

植物学报 ›› 2024, Vol. 59 ›› Issue (2): 291-301.DOI: 10.11983/CBB23143 cstr: 32102.14.CBB23143
夏婧1,†, 饶玉春2,†, 曹丹芸1, 王逸1, 柳林昕1, 徐雅婷1, 牟望舒1,*( ), 薛大伟1,*(
), 薛大伟1,*( )
)
收稿日期:2023-10-29
接受日期:2024-01-30
出版日期:2024-03-10
发布日期:2024-03-10
通讯作者:
* E-mail: 作者简介:† 共同第一作者
基金资助:
Jing Xia1,†, Yuchun Rao2,†, Danyun Cao1, Yi Wang1, Linxin Liu1, Yating Xu1, Wangshu Mou1,*( ), Dawei Xue1,*(
), Dawei Xue1,*( )
)
Received:2023-10-29
Accepted:2024-01-30
Online:2024-03-10
Published:2024-03-10
Contact:
* E-mail: About author:† These authors contributed equally to this paper
摘要: 乙烯在调控水稻(Oryza sativa)生长发育及胁迫响应中具有重要作用。乙烯生物合成的第1步是甲硫氨酸转化为S-腺苷甲硫氨酸(SAM), 然后在ACC合酶(ACS)的催化下合成乙烯前体物质ACC, 最后通过ACC氧化酶(ACO)生成乙烯。该文综述了水稻乙烯生物合成途径中2个关键酶OsACS和OsACO在转录及翻译后的调控机制, 提出了一些未解决的问题, 并展望了未来的研究方向, 以期加深人们对乙烯生物合成复杂机制的理解。
夏婧, 饶玉春, 曹丹芸, 王逸, 柳林昕, 徐雅婷, 牟望舒, 薛大伟. 水稻中乙烯生物合成关键酶OsACS和OsACO调控机制研究进展. 植物学报, 2024, 59(2): 291-301.
Jing Xia, Yuchun Rao, Danyun Cao, Yi Wang, Linxin Liu, Yating Xu, Wangshu Mou, Dawei Xue. Research Progress on the Regulatory Mechanisms of OsACS and OsACO in Rice Ethylene Biosynthesis. Chinese Bulletin of Botany, 2024, 59(2): 291-301.

图2 水稻(Os)、大麦(Hv)、拟南芥(At)和番茄(Sl) 1-甲基环丙烷-1-羧酸合酶(ACS)蛋白序列的系统发生树
Figure 2 Phylogenetic tree of the ACC synthase (ACS) protein sequences of Oryza sativa (Os), Hordeum vulgare (Hv), Arabidopsis thaliana (At) and Solanum lycopersicum (Sl)
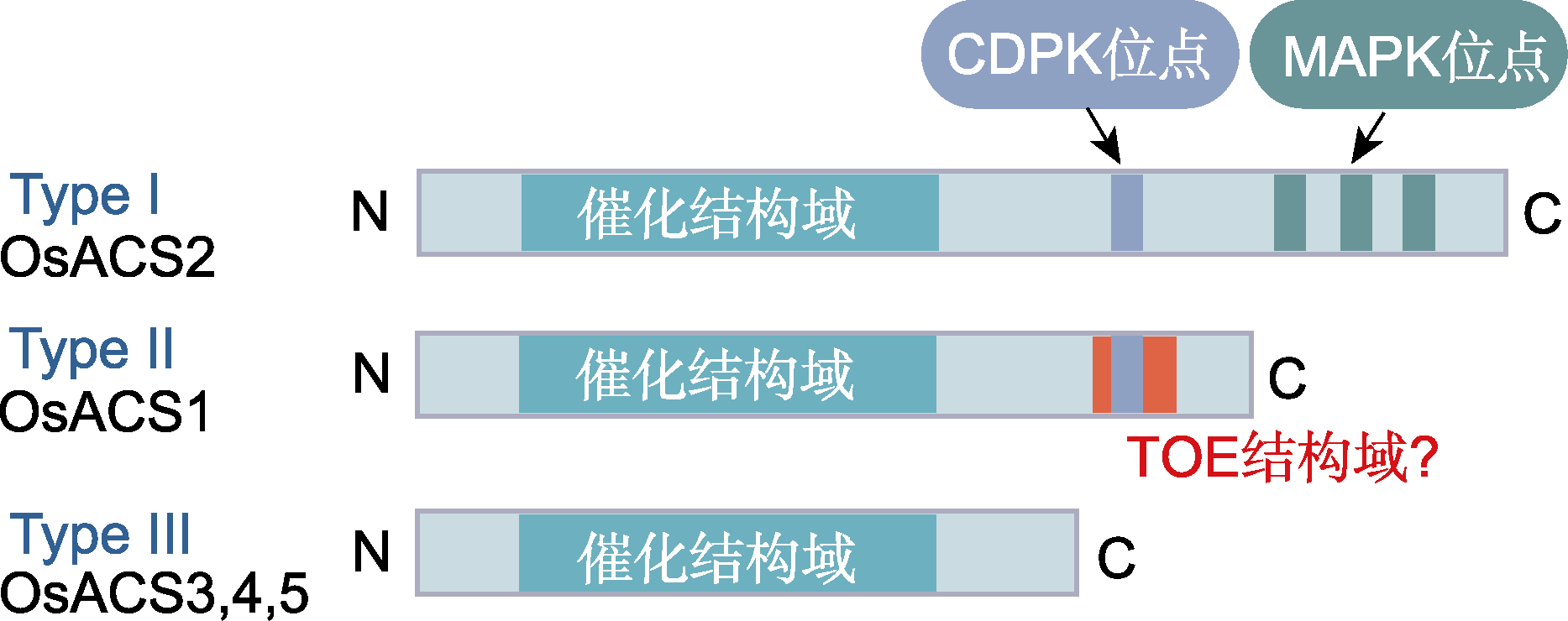
图3 水稻3种亚型的ACS蛋白结构(参考Lee and Yoon, 2018) CDPK: 钙依赖蛋白激酶; MAPK: 丝裂原活化蛋白激酶。ACS同图2。
Figure 3 The structure of three types of ACS proteins in rice (refer to Lee and Yoon, 2018) CDPK: Calcium-dependent protein kinase; MAPK: Mitogen- activated protein kinase. ACS is the same as shown in Figure 2.

