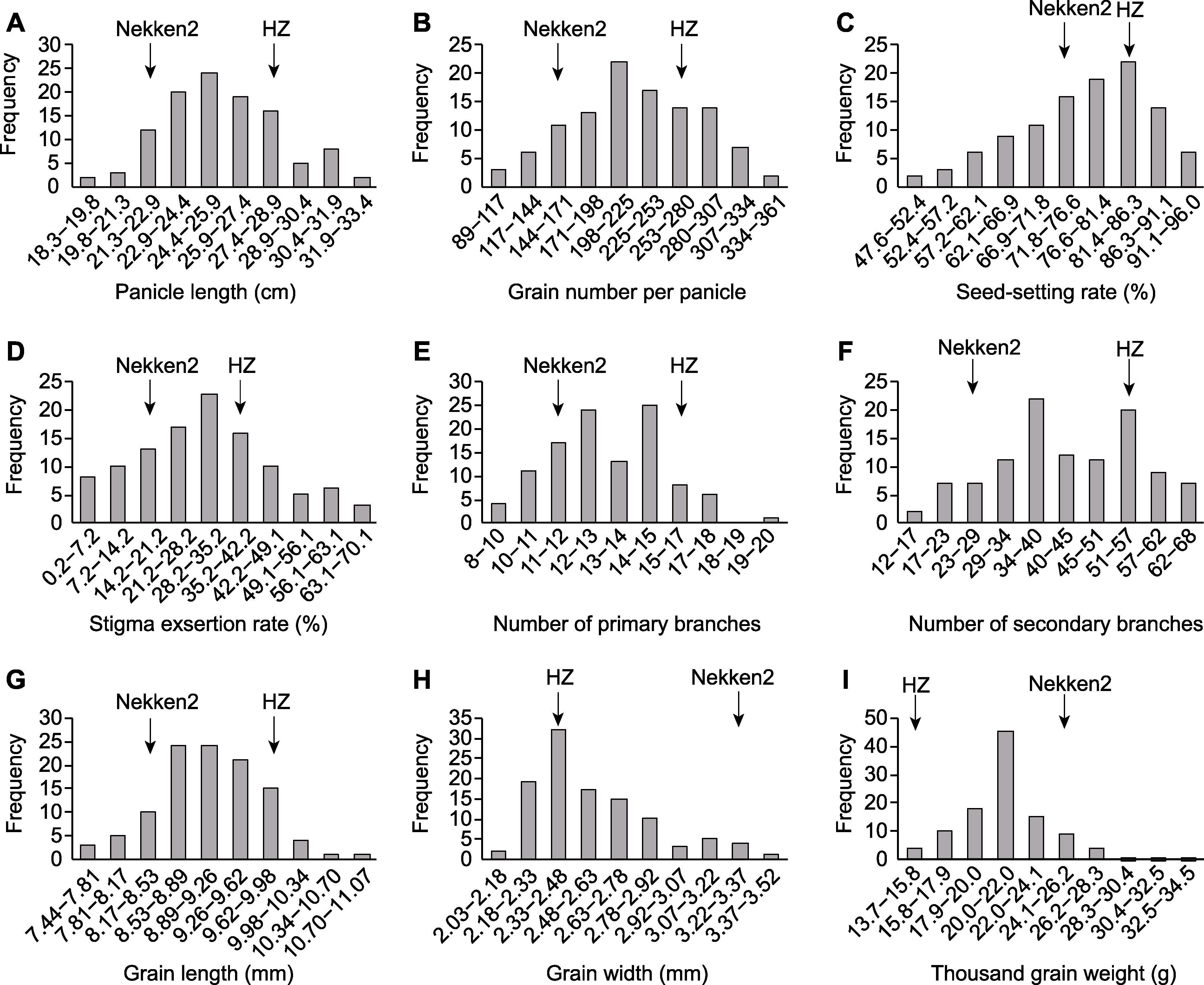
图1 水稻重组自交系穗部性状分布 (A) 穗长; (B) 每穗粒数; (C) 结实率; (D) 柱头外露率; (E) 一次枝梗数; (F) 二次枝梗数; (G) 粒长; (H) 粒宽; (I) 千粒重
Figure 1 Distribution of panicle traits in recombinant inbred lines of rice (A) Panicle length; (B) Grain number per panicle; (C) Seed-setting rate; (D) Stigma exsertion rate; (E) Number of primary branches; (F) Number of secondary branches; (G) Grain length; (H) Grain width; (I) Thousand grain weight

图2 父本华占与母本热研2号表型 (A) 成熟期水稻植株表型(bar=10 cm); (B) 成熟期水稻穗部对比(bar=2 cm)
Figure 2 Phenotypes of the male parent HZ and the female parent Nekken2 (A) Phenotype of mature rice plants (bar=10 cm); (B) Comparison of mature rice panicles (bar=2 cm)
| Traits | QTL | Chromosome | Physical distance (bp) | Position of support (cM) | Limit of detection (LOD) |
|---|---|---|---|---|---|
| Panicle length | qPL3.1 | 3 | 4514694-9883047 | 19.35-42.37 | 5.45 |
| qPL5.1 | 5 | 16473540-17216667 | 70.62-73.80 | 2.54 | |
| qPL11.1 | 11 | 17453046-17570705 | 74.82-75.32 | 2.29 | |
| qPL11.2 | 11 | 26791773-26969035 | 114.85-115.61 | 2.61 | |
| Grain number per panicle | qGNPP3.1 | 3 | 6470188-15088477 | 27.74-64.68 | 2.21 |
| qGNPP5.1 | 5 | 16020128-18858682 | 68.67-80.84 | 2.36 | |
| qGNPP5.2 | 5 | 27007193-27230204 | 115.77-116.73 | 2.41 | |
| Seed-setting rate | qSSR1.1 | 1 | 7203302-7421463 | 30.88-31.81 | 2.41 |
| qSSR2.1 | 2 | 30004559-34415035 | 128.62-147.53 | 3.47 | |
| qSSR10.1 | 10 | 5567413-15675357 | 23.87-67.20 | 2.78 | |
| Stigma exsertion rate | qSER5.1 | 5 | 926888-4584013 | 3.97-19.65 | 3.25 |
| qSER10.1 | 10 | 10115736-11156109 | 43.36-47.82 | 2.41 | |
| Number of primary branches | qNPB1.1 | 1 | 25791086-25812159 | 110.56-110.65 | 2.24 |
| qNPB5.1 | 5 | 5350624-6230055 | 22.94-26.71 | 2.66 | |
| Number of secondary branches | qNSB2.1 | 2 | 34269184-34415035 | 146.90-147.16 | 2.25 |
| qNSB3.1 | 3 | 5859763-7034710 | 25.00-30.16 | 3.18 | |
| qNSB5.1 | 5 | 14979844-15200902 | 64.21-65.16 | 2.13 | |
| qNSB5.2 | 5 | 15944247-17819466 | 68.35-76.39 | 3.06 | |
| qNSB6.1 | 6 | 4263281-5187517 | 18.28-22.24 | 2.88 | |
| qNSB11.1 | 11 | 4416803-4813122 | 18.93-20.63 | 2.47 | |
| Grain length | qGL3.1 | 3 | 24904943-26519283 | 106.76-113.68 | 2.26 |
| qGL6.1 | 6 | 23905310-24717124 | 102.48-105.96 | 2.14 | |
| Grain width | qGW2.1 | 2 | 7512377-21714903 | 32.20-93.09 | 3.61 |
| qGW3.1 | 3 | 10039327-11005670 | 43.04-47.18 | 2.24 | |
| qGW4.1 | 4 | 19218278-19642144 | 82.38-84.20 | 3.28 | |
| qGW5.1 | 5 | 3697712-6833061 | 15.85-29.29 | 3.46 | |
| qGW5.2 | 5 | 28202582-29177297 | 120.90-125.16 | 3.27 | |
| qGW6.1 | 6 | 4227455-6069227 | 18.12-26.02 | 3.77 | |
