

植物学报 ›› 2025, Vol. 60 ›› Issue (3): 460-471.DOI: 10.11983/CBB24106 cstr: 32102.14.CBB24106
收稿日期:2024-07-17
接受日期:2024-12-14
出版日期:2025-05-10
发布日期:2024-12-27
通讯作者:
*高飞, 博士, 湖南农业大学农学院副教授, 硕士生导师, 湖南省“百人计划”青年学者。长期从事植物铁吸收、转运、存储及其分子调控机制研究。利用多组学、遗传学、生物化学与分子生物学等手段挖掘鉴定相关关键基因, 揭示模式植物拟南芥以及水稻铁稳态调控的分子机理。近年来, 主持国家自然科学基金等省部级项目4项。以第一作者或通讯作者在The Plant Cell、Trends in Plant Science和Journal of Experimental Botany等国内外权威学术期刊发表论文10余篇。E-mail: gaofei@hunau.edu.cn
基金资助:Received:2024-07-17
Accepted:2024-12-14
Online:2025-05-10
Published:2024-12-27
Contact:
*E-mail: gaofei@hunau.edu.cn
摘要: 香豆素类化合物是一类以苯并吡喃酮为母环结构的酚类化合物, 可分为简单香豆素和复杂香豆素, 广泛存在于自然界的高等植物中。研究表明, 缺铁条件下, 植物根部分泌的简单香豆素类化合物能够促进铁离子的吸收。该文对近年来发现和鉴定的植物缺铁诱导型香豆素合成及调控相关基因研究进展进行综述, 进一步详细阐述缺铁诱导型香豆素的生物合成、储存、分泌及其调控机制, 探讨其促进植物铁吸收的分子机制。同时, 展望该领域未来的研究方向。
周婧, 高飞. 植物缺铁诱导型香豆素合成及其在铁吸收中的功能研究进展. 植物学报, 2025, 60(3): 460-471.
Zhou Jing, Gao Fei. Advances in Iron Deficiency-induced Coumarin Biosynthesis and Their Functions in Iron Absorption in Plants. Chinese Bulletin of Botany, 2025, 60(3): 460-471.
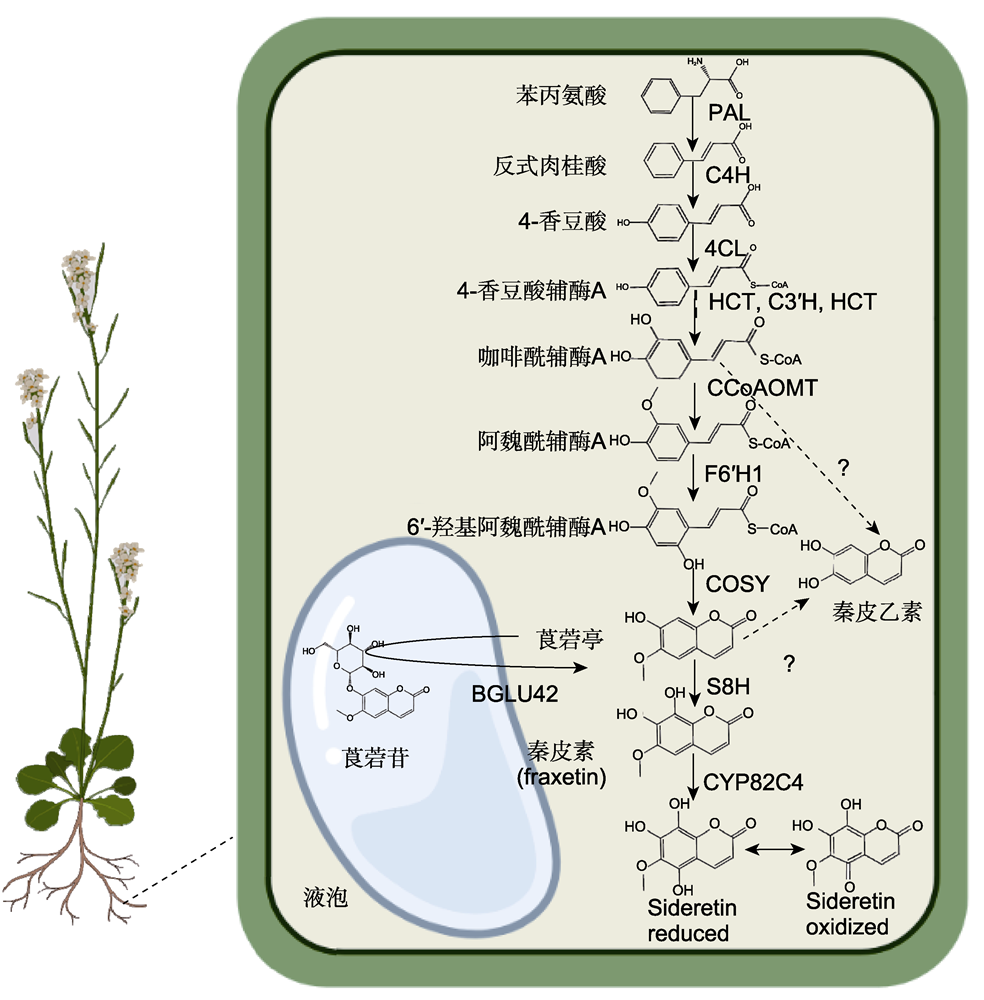
图1 拟南芥缺铁诱导型香豆素类化合物生物合成途径 PAL: 苯丙氨酸解氨酶; C4H: 肉桂酸-4-羟化酶; 4CL: 4-香豆酸:辅酶A连接酶; HCT: 羟基肉桂酰辅酶A莽草酸/奎宁酸羟基肉桂酰转移酶; C3′H: 4-香豆酰莽草酸/奎宁酸3′-羟化酶; CCoAOMT: 咖啡酰辅酶A O-甲基转移酶; F6′H1: 阿魏酰辅酶A 6′-羟化酶; COSY: 香豆素合成酶; S8H: 莨菪亭8-羟化酶; CYP82C4: 细胞色素P450家族酶
Figure 1 Biosynthesis pathways of iron deficiency-induced coumarins in Arabidopsis PAL: Phenylalanine ammonia-lyase; C4H: Cinnamate-4-hydroxylase; 4CL: 4-coumarate:CoA ligases; HCT: Hydroxycinnamoyl-CoA shikimate/quinate hydroxycinnamoyl transferase; C3′H: 4-coumaroyl shikimate/quinate 3′-hydroxylase; CCoAOMT: Caffeoyl-CoA O-methyltransferase; F6′H1: Feruloyl-CoA 6′-hydroxylase; COSY: Coumarin synthase; S8H: Scopoletin 8-hydroxylase; CYP82C4: Cytochrome P450 family enzymes