图4 水稻(Os)、拟南芥(At)和番茄(Sl)中Type II型ACS的C端氨基酸序列比对 基本不变的TOE基序结构WVFRLSF/W序列标为红色, OsACS1中可能存在的保守TOE基序用红色方框标出。ACS同图2。
Figure 4 Sequence alignment of C-terminal amino acids of Type II ACSs from Oryza sativa (Os), Arabidopsis thaliana (At) and Solanum lycopersicum (Sl) The near invariant TOE motif sequence WVFRLSF/W are shown in red, while the possible conserved TOE motif in OsACS1 is marked with red box. ACS is the same as shown in Figure 2.
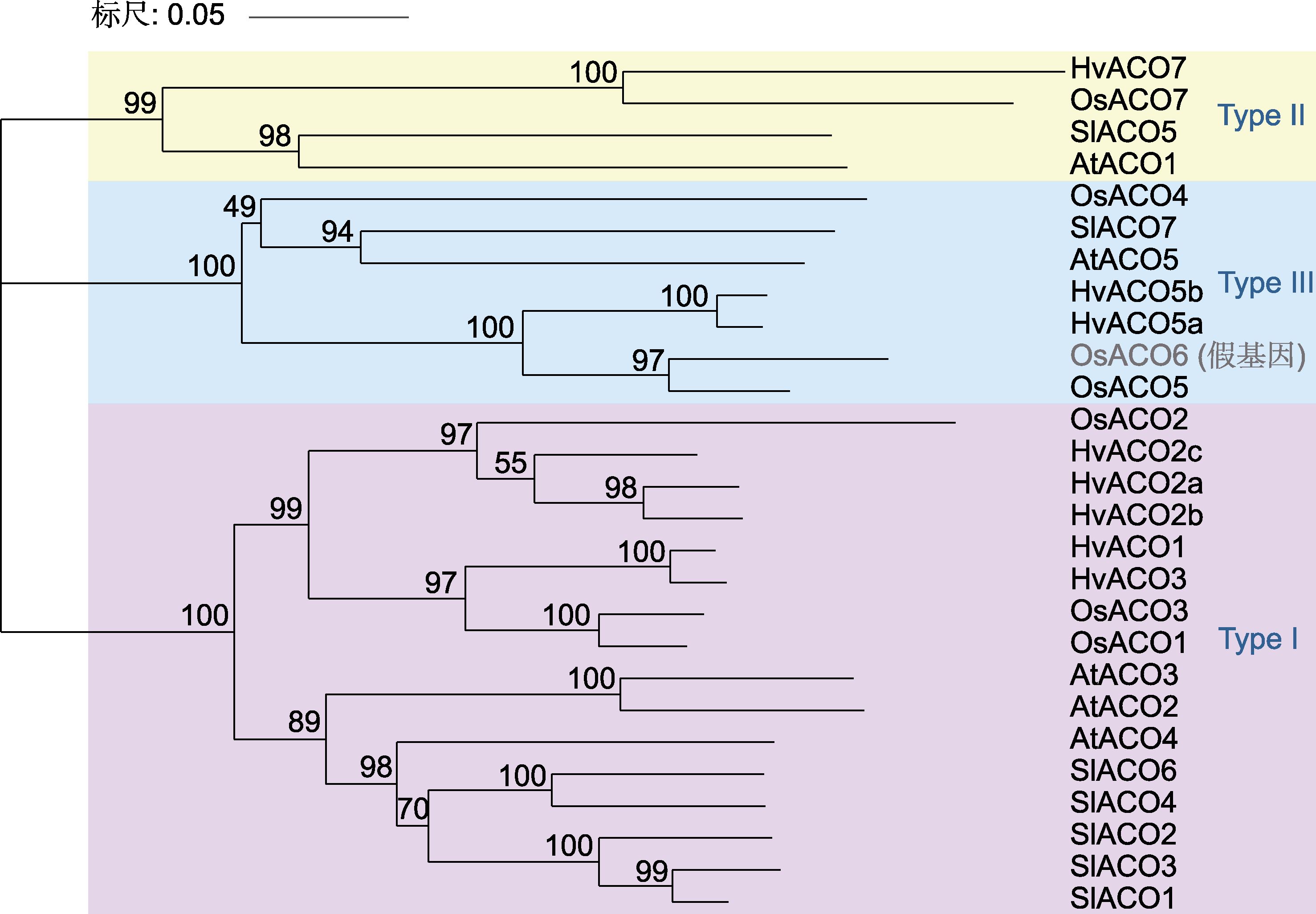
