

植物学报 ›› 2023, Vol. 58 ›› Issue (3): 433-439.DOI: 10.11983/CBB22055 cstr: 32102.14.CBB22055
孙尚1, 胡颖颖1, 韩阳朔2, 薛超2, 龚志云1,2( )
)
收稿日期:2022-03-24
接受日期:2022-08-04
出版日期:2023-05-01
发布日期:2023-05-17
通讯作者:
*E-mail: zygong@yzu.edu.cn
基金资助:
Shang Sun1, Yingying Hu1, Yangshuo Han2, Chao Xue2, Zhiyun Gong1,2( )
)
Received:2022-03-24
Accepted:2022-08-04
Online:2023-05-01
Published:2023-05-17
Contact:
*E-mail: zygong@yzu.edu.cn
摘要: 染色体制备与识别技术是遗传学研究的重要手段, 而寡核苷酸荧光原位杂交(oligo-FISH)是近年来兴起的染色体识别技术。灵活高效的探针是荧光原位杂交过程中的关键因素。传统的单链寡核苷酸探针标记过程复杂, 且获得单个探针的成本较高。在单链寡核苷酸探针的基础上进行改良, 利用靶向全染色体(片段)的特异性引物进行扩增, 将获得产物纯化, 即可得到目的探针, 简化了探针标记过程, 降低了成本, 并提高了标记效率。该文详述了水稻(Oryza sativa)改良后的双链寡核苷酸探针文库的合成及标记方法、有丝分裂时期染色体制片和探针杂交过程。通过设计梯度实验发现水稻中寡核苷酸荧光原位杂交技术染色体和寡核苷酸探针的最佳变性时间与温度分别为85°C 3分钟30秒及90°C 6分钟。该研究在水稻中建立染色体双链寡核苷酸荧光原位杂交技术, 可为多种植物染色体制备与精准识别提供有力的工具。
孙尚, 胡颖颖, 韩阳朔, 薛超, 龚志云. 水稻染色体双链寡核苷酸荧光原位杂交技术. 植物学报, 2023, 58(3): 433-439.
Shang Sun, Yingying Hu, Yangshuo Han, Chao Xue, Zhiyun Gong. Double-stranded Labelled Oligo-FISH in Rice Chromosomes. Chinese Bulletin of Botany, 2023, 58(3): 433-439.
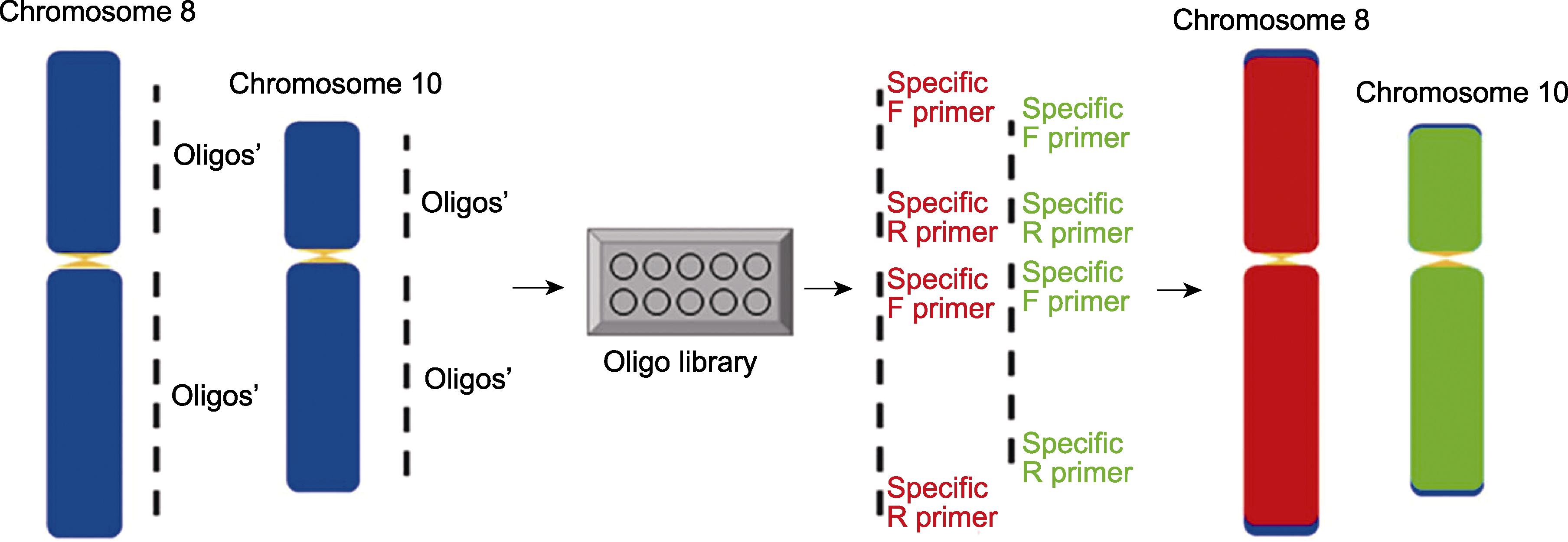
图1 在水稻中建立多重PCR扩增的双链寡核苷酸涂染探针 蓝色矩形代表染色体; 黄色三角代表着丝粒; 红色矩形代表由TAMRA标记的8号染色体涂染的oligos信号; 绿色矩形代表由FAM标记的10号染色体涂染的oligos信号。F和R分别代表不同染色体区段扩增的特异正向和反向引物。虚线代表筛选出的在两端加接头的特异oligos合集。
Figure 1 Establishment of double-stranded oligo-painting probes based on multiplex PCR in rice The blue rectangles are chromosomes; the small yellow triangles are centromeres; the red rectangles represent TAMRA-labelled chromosome 8 oligo signals; the green rectangles represent FAM-labelled chromosome 10 oligo signals. Specific forward and reverse primers for the amplification of different chromosomal segments represented by F and R, respectively. The dashed line represents the selected specific oligo set with connectors at both ends.