| 功能分类 | 基因 | 功能 | 缺铁应答 | 参考文献 |
|---|---|---|---|---|
| 香豆素前体合成 | PAL | 反式肉桂酸合成 | 诱导型 | Rodríguez-Celma et al., |
| C4H | 4-香豆酸合成 | 诱导型 | Rodríguez-Celma et al., | |
| 4CL1/2 | 香豆酰辅酶A合成 | 诱导型 | Rodríguez-Celma et al., | |
| HCT | 咖啡酰辅酶A合成 | 诱导型 | Rodríguez-Celma et al., | |
| CCoAOMT1 | 阿魏酰辅酶A合成 | 诱导型 | Rodríguez-Celma et al., | |
| 香豆素合成 | F6H'1 | 6'-羟基阿魏酰辅酶A合成 | 诱导型 | Kai et al., |
| COSY | 莨菪亭合成 | 诱导型 | Vanholme et al., | |
| S8H | 秦皮素合成 | 诱导型 | Siwinska et al., | |
| CYP82C4 | Sideretin合成 | 诱导型 | Murgia et al., | |
| 香豆素分泌 | PDR9 | 香豆素类化合物外排分泌 | 诱导型 | Rodríguez-Celma et al., |
| BGLU42 | 莨菪苷去糖基化 | 诱导型 | Ziegler et al., | |
| 调控 | FIT | 正调控香豆素合成 | 诱导型 | Schmid et al., |
| MYB72 | 正调控香豆素合成 | 诱导型 | Stringlis et al., | |
| MYB63 | 正调控香豆素合成 | ? | DeLoose et al., | |
| bHLH121 | 正调控香豆素合成 | 组成型 | Gao et al., | |
| MYB15 | 正调控香豆素合成 | ? | Schwarz and Bauer, | |
| KFB1 | 正调控香豆素合成 | 诱导型 | Zhang et al., | |
| KFB20 | 正调控香豆素合成 | 诱导型 | Zhang et al., | |
| KFB50 | 正调控香豆素合成 | 诱导型 | Zhang et al., |
表1 缺铁诱导型香豆素类化合物合成与调控相关基因
Table 1 Genes responsible for the biosynthesis and regulation of iron deficiency-induced coumarins
| 功能分类 | 基因 | 功能 | 缺铁应答 | 参考文献 |
|---|---|---|---|---|
| 香豆素前体合成 | PAL | 反式肉桂酸合成 | 诱导型 | Rodríguez-Celma et al., |
| C4H | 4-香豆酸合成 | 诱导型 | Rodríguez-Celma et al., | |
| 4CL1/2 | 香豆酰辅酶A合成 | 诱导型 | Rodríguez-Celma et al., | |