图5 水稻(Os)、大麦(Hv)、拟南芥(At)和番茄(Sl) 1-甲基环丙烷-1-羧酸氧化酶(ACO)蛋白序列的系统发生树
Figure 5 Phylogenetic tree for ACC oxidase (ACO) protein sequences of Oryza sativa (Os), Hordeum vulgare (Hv), Arabidopsis thaliana (At) and Solanum lycopersicum (Sl)
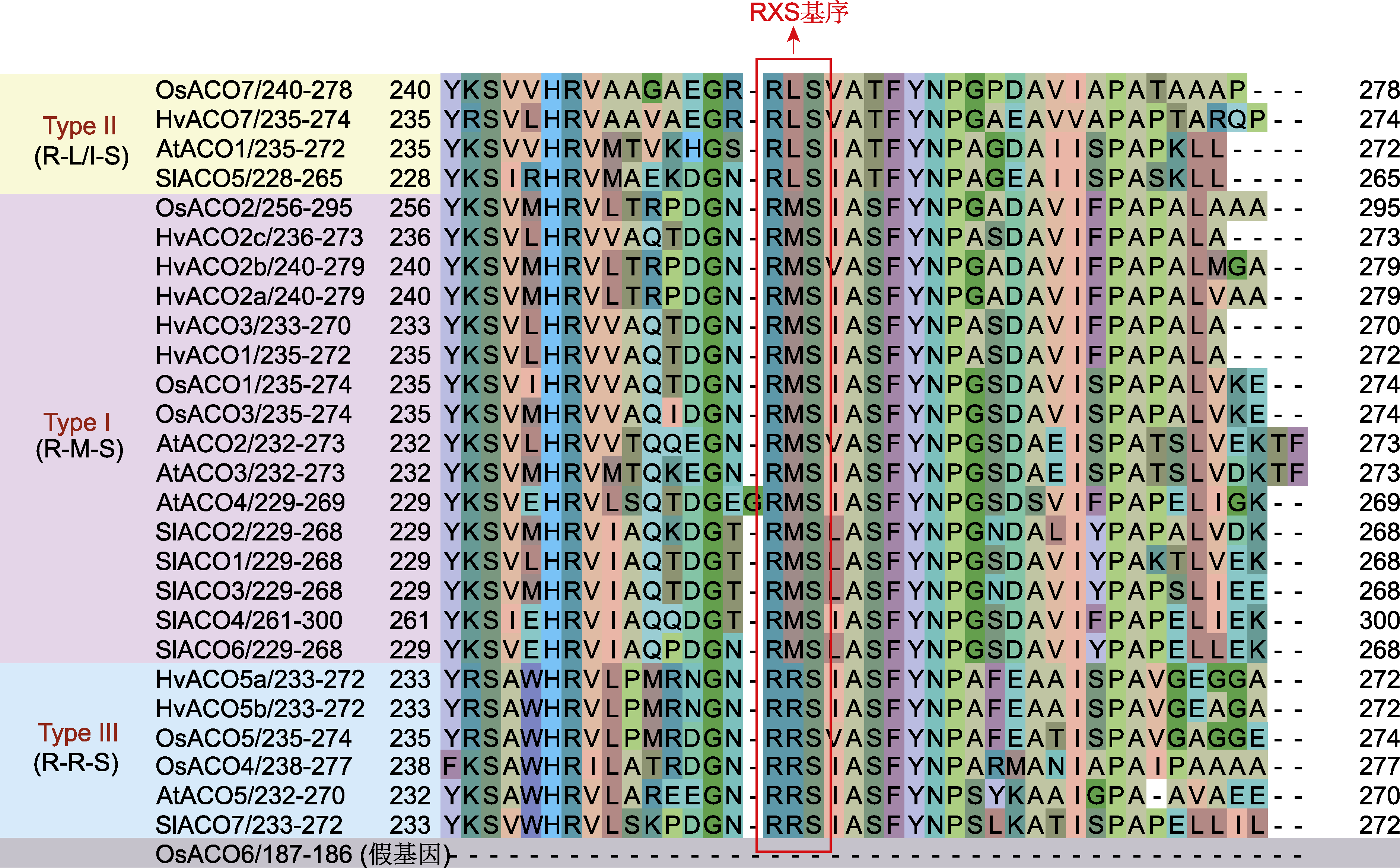
图6 水稻(Os)、大麦(Hv)、拟南芥(At)和番茄(Sl) ACO蛋白序列比对 ACO同图5。
Figure 6 ACO protein sequence alignment of Oryza sativa (Os), Hordeum vulgare (Hv), Arabidopsis thaliana (At) and Solanum lycopersicum (Sl) ACO is the same as shown in Figure 5.