| Thousand grain weight | qTGW1.1 | 1 | 6064691-6235269 | 26.00-26.73 | 2.80 |
| qTGW2.1 | 2 | 9429264-21032980 | 40.42-90.16 | 5.28 | |
| qTGW5.1 | 5 | 6526040-6673100 | 27.98-28.61 | 2.11 |
表2 水稻穗部性状QTL分析
Table 2 QTL analysis of rice panicle traits
| Traits | QTL | Chromosome | Physical distance (bp) | Position of support (cM) | Limit of detection (LOD) |
|---|---|---|---|---|---|
| Panicle length | qPL3.1 | 3 | 4514694-9883047 | 19.35-42.37 | 5.45 |
| qPL5.1 | 5 | 16473540-17216667 | 70.62-73.80 | 2.54 | |
| qPL11.1 | 11 | 17453046-17570705 | 74.82-75.32 | 2.29 | |
| qPL11.2 | 11 | 26791773-26969035 | 114.85-115.61 | 2.61 | |
| Grain number per panicle | qGNPP3.1 | 3 | 6470188-15088477 | 27.74-64.68 | 2.21 |
| qGNPP5.1 | 5 | 16020128-18858682 | 68.67-80.84 | 2.36 | |
| qGNPP5.2 | 5 | 27007193-27230204 | 115.77-116.73 | 2.41 | |
| Seed-setting rate | qSSR1.1 | 1 | 7203302-7421463 | 30.88-31.81 | 2.41 |
| qSSR2.1 | 2 | 30004559-34415035 | 128.62-147.53 | 3.47 | |
| qSSR10.1 | 10 | 5567413-15675357 | 23.87-67.20 | 2.78 | |
| Stigma exsertion rate | qSER5.1 | 5 | 926888-4584013 | 3.97-19.65 | 3.25 |
| qSER10.1 | 10 | 10115736-11156109 | 43.36-47.82 | 2.41 | |
| Number of primary branches | qNPB1.1 | 1 | 25791086-25812159 | 110.56-110.65 | 2.24 |
| qNPB5.1 | 5 | 5350624-6230055 | 22.94-26.71 | 2.66 | |
| Number of secondary branches | qNSB2.1 | 2 | 34269184-34415035 | 146.90-147.16 | 2.25 |
| qNSB3.1 | 3 | 5859763-7034710 | 25.00-30.16 | 3.18 | |
| qNSB5.1 | 5 | 14979844-15200902 | 64.21-65.16 | 2.13 | |
| qNSB5.2 | 5 | 15944247-17819466 | 68.35-76.39 | 3.06 | |
| qNSB6.1 | 6 | 4263281-5187517 | 18.28-22.24 | 2.88 | |
| qNSB11.1 | 11 | 4416803-4813122 | 18.93-20.63 | 2.47 | |
| Grain length | qGL3.1 | 3 | 24904943-26519283 | 106.76-113.68 | 2.26 |
| qGL6.1 | 6 | 23905310-24717124 | 102.48-105.96 | 2.14 | |
| Grain width | qGW2.1 | 2 | 7512377-21714903 | 32.20-93.09 | 3.61 |
| qGW3.1 | 3 | 10039327-11005670 | 43.04-47.18 | 2.24 | |
| qGW4.1 | 4 | 19218278-19642144 | 82.38-84.20 | 3.28 | |
| qGW5.1 | 5 | 3697712-6833061 | 15.85-29.29 | 3.46 | |
| qGW5.2 | 5 | 28202582-29177297 | 120.90-125.16 | 3.27 | |
| qGW6.1 | 6 | 4227455-6069227 | 18.12-26.02 | 3.77 | |
| Thousand grain weight | qTGW1.1 | 1 | 6064691-6235269 | 26.00-26.73 | 2.80 |
| qTGW2.1 | 2 | 9429264-21032980 | 40.42-90.16 | 5.28 | |
| qTGW5.1 | 5 | 6526040-6673100 | 27.98-28.61 | 2.11 |
| Traits | Gene ID | Function |
|---|---|---|
| Panicle length | LOC_Os03g08900 | Multi antimicrobial extrusion protein |
| LOC_Os03g10210 | Helix-turn-helix motif | |
| LOC_Os03g10520 | Radical SAM enzyme | |