| HCT | 咖啡酰辅酶A合成 | 诱导型 | Rodríguez-Celma et al., | |
| CCoAOMT1 | 阿魏酰辅酶A合成 | 诱导型 | Rodríguez-Celma et al., | |
| 香豆素合成 | F6H'1 | 6'-羟基阿魏酰辅酶A合成 | 诱导型 | Kai et al., |
| COSY | 莨菪亭合成 | 诱导型 | Vanholme et al., | |
| S8H | 秦皮素合成 | 诱导型 | Siwinska et al., | |
| CYP82C4 | Sideretin合成 | 诱导型 | Murgia et al., | |
| 香豆素分泌 | PDR9 | 香豆素类化合物外排分泌 | 诱导型 | Rodríguez-Celma et al., |
| BGLU42 | 莨菪苷去糖基化 | 诱导型 | Ziegler et al., | |
| 调控 | FIT | 正调控香豆素合成 | 诱导型 | Schmid et al., |
| MYB72 | 正调控香豆素合成 | 诱导型 | Stringlis et al., | |
| MYB63 | 正调控香豆素合成 | ? | DeLoose et al., | |
| bHLH121 | 正调控香豆素合成 | 组成型 | Gao et al., | |
| MYB15 | 正调控香豆素合成 | ? | Schwarz and Bauer, | |
| KFB1 | 正调控香豆素合成 | 诱导型 | Zhang et al., | |
| KFB20 | 正调控香豆素合成 | 诱导型 | Zhang et al., | |
| KFB50 | 正调控香豆素合成 | 诱导型 | Zhang et al., |
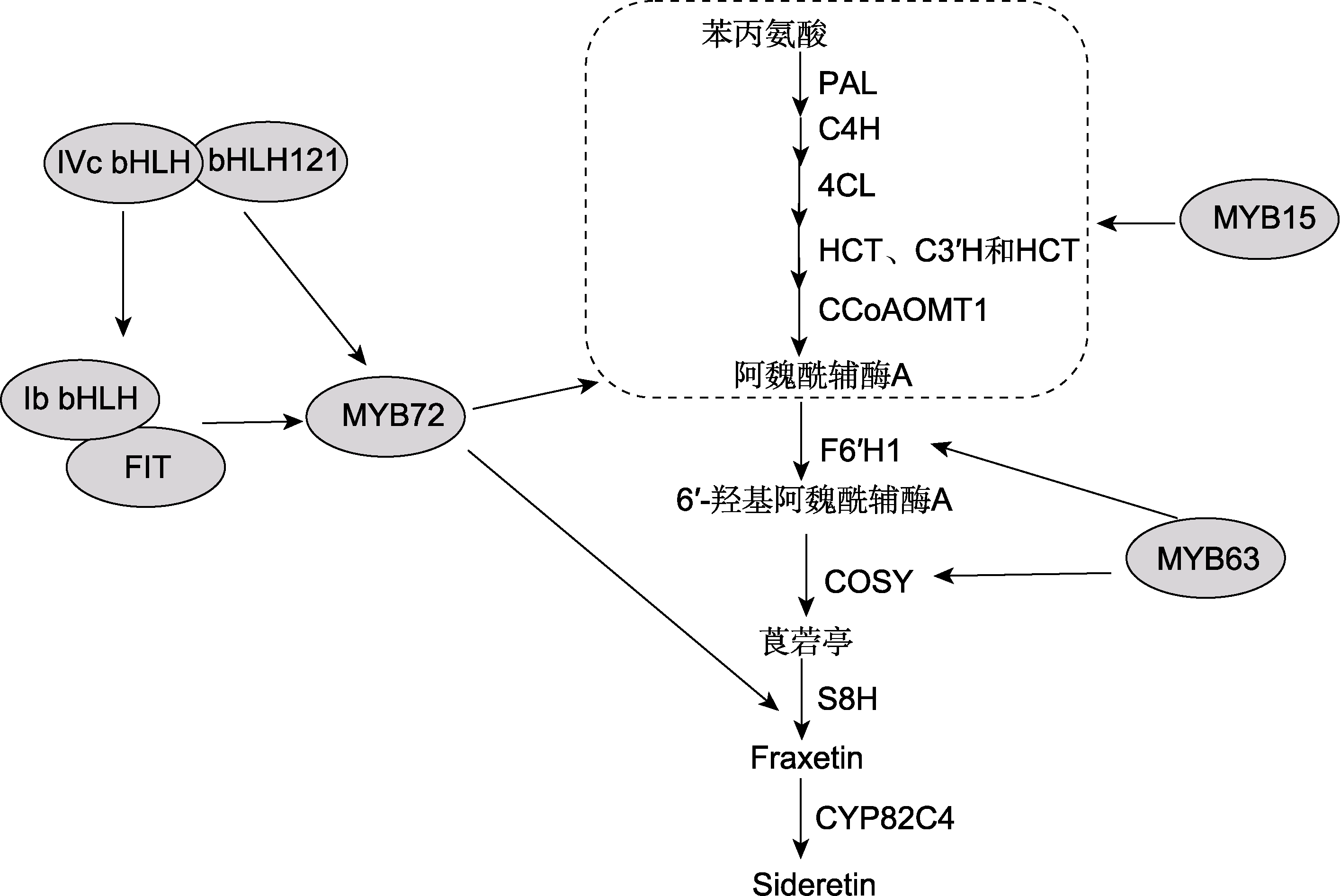
图2 拟南芥缺铁诱导型香豆素类化合物合成调控 PAL、C4H、4CL、HCT、C3′H、CCoAOMT1、F6′H1、COSY、S8H和CYP82C4同图1。IVc bHLH: 拟南芥IVc bHLH亚家族转录因子; Ib bHLH: 拟南芥Ib bHLH亚家族转录因子; FIT: 拟南芥bHLH29转录因子
Figure 2 Regulation of iron deficiency-induced coumarin biosynthesis in Arabidopsis The legends of PAL, C4H, 4CL, HCT, C3′H, CCoAOMT1, F6′H1, COSY, S8H, and CYP82C4 are the same as shown in Figure 1. IVc bHLH: Arabidopsis IVc bHLH subfamily transcription factors; Ib bHLH: Arabidopsis Ib bHLH subfamily transcription factors; FIT: Arabidopsis bHLH29 transcription factor
| [1] | Ahn YO, Shimizu BI, Sakata K, Gantulga D, Zhou Z, Bevan DR, Esen A (2010). Scopolin-hydrolyzing β-glucosidases in roots of Arabidopsis. Plant Cell Physiol 51, 132-143. |