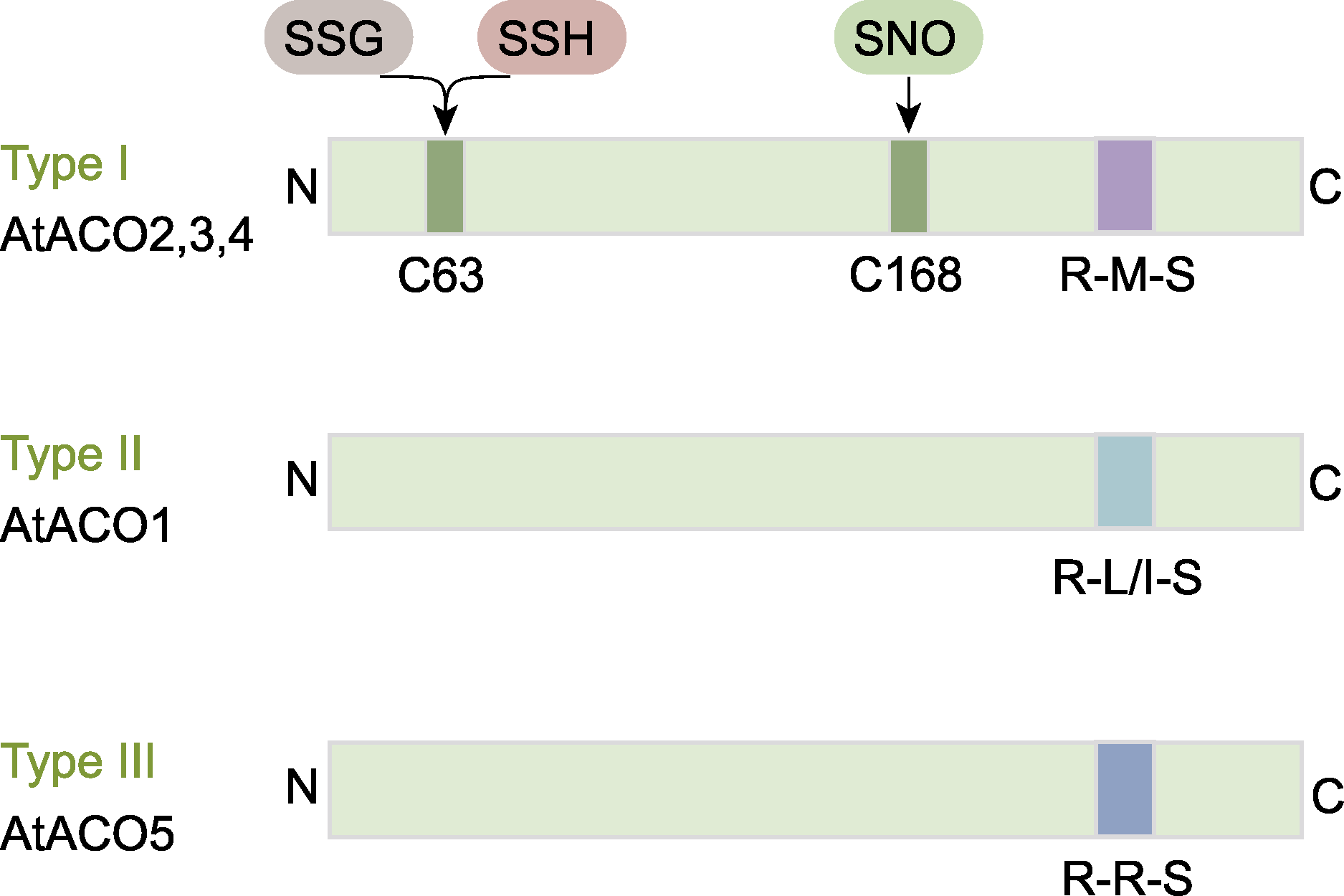
图7 拟南芥3种亚型的ACO蛋白结构(参考Pattyn et al., 2021) C63位点的翻译后修饰包括S-谷胱甘肽化(SSG)和S-硫巯基化(SSH); C168位点的修饰包括S-亚硝基化(SNO)。ACO同图5。
Figure 7 The structure of three types of ACO proteins in Arabidopsis thaliana (refer to Pattyn et al., 2021) Post-translational modifications at C63 include S-glutathionylation (SSG) and S-sulfhydration (SSH), while modifications at C168 involve S-nitrosylation (SNO). ACO is the same as shown in Figure 5.

图8 AtACO2与OsACOs蛋白序列比对 蓝色星号表示拟南芥AtACO2中2个保守的半胱氨酸残基位点C63和C168; 红色方框表示水稻OsACOs与拟南芥保守的半胱氨酸残基位点为C168。
Figure 8 AtACO2 and OsACOs protein sequence alignment The blue stars represent the two conserved cysteine residue sites (C63 and C168) in AtACO2, and red box indicates the conserved cysteine residues (C168) of the OsACOs.
| [1] | Ahmadizadeh M, Chen JT, Hasanzadeh S, Ahmar S, Heidari P (2020). Insights into the genes involved in the ethylene biosynthesis pathway in Arabidopsis thaliana and Oryza sativa. J Genet Engin Biotechnol 18, 62. |
| [2] |
Althiab-Almasaud R, Sallanon H, Chang CR, Chervin C (2021). 1-aminocyclopropane-1-carboxylic acid stimulates tomato pollen tube growth independently of ethylene receptors. Physiol Plant 173, 2291-2297.