| LOC_Os03g10620 | Strigolactone receptor | |
| LOC_Os03g11670 | PPR domain containing protein | |
| LOC_Os03g12120 | NAC domain | |
| Grain number per panicle | LOC_Os05g30890 | CYP450 |
| LOC_Os05g31040 | Cytokinin oxidase/dehydrogenase | |
| LOC_Os05g31280 | Gibberellin regulated protein | |
| LOC_Os05g31380 | Transcription factor GRAS domain containing protein | |
| LOC_Os05g31740 | Cytochrome P450 86A2 | |
| Seed-setting rate | LOC_Os02g50240 | Glutamine synthetase |
| LOC_Os02g50740 | WD40 repeat containing protein | |
| LOC_Os02g52990 | Auxin responsive SAUR protein family protein | |
| LOC_Os02g53680 | Replication protein A | |
| LOC_Os02g54910 | WD40 repeat containing protein | |
| LOC_Os02g56140 | MYC-type, bHLH domain | |
| Stigma exsertion rate | LOC_Os05g02870 | Similar to stigma/style ABC transporter |
| LOC_Os05g04160 | Pentatricopeptide containing protein | |
| LOC_Os05g04770 | F-box domain, cyclin-like domain containing protein | |
| LOC_Os05g05280 | RING-type E3 ubiquitin ligase | |
| LOC_Os05g05490 | Pentatricopeptide | |
| LOC_Os05g06150 | Pentatricopeptide repeat domain containing protein | |
| LOC_Os05g08350 | F-box domain, cyclin-like domain containing protein | |
| Number of primary branches | LOC_Os05g09740 | HAD superfamily phosphatase |
| LOC_Os05g10210 | HAD superfamily phosphatase | |
| LOC_Os05g10370 | Acid phosphatase | |
| LOC_Os05g10690 | Similar to transcription factor MYBS2 | |
| LOC_Os05g10770 | Histone H3K4 demethylase | |
| Number of secondary branches | LOC_Os03g11614 | MADS-domain transcription factor |
| LOC_Os03g11700 | PPR repeat containing protein | |
| LOC_Os03g11790 | Cyclin-like F-box domain containing protein | |
| LOC_Os03g12430 | PPR repeat containing protein | |
| LOC_Os03g12940 | Cyclin-like F-box domain containing protein | |
| LOC_Os05g28720 | Pentatricopeptide repeat domain containing protein | |
| Thousand grain weight | LOC_Os02g17880 | Glycosyl hydrolases family 16 |
| LOC_Os02g18820 | WD40/YVTN repeat-like domain containing protein | |
| LOC_Os02g19820 | RAG1-activating protein 1 homologue domain containing protein | |
| LOC_Os02g29530 | Glycosyl transferase family 8 | |
| LOC_Os02g30850 | CC-type glutaredoxin | |
| LOC_Os02g32370 | Inositol polyphosphate kinase | |
| LOC_Os05g11700 | Pentatrico peptide repeat (PPR) protein | |
| Grain length | LOC_Os03g44430 | Ubiquitin carboxy-terminal hydrolase family 1 |
| LOC_Os03g44500 | Ser/Thr phosphatase; cryptic inhibitor of cytokinin phosphorelay | |
| LOC_Os03g45830 | Auxin responsive SAUR protein domain containing protein | |