| [2] | Bauer P, Ling HQ, Guerinot ML (2007). FIT, the FER-like iron deficiency induced transcription factor in Arabidopsis. Plant Physiol Biochem 45, 260-261. |
| [3] | Briat JF, Dubos C, Gaymard F (2015). Iron nutrition, biomass production, and plant product quality. Trends Plant Sci 20, 33-40. |
| [4] | Brown JC, Ambler JE (1973). “Reductants” released by roots of Fe-deficient soybeans. Agron J 65, 311-314. |
| [5] |
Brumbarova T, Bauer P, Ivanov R (2015). Molecular mechanisms governing Arabidopsis iron uptake. Trends Plant Sci 20, 124-133.
DOI PMID |
| [6] | Carvalhais LC, Dennis PG, Fedoseyenko D, Hajirezaei MR, Borriss R, von Wirén N (2011). Root exudation of sugars, amino acids, and organic acids by maize as affected by nitrogen, phosphorus, potassium, and iron deficiency. J Plant Nutr Soil Sci 174, 3-11. |
| [7] | Chezem WR, Memon A, Li FS, Weng JK, Clay NK (2017). SG2-type R2R3-MYB transcription factor MYB15 controls defense-induced lignification and basal immunity in Arabidop-sis. Plant Cell 29, 1907-1926. |
| [8] |
Chutia R, Abel S, Ziegler J (2019). Iron and phosphate deficiency regulators concertedly control coumarin profiles in Arabidopsis thaliana roots during iron, phosphate, and combined deficiencies. Front Plant Sci 10, 113.
DOI PMID |
| [9] | Colangelo EP, Guerinot ML (2004). The essential basic helix-loop-helix protein FIT1 is required for the iron deficiency response. Plant Cell 16, 3400-3412. |
| [10] |
de Brito Francisco R, Martinoia E (2018). The vacuolar transportome of plant specialized metabolites. Plant Cell Physiol 59, 1326-1336.