DOI PMID |
| [3] |
Argueso CT, Hansen M, Kieber JJ (2007). Regulation of ethylene biosynthesis. J Plant Growth Regul 26, 92-105.
DOI URL |
| [4] |
Aroca A, Benito JM, Gotor C, Romero LC (2017). Persulfidation proteome reveals the regulation of protein function by hydrogen sulfide in diverse biological processes in Arabidopsis. J Exp Bot 68, 4915-4927.
DOI PMID |
| [5] | Booker MA, DeLong A (2015). Producing the ethylene signal: regulation and diversification of ethylene biosynthetic enzymes. Plant Physiol 169, 42-50. |
| [6] |
Chae HS, Faure F, Kieber JJ (2003). The eto1, eto2, and eto3 mutations and cytokinin treatment increase ethylene biosynthesis in Arabidopsis by increasing the stability of ACS protein. Plant Cell 15, 545-559.
PMID |
| [7] |
Chen J, Wang X, Zhang WW, Zhang SQ, Zhao FJ (2020). Protein phosphatase 2A alleviates cadmium toxicity by modulating ethylene production in Arabidopsis thaliana. Plant Cell Environ 43, 1008-1022.
DOI URL |
| [8] |
Christians MJ, Gingerich DJ, Hansen M, Binder BM, Kieber JJ, Vierstra RD (2009). The BTB ubiquitin ligases ETO1, EOL1 and EOL2 act collectively to regulate ethylene biosynthesis in Arabidopsis by controlling type-2 ACC synthase levels. Plant J 57, 332-345.
DOI URL |
| [9] |
Datta R, Kumar D, Sultana A, Hazra S, Bhattacharyya D, Chattopadhyay S (2015). Glutathione regulates 1-aminocyclopropane-1-carboxylate synthase transcription via WRKY33 and 1-aminocyclopropane-1-carboxylate oxidase by modulating messenger RNA stability to induce ethylene synthesis during stress. Plant Physiol 169, 2963-2981.
DOI PMID |
| [10] | Dilley DR, Wang ZY, Kadirjan-Kalbach DK, Ververidis F, Beaudry R, Padmanabhan K (2013). 1-aminocyclopropane-1-carboxylic acid oxidase reaction mechanism and putative post-translational activities of the ACCO protein. AoB Plants 5, plt031. |
| [11] |
Du H, Wu N, Cui F, You L, Li XH, Xiong LZ (2014). A homolog of ETHYLENE OVERPRODUCER, OsETOL1, differentially modulates drought and submergence tolerance in rice. Plant J 78, 834-849.
DOI URL |
| [12] | Freeman AK, Morrison DK (2011). 14-3-3 proteins: diverse functions in cell proliferation and cancer progression. Semi Cell Devel Biol 22, 681-687. |
| [13] | Han L, Li GJ, Yang KY, Mao GH, Wang RG, Liu YD, Zhang SQ (2010). Mitogen-activated protein kinase 3 and 6 regulate Botrytis cinerea-induced ethylene production in Arabidopsis. Plant J 64, 114-127. |
| [14] | Houben M, Van de Poel B(2019). 1-aminocyclopropane- 1-carboxylic acid oxidase (ACO): the enzyme that makes the plant hormone ethylene. Front Plant Sci 10, 695. |
| [15] | Hu JL, Huang XH, Chen LC, Sun XW, Lu CM, Zhang LX, Wang YC, Zuo JR (2015). Site-specific nitrosoproteomic identification of endogenously S-nitrosylated proteins in Arabidopsis. Plant Physiol 167, 1731-1746. |
| [16] | Iwai T, Miyasaka A, Seo S, Ohashi Y (2006). Contribution of ethylene biosynthesis for resistance to blast fungus infection in young rice plants. Plant Physiol 142, 1202-1215. |
| [17] |
Iwamoto M, Takano M (2011). Phytochrome-regulated EBL1 contributes to ACO1 upregulation in rice. Biotechnol Lett 33, 173-178.
DOI PMID |
| [18] |
Jia HL, Chen SS, Liu D, Liesche J, Shi C, Wang J, Ren MJ, Wang XF, Yang J, Shi W, Li JS (2018). Ethylene-induced hydrogen sulfide negatively regulates ethylene biosynthesis by persulfidation of ACO in tomato under osmotic stress. Front Plant Sci 9, 1517.
DOI PMID |
| [19] | Joo S, Liu YD, Lueth A, Zhang SQ (2008). MAPK phosphorylation-induced stabilization of ACS6 protein is mediated by the non-catalytic C-terminal domain, which also contains the cis-determinant for rapid degradation by the 26S proteasome pathway. Plant J 54, 129-140. |
| [20] |
Kawai Y, Ono E, Mizutani M (2014). Evolution and diversity of the 2-oxoglutarate-dependent dioxygenase superfamily in plants. Plant J 78, 328-343.
DOI URL |
| [21] |
Lee HY, Chen YC, Kieber JJ, Yoon GM (2017). Regulation of the turnover of ACC synthases by phytohormones and heterodimerization in Arabidopsis. Plant J 91, 491-504.