| LOC_Os03g46180 | Plant lipid transfer protein/seed storage/trypsin-alpha amylase inhibitor domain containing protein | |
| Grain length | LOC_Os03g46560 | RPD1 |
| Grain width | LOC_Os02g30910 | Sugar transporter |
| LOC_Os06g10950 | Similar to glycosyltransferase family 37 | |
| LOC_Os06g10960 | Similar to glycosyltransferase family 37 | |
| LOC_Os06g11270 | UDP-glucuronosyl | |
| LOC_Os06g09270 | Hypro1 |
表3 水稻穗部性状相关候选基因的功能
Table 3 The functions of candidate genes associated with rice panicle traits
| Traits | Gene ID | Function |
|---|---|---|
| Panicle length | LOC_Os03g08900 | Multi antimicrobial extrusion protein |
| LOC_Os03g10210 | Helix-turn-helix motif | |
| LOC_Os03g10520 | Radical SAM enzyme | |
| LOC_Os03g10620 | Strigolactone receptor | |
| LOC_Os03g11670 | PPR domain containing protein | |
| LOC_Os03g12120 | NAC domain | |
| Grain number per panicle | LOC_Os05g30890 | CYP450 |
| LOC_Os05g31040 | Cytokinin oxidase/dehydrogenase | |
| LOC_Os05g31280 | Gibberellin regulated protein | |
| LOC_Os05g31380 | Transcription factor GRAS domain containing protein | |
| LOC_Os05g31740 | Cytochrome P450 86A2 | |
| Seed-setting rate | LOC_Os02g50240 | Glutamine synthetase |
| LOC_Os02g50740 | WD40 repeat containing protein | |
| LOC_Os02g52990 | Auxin responsive SAUR protein family protein | |
| LOC_Os02g53680 | Replication protein A | |
| LOC_Os02g54910 | WD40 repeat containing protein | |
| LOC_Os02g56140 | MYC-type, bHLH domain | |
| Stigma exsertion rate | LOC_Os05g02870 | Similar to stigma/style ABC transporter |
| LOC_Os05g04160 | Pentatricopeptide containing protein | |
| LOC_Os05g04770 | F-box domain, cyclin-like domain containing protein | |
| LOC_Os05g05280 | RING-type E3 ubiquitin ligase | |
| LOC_Os05g05490 | Pentatricopeptide | |
| LOC_Os05g06150 | Pentatricopeptide repeat domain containing protein | |
| LOC_Os05g08350 | F-box domain, cyclin-like domain containing protein | |
| Number of primary branches | LOC_Os05g09740 | HAD superfamily phosphatase |
| LOC_Os05g10210 | HAD superfamily phosphatase | |
| LOC_Os05g10370 | Acid phosphatase | |
| LOC_Os05g10690 | Similar to transcription factor MYBS2 | |
| LOC_Os05g10770 | Histone H3K4 demethylase | |
| Number of secondary branches | LOC_Os03g11614 | MADS-domain transcription factor |
| LOC_Os03g11700 | PPR repeat containing protein | |
| LOC_Os03g11790 | Cyclin-like F-box domain containing protein | |
| LOC_Os03g12430 | PPR repeat containing protein | |
| LOC_Os03g12940 | Cyclin-like F-box domain containing protein | |
| LOC_Os05g28720 | Pentatricopeptide repeat domain containing protein | |