DOI PMID |
| [11] | DeLoose M, Cho H, Bouain N, Choi I, Prom-U-Thai C, Shahzad Z, Zheng LQ, Rouached H (2024). PDR9 allelic variation and MYB63 modulate nutrient-dependent coumarin homeostasis in Arabidopsis. Plant J 117, 1716-1727. |
| [12] | Dong NQ, Lin HX (2021). Contribution of phenylpropanoid metabolism to plant development and plant-environment interactions. J Integr Plant Biol 63, 180-209. |
| [13] | Ducos E, Fraysse ÅS, Boutry M (2005). NtPDR3, an iron-deficiency inducible ABC transporter in Nicotiana taba-cum. FEBS Lett 579, 6791-6795. |
| [14] |
Fourcroy P, Sisó-Terraza P, Sudre D, Savirón M, Reyt G, Gaymard F, Abadía A, Abadia J, Álvarez-Fernández A, Briat JF (2014). Involvement of the ABCG37 transporter in secretion of scopoletin and derivatives by Arabidopsis roots in response to iron deficiency. New Phytol 201, 155-167.
DOI PMID |
| [15] |
Fourcroy P, Tissot N, Gaymard F, Briat JF, Dubos C (2016). Facilitated Fe nutrition by phenolic compounds excreted by the Arabidopsis ABCG37/PDR9 transporter requires the IRT1/FRO2 high-affinity root Fe2+ transport system. Mol Plant 9, 485-488.
DOI PMID |
| [16] |
Gao F, Dubos C (2021). Transcriptional integration of plant responses to iron availability. J Exp Bot 72, 2056-2070.
DOI PMID |
| [17] | Gao F, Robe K, Bettembourg M, Navarro N, Rofidal V, Santoni V, Gaymard F, Vignols F, Roschzttardtz H, Izquierdo E, Dubos C (2020a). The transcription factor bHLH121 interacts with bHLH105 (ILR3) and its closest homologs to regulate iron homeostasis in Arabidopsis. Plant Cell 32, 508-524. |
| [18] | Gao F, Robe K, Dubos C (2020b). Further insights into the role of bHLH121 in the regulation of iron homeostasis in Arabidopsis thaliana. Plant Signal Behav 15, 1795582. |
| [19] |
Gao F, Robe K, Gaymard F, Izquierdo E, Dubos C (2019). The transcriptional control of iron homeostasis in plants: a tale of bHLH transcription factors? Front Plant Sci 10, 6.
DOI PMID |
| [20] |
Guerinot ML, Yi Y (1994). Iron: nutritious, noxious, and not readily available. Plant Physiol 104, 815-820.
DOI PMID |
| [21] |
Hänsch R, Mendel RR (2009). Physiological functions of mineral micronutrients (Cu, Zn, Mn, Fe, Ni, Mo, B, Cl). Curr Opin Plant Biol 12, 259-266.
DOI PMID |
| [22] | Hether NH, Olsen RA, Jackson LL (1984). Chemical identification of iron reductants exuded by plant roots. J Plant Nutr 7, 667-676. |
| [23] |
Jin CW, You GY, He YF, Tang CX, Wu P, Zheng SJ (2007). Iron deficiency-induced secretion of phenolics facilitates the reutilization of root apoplastic iron in red clover. Plant Physiol 144, 278-285.
DOI PMID |
| [24] | Kai K, Mizutani M, Kawamura N, Yamamoto R, Tamai M, Yamaguchi H, Sakata K, Shimizu BI (2008). Scopoletin is biosynthesized via ortho-hydroxylation of feruloyl CoA by a 2-oxoglutarate-dependent dioxygenase in Arabidopsis thaliana. Plant J 55, 989-999. |
| [25] |
Knoblauch M, Vendrell M, De Leau E, Paterlini A, Knox K, Ross-Elliot T, Reinders A, Brockman SA, Ward J, Oparka K (2015). Multispectral phloem-mobile probes: properties and applications. Plant Physiol 167, 1211-1220.
DOI PMID |
| [26] |
Kobayashi T, Nishizawa NK (2012). Iron uptake, translocation, and regulation in higher plants. Annu Rev Plant Biol 63, 131-152.
DOI PMID |
| [27] | Lan P, Li WF, Wen TN, Shiau JY, Wu YC, Lin W, Schmidt W (2011). iTRAQ protein profile analysis of Arabidopsis roots reveals new aspects critical for iron homeostasis. Plant Physiol 155, 821-834. |
| [28] | Lefèvre F, Fourmeau J, Pottier M, Baijot A, Cornet T, Abadía J, Álvarez-Fernández A, Boutry M (2018). The Nicotiana tabacum ABC transporter NtPDR3 secretes O- methylated coumarins in response to iron deficiency. J Exp Bot 69, 4419-4431. |
| [29] |
Leiková A, Giehl RFH, Hartmann A, Fargaiová A, von Wirén N (2017). Heavy metals induce iron deficiency responses at different hierarchic and regulatory levels. Plant Physiol 174, 1648-1668.