DOI URL |
| [22] | Lee HY, Park HL, Park C, Chen YC, Yoon GM (2021). Reciprocal antagonistic regulation of E3 ligases controls ACC synthase stability and responses to stress. Proc Natl Acad Sci USA 34, e2011900118. |
| [23] | Lee HY, Yoon GM (2018). Regulation of ethylene biosynthesis by phytohormones in etiolated rice (Oryza sativa L.) seedlings. Mol Cells 41, 311-319. |
| [24] | Li CH, Wang G, Zhao JL, Zhang LQ, Ai LF, Han YF, Sun DY, Zhang SW, Sun Y (2014). The receptor-like kinase SIT1 mediates salt sensitivity by activating MAPK3/6 and regulating ethylene homeostasis in rice. Plant Cell 26, 2538-2553. |
| [25] | Li DD, Flores-Sandoval E, Ahtesham U, Coleman A, Clay JM, Bowman JL, Chang CR (2020). Ethylene-independent functions of the ethylene precursor ACC in Marchantia polymorpha. Nat Plants 6, 1335-1344. |
| [26] |
Liu H, Dong SY, Gu FW, Liu W, Yang GL, Huang M, Xiao WM, Liu YZ, Guo T, Wang H, Chen ZQ, Wang JF (2017). NBS-LRR protein Pik-H4 interacts with OsBIHD1 to balance rice blast resistance and growth by coordinating ethylene-brassinosteroid pathway. Front Plant Sci 8, 127.
DOI PMID |
| [27] | Liu HQ, Zou YJ, Li XF, Wu L, Guo GQ (2021). Stablization of ACOs by NatB mediated N-terminal acetylation is required for ethylene homeostasis. BMC Plant Biol 21, 320. |
| [28] |
Liu MH, Wei JW, Liu W, Gong B (2023). S-nitrosylation of ACO homolog 4 improves ethylene synthesis and salt tolerance in tomato. New Phytol 239, 159-173.
DOI URL |
| [29] |
Ludwików A, Cieśla A, Kasprowicz-Maluśki A, Mituła F, Tajdel M, Gałgański L, Ziółkowski PA, Kubiak P, Małecka A, Piechalak A, Szabat M, Górska A, Dąbrowski M, Ibragimow I, Sadowski J (2014). Arabidopsis protein phosphatase 2C ABI1 interacts with Type I ACC synthases and is involved in the regulation of ozone-induced ethylene biosynthesis. Mol Plant 7, 960-976.
DOI PMID |
| [30] |
Lyzenga WJ, Booth JK, Stone SL (2012). The Arabidopsis RING-type E3 ligase XBAT32 mediates the proteasomal degradation of the ethylene biosynthetic enzyme, 1-aminocyclopropane-1-carboxylate synthase 7. Plant J 71, 23-34.
DOI URL |
| [31] |
Matsushima R, Maekawa M, Kusano M, Tomita K, Kondo H, Nishimura H, Crofts N, Fujita N, Sakamoto W (2016). Amyloplast membrane protein SUBSTANDARD STARCH GRAIN6 controls starch grain size in rice endosperm. Plant Physiol 170, 1445-1459.
DOI PMID |
| [32] | Mou WS, Kao YT, Michard E, Simon AA, Li DD, Wudick MM, Lizzio MA, Feijó JA, Chang CR (2020). Ethylene-independent signaling by the ethylene precursor ACC in Arabidopsis ovular pollen tube attraction. Nat Commun 11, 4082. |
| [33] | Ouyang S, Zhu W, Hamilton J, Lin HN, Campbell M, Childs K, Thibaud-Nissen F, Malek RL, Lee Y, Zheng L, Orvis J, Haas B, Wortman J, Buell CR (2007). The TIGR rice genome annotation resource: improvements and new features. Nucleic Acids Res 35, D883-D887. |
| [34] | Park C, Lee HY, Yoon GM (2021). The regulation of ACC synthase protein turnover: a rapid route for modulating plant development and stress responses. Curr Opin Plant Biol 63, 102046. |
| [35] | Pattyn J, Vaughan-Hirsch J, Van de Poel B (2021). The regulation of ethylene biosynthesis: a complex multilevel control circuitry. New Phytol 229, 770-782. |
| [36] |
Qin H, Wang J, Chen XB, Wang FF, Peng P, Zhou Y, Miao YC, Zhang YQ, Gao YD, Qi YD, Zhou JH, Huang RF (2019). Rice OsDOF15 contributes to ethylene-inhibited primary root elongation under salt stress. New Phytol 223, 798-813.
DOI PMID |
| [37] |
Rauf M, Arif M, Fisahn J, Xue GP, Balazadeh S, Mueller-Roeber B (2013). NAC transcription factor SPEEDY HYPONASTIC GROWTH regulates flooding-induced leaf movement in Arabidopsis. Plant Cell 25, 4941-4955.
DOI URL |
| [38] | Stange LMC, Osborne DJ (1989). Contrary effects of ethylene and ACC on cell growth in the liverwort Riella helicophylla. In: Clijsters H, Proft M, Marcelle R, Poucke M, eds. Biochemical and Physiological Aspects of Ethylene Production in Lower and Higher Plants. Dordrecht: Springer. pp. 341-348. |
| [39] |
Tan ST, Xue HW (2014). Casein kinase 1 regulates ethylene synthesis by phosphorylating and promoting the turnover of ACS5. Cell Rep 9, 1692-1702.