| Thousand grain weight | LOC_Os02g17880 | Glycosyl hydrolases family 16 |
| LOC_Os02g18820 | WD40/YVTN repeat-like domain containing protein | |
| LOC_Os02g19820 | RAG1-activating protein 1 homologue domain containing protein | |
| LOC_Os02g29530 | Glycosyl transferase family 8 | |
| LOC_Os02g30850 | CC-type glutaredoxin | |
| LOC_Os02g32370 | Inositol polyphosphate kinase | |
| LOC_Os05g11700 | Pentatrico peptide repeat (PPR) protein | |
| Grain length | LOC_Os03g44430 | Ubiquitin carboxy-terminal hydrolase family 1 |
| LOC_Os03g44500 | Ser/Thr phosphatase; cryptic inhibitor of cytokinin phosphorelay | |
| LOC_Os03g45830 | Auxin responsive SAUR protein domain containing protein | |
| LOC_Os03g46180 | Plant lipid transfer protein/seed storage/trypsin-alpha amylase inhibitor domain containing protein | |
| Grain length | LOC_Os03g46560 | RPD1 |
| Grain width | LOC_Os02g30910 | Sugar transporter |
| LOC_Os06g10950 | Similar to glycosyltransferase family 37 | |
| LOC_Os06g10960 | Similar to glycosyltransferase family 37 | |
| LOC_Os06g11270 | UDP-glucuronosyl | |
| LOC_Os06g09270 | Hypro1 |
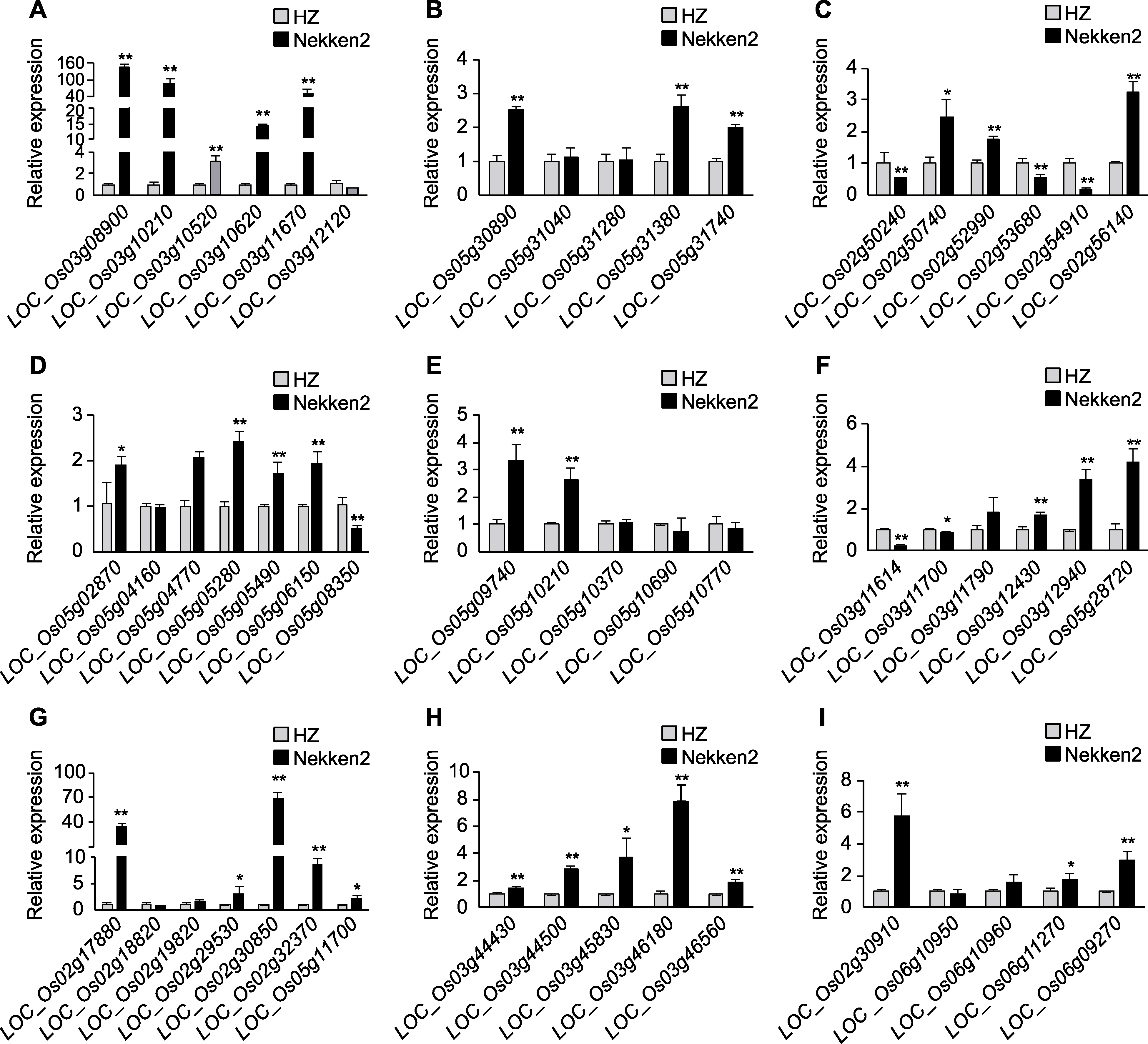
图4 水稻穗部性状候选基因表达差异 (A) 穗长; (B) 每穗粒数; (C) 结实率; (D) 柱头外露率; (E) 一次枝梗数; (F) 二次枝梗数; (G) 千粒重; (H) 粒长; (I) 粒宽。* P<0.05; ** P<0.01
Figure 4 Differences in the expression of candidate genes for panicle traits in rice (A) Panicle length; (B) Grain number per panicle; (C) Seed-setting rate; (D) Stigma exsertion rate; (E) Number of primary branches; (F) Number of secondary branches; (G) Thousand grain weight; (H) Grain length; (I) Grain width. * P<0.05; ** P<0.01
| [1] | Ashikari M, Sakakibara H, Lin SY, Yamamoto T, Takashi T, Nishimura A, Angeles ER, Qian Q, Kitano H, Matsuoka M (2005). Cytokinin oxidase regulates rice grain production. Science 309, 741-745. |
| [2] |
Chang YX, Gong L, Yuan WY, Li XW, Chen GX, Li XH, Zhang QF, Wu CY (2009). Replication protein A (RPA1a) is required for meiotic and somatic DNA repair but is dispensable for DNA replication and homologous recombination in rice. Plant Physiol 151, 2162-2173.
DOI PMID |
| [3] | Chen YNJ, Zhang XZ, Tai DW, Zhang PS, Gao GJ, Zhang QL, He YQ (2022). Mapping QTLs for rice stigma exsertion rate using recombinant inbred line population. Mol Plant Breed 20, 1198-1205. (in Chinese) |
| 陈应南君, 张效忠, 台德卫, 张培申, 高冠军, 张庆路, 何予卿 (2022). 利用重组自交系群体定位水稻柱头外露率QTL. 分子植物育种 20, 1198-1205. | |
| [4] |
Duan JB, Yu H, Yuan K, Liao ZB, Meng XB, Jing YH, Liu GF, Chu JF, Li JY (2019). Strigolactone promotes cytokinin degradation through transcriptional activation of CYTOKININ OXIDASE/DEHYDROGENASE 9 in rice. Proc Natl Acad Sci USA 116, 14319-14324.