DOI PMID |
| [30] | Li LM, Wu LH, Ma GR (2010). The progress on iron-absorbing mechanism and related gene in plant. Chin J Soil Sci 41, 994-999. (in Chinese) |
| 李利敏, 吴良欢, 马国瑞 (2010). 植物吸收铁机理及其相关基因研究进展. 土壤通报 41, 994-999. | |
| [31] | Murgia I, Tarantino D, Soave C, Morandini P (2011). Arabidopsis CYP82C4 expression is dependent on Fe availability and circadian rhythm, and correlates with genes involved in the early Fe deficiency response. J Plant Physiol 168, 894-902. |
| [32] | Paffrath V, Tandron Moya YA, Weber G, von Wirén N, Giehl RFH (2024). A major role of coumarin-dependent ferric iron reduction in strategy I-type iron acquisition in Arabidopsis. Plant Cell 36, 642-664. |
| [33] |
Pan IC, Tsai HH, Cheng YT, Wen TN, Buckhout TJ, Schmidt W (2015). Post-transcriptional coordination of the Arabidopsis iron deficiency response is partially dependent on the E3 ligases RING DOMAIN LIGASE1 (RGLG1) and RING DOMAIN LIGASE2 (RGLG2). Mol Cell Proteomics 14, 2733-2752.
DOI PMID |
| [34] |
Rajniak J, Giehl RFH, Chang E, Murgia I, von Wirén N, Sattely ES (2018). Biosynthesis of redox-active metabolites in response to iron deficiency in plants. Nat Chem Biol 14, 442-450.
DOI PMID |
| [35] |
Riaz N, Guerinot ML (2021). All together now: regulation of the iron deficiency response. J Exp Bot 72, 2045-2055.
DOI PMID |
| [36] | Robe K, Conejero G, Gao F, Lefebvre-Legendre L, Sylvestre-Gonon E, Rofidal V, Hem S, Rouhier N, Barberon M, Hecker A, Gaymard F, Izquierdo E, Dubos C (2021a). Coumarin accumulation and trafficking in Arabidopsis thaliana: a complex and dynamic process. New Phytol 229, 2062-2079. |
| [37] | Robe K, Izquierdo E, Vignols F, Rouached H, Dubos C (2021b). The coumarins: secondary metabolites playing a primary role in plant nutrition and health. Trends Plant Sci 26, 248-259. |
| [38] | Rodríguez-Celma J, Lin WD, Fu GM, Abadía J, Lopez-Millan AF, Schmidt W (2013). Mutually exclusive alterations in secondary metabolism are critical for the uptake of insoluble iron compounds by Arabidopsis and Medicago truncatula. Plant Physiol 162, 1473-1485. |
| [39] |
Rodríguez-Celma J, Tsai YH, Wen TN, Wu YC, Curie C, Schmidt W (2016). Systems-wide analysis of manganese deficiency-induced changes in gene activity of Arabidopsis roots. Sci Rep 6, 35846.
DOI PMID |
| [40] | Rodríguez-Celma J, Vázquez-Reina S, Orduna J, Abadía A, Abadía J, Álvarez-Fernández A, López-Millán AF (2011). Characterization of flavins in roots of Fe-deficient strategy I plants, with a focus on Medicago truncatula. Plant Cell Physiol 52, 2173-2189. |
| [41] | Römheld V, Marschner H (1981). Iron deficiency stress induced morphological and physiological changes in root tips of sunflower. Physiol Plant 53, 354-360. |
| [42] | Römheld V, Marschner H (1983). Mechanism of iron uptake by peanut plants: I. FeIII reduction, chelate splitting, and release of phenolics. Plant Physiol 71, 949-954. |
| [43] | Römheld V, Marschner H (1986). Evidence for a specific uptake system for iron phytosiderophores in roots of gras- ses. Plant Physiol 80, 175-180. |
| [44] | Schmid NB, Giehl RFH, Döll S, Mock HP, Strehmel N, Scheel D, Kong XL, Hider RC, von Wirén N (2014). Feruloyl-CoA 6′-hydroxylase1-dependent coumarins mediate iron acquisition from alkaline substrates in Arabidopsis. Plant Physiol 164, 160-172. |
| [45] | Schmidt W (1999). Mechanisms and regulation of reduction-based iron uptake in plants. New Phytol 141, 1-26. |
| [46] |
Schmidt W (2003). Iron solutions: acquisition strategies and signaling pathways in plants. Trends Plant Sci 8, 188-193.
DOI PMID |
| [47] |
Schwarz B, Bauer P (2020). FIT, a regulatory hub for iron deficiency and stress signaling in roots, and FIT-dependent and independent gene signatures. J Exp Bot 71, 1694-1705.