DOI URL |
| [40] |
Tsolakidou MD, Pantelides LS, Tzima AK, Kang S, Paplomatas EJ, Tsaltas D (2019). Disruption and overexpression of the gene encoding ACC (1-aminocyclopropane-1-carboxylic acid) deaminase in soil-borne fungal pathogen Verticillium dahliae revealed the role of ACC as a potential regulator of virulence and plant defense. Mol Plant Microbe Interact 32, 639-653.
DOI URL |
| [41] |
Tsuchisaka A, Theologis A (2004). Unique and overlapping expression patterns among the Arabidopsis 1-amino- cyclopropane-1-carboxylate synthase gene family members. Plant Physiol 136, 2982-3000.
DOI PMID |
| [42] |
Uji T, Endo H, Mizuta H (2020). Sexual reproduction via a 1-aminocyclopropane-1-carboxylic acid-dependent pathway through redox modulation in the marine red alga Pyropia yezoensis (Rhodophyta). Front Plant Sci 11, 60.
DOI URL |
| [43] |
Van de Poel B, Bulens I, Hertog MLATM, Nicolai BM, Geeraerd AH (2014a). A transcriptomics-based kinetic model for ethylene biosynthesis in tomato (Solanum lycopersicum) fruit: development, validation and exploration of novel regulatory mechanisms. New Phytol 202, 952-963.
DOI URL |
| [44] | Van de Poel B, Bulens I, Markoula A, Hertog MLATM, Dreesen R, Wirtz M, Vandoninck S, Oppermann Y, Keulemans J, Hell R, Waelkens E, De Proft MP, Sauter M, Nicolai BM, Geeraerd AH (2012). Targeted systems biology profiling of tomato fruit reveals coordination of the Yang cycle and a distinct regulation of ethylene biosynthesis during postclimacteric ripening. Plant Physiol 160, 1498-1514. |
| [45] |
Van de Poel B, Vandenzavel N, Smet C, Nicolay T, Bulens I, Mellidou I, Vandoninck S, Hertog MLATM, Derua R, Spaepen S, Vanderleyden J, Waelkens E, De Proft MP, Nicolai BM, Geeraerd AH (2014b). Tissue specific analysis reveals a differential organization and regulation of both ethylene biosynthesis and E8 during climacteric ripening of tomato. BMC Plant Biol 14, 11.
DOI |
| [46] | Vanderstraeten L, Depaepe T, Bertrand S, Van Der Straeten D (2019). The ethylene precursor ACC affects early vegetative development independently of ethylene signaling. Front Plant Sci 10, 1591. |
| [47] |
Wang KLC, Yoshida H, Lurin C, Ecker JR (2004). Regulation of ethylene gas biosynthesis by the Arabidopsis ETO1 protein. Nature 428, 945-950.
DOI |
| [48] |
Wang ZQ, Wei XY, Wang YQ, Sun MT, Zhao PY, Wang QN, Yang B, Li J, Jiang YQ (2023). WRKY 29 transcription factor regulates ethylene biosynthesis and response in Arabidopsis. Plant Physiol Biochem 194, 134-145.
DOI URL |
| [49] |
Xiong L, Xiao D, Xu XX, Guo ZX, Wang NN (2014). The non-catalytic N-terminal domain of ACS7 is involved in the post-translational regulation of this gene in Arabidopsis. J Exp Bot 65, 4397-4408.
DOI PMID |
| [50] | Xu SL (2008). Two Leucine-rich Receptor Kinases Mediate Signaling Linking Cell Wall Bio-synthesis and ACC Synthase in Arabidopsis and Possible Downstream Elements in the Pathway. Doctoral dissertation. United States-North Carolina: The University of North Carolina at Chapel Hill. pp. 1-128. |
| [51] |
Yao Y, Du Y, Jiang L, Liu JY (2007). Interaction between ACC synthase 1 and 14-3-3 proteins in rice: a new insight. Biochem (Moscow) 72, 1003-1007.
DOI URL |
| [52] |
Yin J, Zhang XQ, Zhang GS, Wen YY, Liang G, Chen XL (2019). Aminocyclopropane-1-carboxylic acid is a key regulator of guard mother cell terminal division in Arabidopsis thaliana. J Exp Bot 70, 897-908.
DOI PMID |
| [53] |
Yoon GM (2015). New insights into the protein turnover regulation in ethylene biosynthesis. Mol Cells 38, 597-603.
DOI PMID |
| [54] |
Yoon GM, Kieber JJ (2013a). 14-3-3 regulates 1-aminocyclopropane-1-carboxylate synthase protein turnover in Arabidopsis. Plant Cell 25, 1016-1028.
DOI URL |
| [55] |
Yoon GM, Kieber JJ (2013b). ACC synthase and its cognate E3 ligase are inversely regulated by light. Plant Signal Behav 8, e26478.
DOI URL |
| [56] | Yoon J, Cho LH, Yang WZ, Pasriga R, Wu YF, Hong WJ, Bureau C, Wi SJ, Zhang T, Wang RC, Zhang DB, Jung KH, Park KY, Périn C, Zhao YD, An G (2020). Homeobox transcription factor OsZHD2 promotes root meristem activity in rice by inducing ethylene biosynthesis. J Exp Bot 71, 5348-5364. |
| [57] |
Yoshida H, Nagata M, Saito K, Wang KLC, Ecker JR (2005). Arabidopsis ETO1 specifically interacts with and negatively regulates type 2 1-aminocyclopropane-1-carboxylate synthases. BMC Plant Biol 5, 14.