DOI URL |
| [5] | Fan CC, Xing YZ, Mao HL, Lu TT, Han B, Xu CG, Li XH, Zhang QF (2006). GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theor Appl Genet 112, 1164-1171. |
| [6] |
Hao JQ, Wang DK, Wu YB, Huang K, Duan PG, Li N, Xu R, Zeng DL, Dong GJ, Zhang BL, Zhang LM, Inzé D, Qian Q, Li YH (2021). The GW2-WG1-OsbZIP47 pathway controls grain size and weight in rice. Mol Plant 14, 1266-1280.
DOI PMID |
| [7] |
Ikeda M, Miura K, Aya K, Kitano H, Matsuoka M (2013). Genes offering the potential for designing yield-related traits in rice. Curr Opin Plant Biol 16, 213-220.
DOI PMID |
| [8] | Jin JY, Luo YT, Yang HM, Lu T, Ye HF, Xie JY, Wang KX, Chen QY, Fang Y, Wang YX, Rao YC (2023). QTL mapping and expression analysis on candidate genes related to chlorophyll content in rice. Chin Bull Bot 58, 394-403. (in Chinese) |
| 金佳怡, 罗怿婷, 杨惠敏, 芦涛, 叶涵斐, 谢继毅, 王珂欣, 陈芊羽, 方媛, 王跃星, 饶玉春 (2023). 水稻叶绿素含量QTL定位与候选基因表达分析. 植物学报 58, 394-403. | |
| [9] |
Komatsu M, Maekawa M, Shimamoto K, Kyozuka J (2001). The LAX1 and FRIZZY PANICLE 2 genes determine the inflorescence architecture of rice by controlling rachis- branch and spikelet development. Dev Biol 231, 364-373.
DOI PMID |
| [10] | Li JF, Wang BX, Liu Y, Liu JB, Chen TM, Sun ZG, Yang B, Xing YG, Chi M, Xu B, Xu DY (2022). Progress of research in functions of PPR proteins in growth and development of rice. J Plant Genet Resour 23, 358-367. (in Chinese) |
| 李景芳, 王宝祥, 刘艳, 刘金波, 陈庭木, 孙志广, 杨波, 邢运高, 迟铭, 徐波, 徐大勇 (2022). PPR蛋白在水稻生长发育中的功能研究进展. 植物遗传资源学报 23, 358-367. | |
| [11] |
Li RX, Zhou K, Wang DC, Li QL, Xiang AN, Li L, Li MM, Xiang SQ, Ling YH, He GH, Zhao FM (2023). Analysis of QTLs and breeding of secondary substitution lines for panicle traits based on rice chromosome segment substitution line CSSL-Z481. Sci Agric Sin 56, 1228-1247. (in Chinese)
DOI |
|
李儒香, 周恺, 王大川, 李巧龙, 向奥妮, 李璐, 李苗苗, 向思茜, 凌英华, 何光华, 赵芳明 (2023). 水稻CSSL-Z481 代换片段携带的穗部性状 QTL 分析及次级代换系培育. 中国农业科学 56, 1228-1247.
DOI |
|
| [12] |
Li SC, Li WB, Huang B, Cao XM, Zhou XY, Ye SM, Li CB, Gao FY, Zou T, Xie KL, Ren Y, Ai P, Tang YF, Li XM, Deng QM, Wang SQ, Zheng AP, Zhu J, Liu HN, Wang LX, Li P (2013). Natural variation in PTB1 regulates rice seed setting rate by controlling pollen tube growth. Nat Commun 4, 2793.
DOI PMID |
| [13] | Liu EB, Liu Y, Wu GC, Zeng SY, Thi TGT, Liang LJ, Liang YF, Dong ZY, She D, Wang H, Zaid IU, Hong DL (2016a). Identification of a candidate gene for panicle length in rice (Oryza sativa L.) via association and linkage analysis. Front Plant Sci 7, 596. |
| [14] | Liu WZ, Wu C, Fu YP, Hu GC, Si HM, Zhu L, Luan WJ, He ZQ, Sun ZX (2016b). Identification and characterization of HTD2: a novel gene negatively regulating tiller bud outgrowth in rice. Planta 230, 649-658. |
| [15] |
Livak KJ, Schmittgen TD (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-∆∆CT method. Methods 25, 402-408.