DOI PMID |
| [48] | Shen HY, Xiong HC, Guo XT, Zuo YM (2011). Progress of molecular and physiological mechanism of iron uptake and translocation in plants. Plant Nutr Fert Sci 17, 1522-1530. (in Chinese) |
| 申红芸, 熊宏春, 郭笑彤, 左元梅 (2011). 植物吸收和转运铁的分子生理机制研究进展. 植物营养与肥料学报 17, 1522-1530. | |
| [49] | Siwinska J, Siatkowska K, Olry A, Grosjean J, Hehn A, Bourgaud F, Meharg AA, Carey M, Lojkowska E, Ihnatowicz A (2018). Scopoletin 8-hydroxylase: a novel enzyme involved in coumarin biosynthesis and iron-deficiency responses in Arabidopsis. J Exp Bot 69, 1735-1748. |
| [50] | Stringlis IA, Yu KE, Feussner K, de Jonge R, Van Bentum S, Van Verk MC, Berendsen RL, Bakker PA, Feussner I, Pieterse CM (2018). MYB72-dependent coumarin exudation shapes root microbiome assembly to promote plant health. Proc Natl Acad Sci USA 115, E5213- E5222. |
| [51] | Takemoto T, Nomoto K, Fushiya S, Ouchi R, Kusano G, Hikino H, Takagi SI, Matsuura Y, Kakudo M (1978). Structure of mugineic acid, a new amino acid possessing an iron-chelating activity from roots washings of water-cultured Hordeum vulgare L. Proc Jpn Acad Ser B 54, 469-473. |
| [52] | Tsai HH, Rodríguez-Celma J, Lan P, Wu YC, Vélez- Bermúdez IC, Schmidt W (2018). Scopoletin 8-hydroxylase-mediated fraxetin production is crucial for iron mobi- lization. Plant Physiol 177, 194-207. |
| [53] | Tsai HH, Schmidt W (2017). Mobilization of iron by plant- borne coumarins. Trends Plant Sci 22, 538-548. |
| [54] |
Vanholme R, Sundin L, Seetso KC, Kim H, Liu XY, Li J, De Meester B, Hoengenaert L, Goeminne G, Morreel K, Haustraete J, Tsai HH, Schmidt W, Vanholme B, Ralph J, Boerjan W (2019). COSY catalyses trans-cis isomerization and lactonization in the biosynthesis of coumarins. Nat Plants 5, 1066-1075.
DOI PMID |
| [55] |
Vogt T (2010). Phenylpropanoid biosynthesis. Mol Plant 3, 2-20.
DOI PMID |
| [56] |
Waters BM, Amundsen K, Graef G (2018). Gene expression profiling of iron deficiency chlorosis sensitive and tolerant soybean indicates key roles for phenylpropanoids under alkalinity stress. Front Plant Sci 9, 10.
DOI PMID |
| [57] | Welkie GW (2000). Taxonomic distribution of dicotyledonous species capable of root excretion of riboflavin under iron deficiency. J Plant Nutr 23, 1819-1831. |
| [58] | Werner C, Matile P (1985). Accumulation of coumarylglucosides in vacuoles of barley mesophyll protoplasts. J Plant Physiol 118, 237-249. |
| [59] | World Health Organization (2003). Diet, Nutrition and the Prevention of Chronic Diseases. Geneva: WHO. pp. 1-149. |
| [60] | Wu HL, Wang N, Ling HQ (2007). Uptake, translocation and regulation of iron in plants. Chin Bull Bot 24, 779-788. (in Chinese) |
| 吴慧兰, 王宁, 凌宏清 (2007). 植物铁吸收、转运和调控的分子机制研究进展. 植物学通报 24, 779-788. | |
| [61] |
Xu WJ, Dubos C, Lepiniec L (2015). Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes. Trends Plant Sci 20, 176-185.
DOI PMID |
| [62] | Yang TJW, Lin WD, Schmidt W (2010). Transcriptional profiling of the Arabidopsis iron deficiency response reveals conserved transition metal homeostasis networks. Plant Physiol 152, 2130-2141. |
| [63] |
Zamioudis C, Hanson J, Pieterse CMJ (2014). β-glucosidase BGLU42 is a MYB72-dependent key regulator of rhizobacteria-induced systemic resistance and modulates iron deficiency responses in Arabidopsis roots. New Phytol 204, 368-379.