PMID |
| [58] |
Yoshida H, Wang KLC, Chang CM, Mori K, Uchida E, Ecker JR (2006). The ACC synthase TOE sequence is required for interaction with ETO1 family proteins and destabilization of target proteins. Plant Mol Biol 62, 427-437.
PMID |
| [59] |
Yu JT, Mao CJ, Zhong Q, Yao XF, Li P, Liu CM, Ming F (2021). OsNAC2 is involved in multiple hormonal pathways to mediate germination of rice seeds and establishment of seedling. Front Plant Sci 12, 699303.
DOI URL |
| [60] | Yuan M, Qu LJ, Wang XJ, Qian Q, Yang WC, Wang T, Kong HZ, Jiang GM, Chong K (2014). Research advances on plant science in China in 2013. Chin Bull Bot 49, 347-406. (in Chinese) |
| 袁明, 瞿礼嘉, 王小菁, 钱前, 杨维才, 王台, 孔宏智, 蒋高明, 种康 (2014). 2013年中国植物科学若干领域重要研究进展. 植物学报 49, 347-406. | |
| [61] |
Zhang HW, Zhang JF, Quan RD, Pan XW, Wan LY, Huang RF (2013). EAR motif mutation of rice OsERF3 alters the regulation of ethylene biosynthesis and drought tolerance. Planta 237, 1443-1451.
DOI PMID |
| [62] |
Zhou Y, Xiong Q, Yin CC, Ma B, Chen SY, Zhang JS (2020). Ethylene biosynthesis, signaling, and crosstalk with other hormones in rice. Small Methods 4, 1900278.
DOI URL |
| [1] | 王鸿梅, 袁蔚, 薛芳, 张召聪, 刘坤, 陈四龙. 植物SWEET基因参与逆境胁迫响应及其调控机制[J]. 植物学报, 2025, 60(4): 1-0. |
| [2] | 叶灿, 姚林波, 金莹, 高蓉, 谭琪, 李旭映, 张艳军, 陈析丰, 马伯军, 章薇, 张可伟. 水稻水杨酸代谢突变体高通量筛选方法的建立与应用[J]. 植物学报, 2025, 60(4): 1-0. |
| [3] | 周婧, 高飞. 植物缺铁诱导型香豆素合成及其在铁吸收中的功能研究进展[J]. 植物学报, 2025, 60(3): 460-471. |
| [4] | 熊良林, 梁国鲁, 郭启高, 景丹龙. 基因可变剪接调控植物响应非生物胁迫研究进展[J]. 植物学报, 2025, 60(3): 435-448. |
| [5] | 赵凌, 管菊, 梁文化, 张勇, 路凯, 赵春芳, 李余生, 张亚东. 基于高密度Bin图谱的水稻苗期耐热性QTL定位[J]. 植物学报, 2025, 60(3): 342-353. |
| [6] | 李新宇, 谷月, 徐非非, 包劲松. 水稻胚乳淀粉合成相关蛋白的翻译后修饰研究进展[J]. 植物学报, 2025, 60(2): 256-270. |
| [7] | 李建国, 张怡, 张文君. 水稻根系铁膜形成及对磷吸收的影响[J]. 植物学报, 2025, 60(1): 132-143. |
| [8] | 胡海涛, 武越, 杨玲. 植物NAD(P)+的生物合成及其生物学功能研究进展[J]. 植物学报, 2025, 60(1): 114-131. |
| [9] | 姚瑞枫, 谢道昕. 水稻独脚金内酯信号感知的激活和终止[J]. 植物学报, 2024, 59(6): 873-877. |
| [10] | 李红菊, 杨维才. 微肽大用: 种子脱水调控新机制[J]. 植物学报, 2024, 59(6): 869-872. |
| [11] | 范雪兰, 落艳娇, 徐超群, 郭宝林. 淫羊藿类黄酮生物合成相关基因研究进展[J]. 植物学报, 2024, 59(5): 834-846. |
| [12] | 连锦瑾, 唐璐瑶, 张伊诺, 郑佳兴, 朱超宇, 叶语涵, 王跃星, 商文楠, 傅正浩, 徐昕璇, 吴日成, 路梅, 王长春, 饶玉春. 水稻抗氧化性状遗传位点挖掘及候选基因分析[J]. 植物学报, 2024, 59(5): 738-751. |
| [13] | 黄佳慧, 杨惠敏, 陈欣雨, 朱超宇, 江亚楠, 胡程翔, 连锦瑾, 芦涛, 路梅, 张维林, 饶玉春. 水稻突变体pe-1对弱光胁迫的响应机制[J]. 植物学报, 2024, 59(4): 574-584. |
| [14] | 周俭民. 收放自如的明星战车[J]. 植物学报, 2024, 59(3): 343-346. |
| [15] | 朱超宇, 胡程翔, 朱哲楠, 张芷宁, 汪理海, 陈钧, 李三峰, 连锦瑾, 唐璐瑶, 钟芊芊, 殷文晶, 王跃星, 饶玉春. 水稻穗部性状QTL定位及候选基因分析[J]. 植物学报, 2024, 59(2): 217-230. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||