DOI PMID |
| [16] | McCouch SR, Cho YC, Yano M, Paul E, Blinstrub M, Morishima H, Kinoshita T (1997). Report on QTL nomenclature. Rice Genet Newsletter 14, 11-13. |
| [17] | Pan CY, Ye HF, Zhou WY, Wang S, Li MJ, Lu M, Li SF, Zhu XD, Wang YX, Rao YC, Dai GX (2021). QTL mapping of candidate genes involved in Cd accumulation in rice grain. Chin Bull Bot 56, 25-32. (in Chinese) |
| 潘晨阳, 叶涵斐, 周维永, 王盛, 李梦佳, 路梅, 李三峰, 朱旭东, 王跃星, 饶玉春, 戴高兴 (2021). 水稻籽粒镉积累QTL定位及候选基因分析. 植物学报 56, 25-32. | |
| [18] | Qi P, Lin YS, Song XJ, Shen JB, Huang W, Shan JX, Zhu MZ, Jiang LW, Gao JP, Lin HX (2012). The novel quantitative trait locus GL3.1 controls rice grain size and yield by regulating Cyclin-T1;3. Cell Res 22, 1666-1680. |
| [19] |
Raj SRG, Nadarajah K (2022). QTL and candidate genes: techniques and advancement in abiotic stress resistance breeding of major cereals. Int J Mol Sci 24, 6.
DOI URL |
| [20] |
Song XJ, Huang W, Shi M, Zhu MZ, Lin HX (2007). A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat Genet 39, 623-630.
DOI |
| [21] | Sun YW (2009). QTL Mapping for Agronomic Traits in Chromosome Segment Substitution Lines of Rice. Master’s thesis. Yangzhou: Yangzhou University. pp. 18-35. (in Chinese) |
| 孙亚伟 (2009). 水稻染色体单片段代换系农艺性状分析及QTL定位. 硕士论文. 扬州: 扬州大学. pp. 18-35. | |
| [22] | Wei HP, Lu T, Jia QW, Deng F, Zhu H, Qi ZH, Wang YX, Ye HF, Yin WJ, Fang Y, Mu D, Rao YC (2022). QTL mapping of candidate genes for heading date in rice. Chin Bull Bot 57, 588-595. (in Chinese) |
|
魏和平, 芦涛, 贾绮玮, 邓飞, 朱浩, 岂泽华, 王玉玺, 叶涵斐, 殷文晶, 方媛, 穆丹, 饶玉春 (2022). 水稻抽穗期 QTL定位及候选基因分析. 植物学报 57, 588-595.
DOI |
|
| [23] | Wu XC, Gui JX, Liu T, Peng H, Yan YT, He X, Zhu XY, Zhang HQ, He JW (2023). Genome-wide association study for panicle length in rice. Mol Plant Breed 1-13. http://kns.cnki.net/kcms/detail/46.1068.S.20230703.1558.028.html (online). (in Chinese) |
| 吴心成, 桂金鑫, 刘瑫, 彭慧, 闫蕴韬, 何兮, 祝小雅, 张海清, 贺记外 (2023). 水稻穗长全基因组关联分析. 分子植物育种1-13. http://kns.cnki.net/kcms/detail/46.1068.S.20230703.1558.028.html (网络首发). | |
| [24] |
Xing YZ, Zhang QE (2010). Genetic and molecular bases of rice yield. Annu Rev Plant Biol 61, 421-442.
DOI PMID |
| [25] |
Xue WY, Xing YZ, Weng XY, Zhao Y, Tang WJ, Wang L, Zhou HJ, Yu SB, Xu CG, Li XH, Zhang QF (2008). Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat Genet 40, 761-767.
DOI PMID |
| [26] |
Yan CJ, Zhou JH, Yan S, Chen F, Yeboah M, Tang SZ, Liang GH, Gu MH (2007). Identification and characterization of a major QTL responsible for erect panicle trait in japonica rice (Oryza sativa L.). Theor Appl Genet 115, 1093-1100.
DOI URL |
| [27] | Yan YP, Yu XQ, Ren DY, Qian Q (2023). Genetic mechanisms and breeding utilization of grain number per panicle in rice. Chin Bull Bot 58, 359-372. (in Chinese) |
|
严语萍, 俞晓琦, 任德勇, 钱前 (2023). 水稻穗粒数遗传机制与育种利用. 植物学报 58, 359-372.
DOI |
|
| [28] |
Yang Y, Zhang Y, Li J, Xu P, Wu ZJ, Deng XN, Pu QH, Lv YG, Elgamal WHAS, Maniruzzaman S, Deng W, Zhou JW, Tao DY (2023). Three QTL from Oryza meridionalis could improve panicle architecture in Asian cultivated rice. Rice 16, 22.
DOI PMID |
| [29] |
Yoshida A, Ohmori Y, Kitano H, Taguchi-Shiobara F, Hirano HY (2012). ABERRANT SPIKELET AND PANICLE1, encoding a TOPLESS-related transcriptional co-repressor, is involved in the regulation of meristem fate in rice. Plant J 70, 327-339.
DOI URL |
| [30] | Yoshida A, Sasao M, Yasuno N, Takagi K, Daimon Y, Chen RH, Yamazaki R, Tokunaga H, Kitaguchi Y, Sato Y, Nagamura Y, Ushijima T, Kumamaru T, Iida S, Maekawa M, Kyozuka J (2013). TAWAWA1, a regulator of rice inflorescence architecture, functions through the suppression of meristem phase transition. Proc Natl Acad Sci USA 110, 767-772. |
| [31] |
Zhang L, Qi YZ, Wu MM, Zhao L, Zhao ZC, Lei CL, Hao YY, Yu XW, Sun YL, Zhang X, Guo XP, Ren YL (2021). Mitochondrion-targeted PENTATRICOPEPTIDE REPEAT5 is required for cis-splicing of nad4 intron 3 and endosperm development in rice. Crop J 9, 282-296.