DOI PMID |
| [64] | Zhang XB, Gou MY, Guo CR, Yang H, Liu CJ (2015). Down-regulation of Kelch domain-containing F-box protein in Arabidopsis enhances the production of (poly) phe- nols and tolerance to ultraviolet radiation. Plant Physiol 167, 337-350. |
| [65] | Zhang XB, Gou MY, Liu CJ (2013). Arabidopsis Kelch repeat F-box proteins regulate phenylpropanoid biosynthesis via controlling the turnover of phenylalanine ammonia-lyase. Plant Cell 25, 4994-5010. |
| [66] | Zhao AN, Luo GM, Luo YJ, Song DD, Xia HD, Ren HM, Zhang P (2021). Mechanism of bHLH transcription factors in the regulatory network of plant iron deficiency. J Agric Biotechnol 29, 2427-2435. (in Chinese) |
| 赵安娜, 罗光明, 罗扬婧, 宋丹丹, 夏鸿东, 任洪曼, 张攀 (2021). bHLH转录因子在植物缺铁调控网络中的作用机制. 农业生物技术学报 29, 2427-2435. | |
| [67] |
Ziegler J, Schmidt S, Chutia R, Müller J, Böttcher C, Strehmel N, Scheel D, Abel S (2016). Non-targeted profiling of semi-polar metabolites in Arabidopsis root exudates uncovers a role for coumarin secretion and lignification during the local response to phosphate limitation. J Exp Bot 67, 1421-1432.
DOI PMID |
| [68] | Ziegler J, Schmidt S, Strehmel N, Scheel D, Abel S (2017). Arabidopsis transporter ABCG37/PDR9 contributes primarily highly oxygenated coumarins to root exudation. Sci Rep 7, 3704. |
| [1] | 江亚楠, 徐雨青, 魏毅铤, 陈钧, 张蓉菀, 赵蓓蓓, 林宇翔, 饶玉春. 水稻抗病调控机制研究进展[J]. 植物学报, 2025, 60(5): 1-0. |
| [2] | 粟思琳 唐先宇 陈祎 王婷 夏石头. 系统获得性抗性的转录调控[J]. 植物学报, 2025, 60(5): 1-0. |
| [3] | 王鸿梅, 袁蔚, 薛芳, 张召聪, 刘坤, 陈四龙. 植物SWEET基因参与逆境胁迫响应及其调控机制[J]. 植物学报, 2025, 60(4): 1-0. |
| [4] | 王子韵, 吕燕文, 肖钰, 吴超, 胡新生. 植物基因表达调控与进化机制研究进展[J]. 植物学报, 2025, 60(4): 1-0. |
| [5] | 熊良林, 梁国鲁, 郭启高, 景丹龙. 基因可变剪接调控植物响应非生物胁迫研究进展[J]. 植物学报, 2025, 60(3): 435-448. |
| [6] | 李新宇, 谷月, 徐非非, 包劲松. 水稻胚乳淀粉合成相关蛋白的翻译后修饰研究进展[J]. 植物学报, 2025, 60(2): 256-270. |
| [7] | 胡海涛, 武越, 杨玲. 植物NAD(P)+的生物合成及其生物学功能研究进展[J]. 植物学报, 2025, 60(1): 114-131. |
| [8] | 范雪兰, 落艳娇, 徐超群, 郭宝林. 淫羊藿类黄酮生物合成相关基因研究进展[J]. 植物学报, 2024, 59(5): 834-846. |
| [9] | 陈雯, 周颖盈, 罗平, 崔永一. 被子植物花朵重瓣化分子调控机制[J]. 植物学报, 2024, 59(2): 257-277. |
| [10] | 夏婧, 饶玉春, 曹丹芸, 王逸, 柳林昕, 徐雅婷, 牟望舒, 薛大伟. 水稻中乙烯生物合成关键酶OsACS和OsACO调控机制研究进展[J]. 植物学报, 2024, 59(2): 291-301. |
| [11] | 朱璐, 袁冲, 刘义飞. 植物次生代谢产物生物合成基因簇研究进展[J]. 植物学报, 2024, 59(1): 134-143. |
| [12] | 刘潇潇, 巩迪, 高天鹏, 殷俐娜, 王仕稳. 植物类囊体主要膜脂及其生物合成[J]. 植物学报, 2024, 59(1): 144-155. |
| [13] | 董小云, 魏家萍, 崔俊美, 武泽峰, 郑国强, 李辉, 王莹, 田海燕, 刘自刚. 植物抗冻蛋白研究进展[J]. 植物学报, 2023, 58(6): 966-981. |
| [14] | 苏鲁方, 王萍, 李顺, 蔡燕, 郭丹丹, 刘琴, 刘小云. 植物sirtuin蛋白家族研究进展[J]. 植物学报, 2023, 58(6): 998-1007. |
| [15] | 戴若惠, 钱心妤, 孙静蕾, 芦涛, 贾绮玮, 陆天麒, 路梅, 饶玉春. 水稻叶色调控机制及相关基因研究进展[J]. 植物学报, 2023, 58(5): 799-812. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