DOI |
| [32] |
Zhang XJ, Wang JF, Huang J, Lan HX, Wang CL, Yin CF, Wu YY, Tang HJ, Qian Q, Li JY, Zhang HS (2012). Rare allele of OsPPKL1 associated with grain length causes extra-large grain and a significant yield increase in rice. Proc Natl Acad Sci USA 109, 21534-21539.
DOI PMID |
| [33] | Zhao MZ, Wang YZ, He N, Pang X, Wang LL, Ma ZB, Tang ZQ, Gao H, Zhang LY, Fu L, Wang CH, Liu JG, Zheng WJ (2022). QTL detection for rice grain length and fine mapping of a novel locus qGL6.1. Rice 15, 60. |
| [34] | Zhuang H (2023). QTL Scanning of Rice Panicle Traits and Fine Mapping of qPL-3. Master’s thesis. Yangzhou: Yangzhou University. pp. 16-17. (in Chinese) |
| 庄昊 (2023). 水稻穗部性状的QTL检测及qPL-3的精细定位. 硕士论文. 扬州: 扬州大学. pp. 16-17. |
| [1] | 叶灿, 姚林波, 金莹, 高蓉, 谭琪, 李旭映, 张艳军, 陈析丰, 马伯军, 章薇, 张可伟. 水稻水杨酸代谢突变体高通量筛选方法的建立与应用[J]. 植物学报, 2025, 60(4): 1-0. |
| [2] | 赵凌, 管菊, 梁文化, 张勇, 路凯, 赵春芳, 李余生, 张亚东. 基于高密度Bin图谱的水稻苗期耐热性QTL定位[J]. 植物学报, 2025, 60(3): 342-353. |
| [3] | 李新宇, 谷月, 徐非非, 包劲松. 水稻胚乳淀粉合成相关蛋白的翻译后修饰研究进展[J]. 植物学报, 2025, 60(2): 256-270. |
| [4] | 李建国, 张怡, 张文君. 水稻根系铁膜形成及对磷吸收的影响[J]. 植物学报, 2025, 60(1): 132-143. |
| [5] | 姚瑞枫, 谢道昕. 水稻独脚金内酯信号感知的激活和终止[J]. 植物学报, 2024, 59(6): 873-877. |
| [6] | 连锦瑾, 唐璐瑶, 张伊诺, 郑佳兴, 朱超宇, 叶语涵, 王跃星, 商文楠, 傅正浩, 徐昕璇, 吴日成, 路梅, 王长春, 饶玉春. 水稻抗氧化性状遗传位点挖掘及候选基因分析[J]. 植物学报, 2024, 59(5): 738-751. |
| [7] | 黄佳慧, 杨惠敏, 陈欣雨, 朱超宇, 江亚楠, 胡程翔, 连锦瑾, 芦涛, 路梅, 张维林, 饶玉春. 水稻突变体pe-1对弱光胁迫的响应机制[J]. 植物学报, 2024, 59(4): 574-584. |
| [8] | 周俭民. 收放自如的明星战车[J]. 植物学报, 2024, 59(3): 343-346. |
| [9] | 夏婧, 饶玉春, 曹丹芸, 王逸, 柳林昕, 徐雅婷, 牟望舒, 薛大伟. 水稻中乙烯生物合成关键酶OsACS和OsACO调控机制研究进展[J]. 植物学报, 2024, 59(2): 291-301. |
| [10] | 方妍力, 田传玉, 苏如意, 刘亚培, 王春连, 陈析丰, 郭威, 纪志远. 水稻抗细菌性条斑病基因挖掘与初定位[J]. 植物学报, 2024, 59(1): 1-9. |
| [11] | 朱宝, 赵江哲, 张可伟, 黄鹏. 水稻细胞分裂素氧化酶9参与调控水稻叶夹角发育[J]. 植物学报, 2024, 59(1): 10-21. |
| [12] | 贾绮玮, 钟芊芊, 顾育嘉, 陆天麒, 李玮, 杨帅, 朱超宇, 胡程翔, 李三峰, 王跃星, 饶玉春. 水稻茎秆细胞壁相关组分含量QTL定位及候选基因分析[J]. 植物学报, 2023, 58(6): 882-892. |
| [13] | 戴若惠, 钱心妤, 孙静蕾, 芦涛, 贾绮玮, 陆天麒, 路梅, 饶玉春. 水稻叶色调控机制及相关基因研究进展[J]. 植物学报, 2023, 58(5): 799-812. |
| [14] | 田传玉, 方妍力, 沈晴, 王宏杰, 陈析丰, 郭威, 赵开军, 王春连, 纪志远. 2019-2021年我国南方稻区白叶枯病菌的毒力与遗传多样性调查研究[J]. 植物学报, 2023, 58(5): 743-749. |
| [15] | 孙尚, 胡颖颖, 韩阳朔, 薛超, 龚志云. 水稻染色体双链寡核苷酸荧光原位杂交技术[J]. 植物学报, 2023, 58(3): 433-439. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||