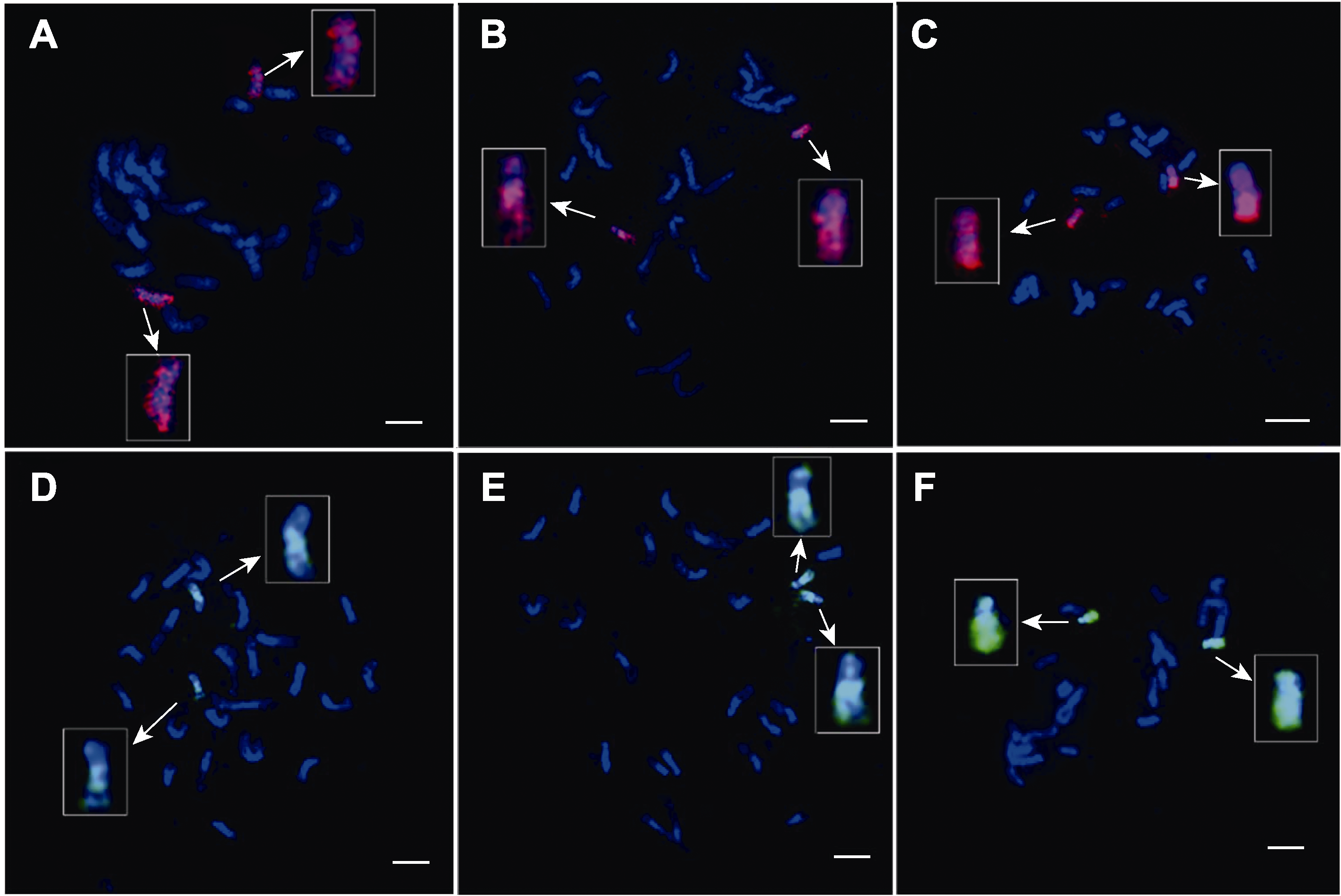
图2 双链寡核苷酸荧光原位杂交技术中水稻染色体变性时间的探索 (A)-(C) 实验过程中水稻染色体变性时间为1分30秒、2分30秒和3分30秒时, 由TAMRA标记的8号全染色体的oligos探针信号的呈现效果; (D)-(F) 实验过程中水稻染色体变性时间为1分30秒、2分30秒和3分30秒时, 由FAM标记的10号全染色体的oligos探针信号的呈现效果。白色箭头示放大后的涂染染色体。染色体由DAPI复染。Bars=5 μm
Figure 2 Exploring the denaturation time of rice chromosomes in double-stranded oligo-FISH (A)-(C) During the experiment, the effects of oligo signals labelled by TAMRA on chromosome 8 while the denaturation time at 1 min 30 s, 2 min 30 s and 3 min 30 s, respectively. (D)-(F) During the experiment, the effects of oligo signals labelled by FAM on chromosome 10 while the denaturation time at 1 min 30 s, 2 min 30 s and 3 min 30 s, respectively. The white arrow points to the painting-chromosomes after magnification. Chromosomes were counterstained with DAPI. Bars=5 μm
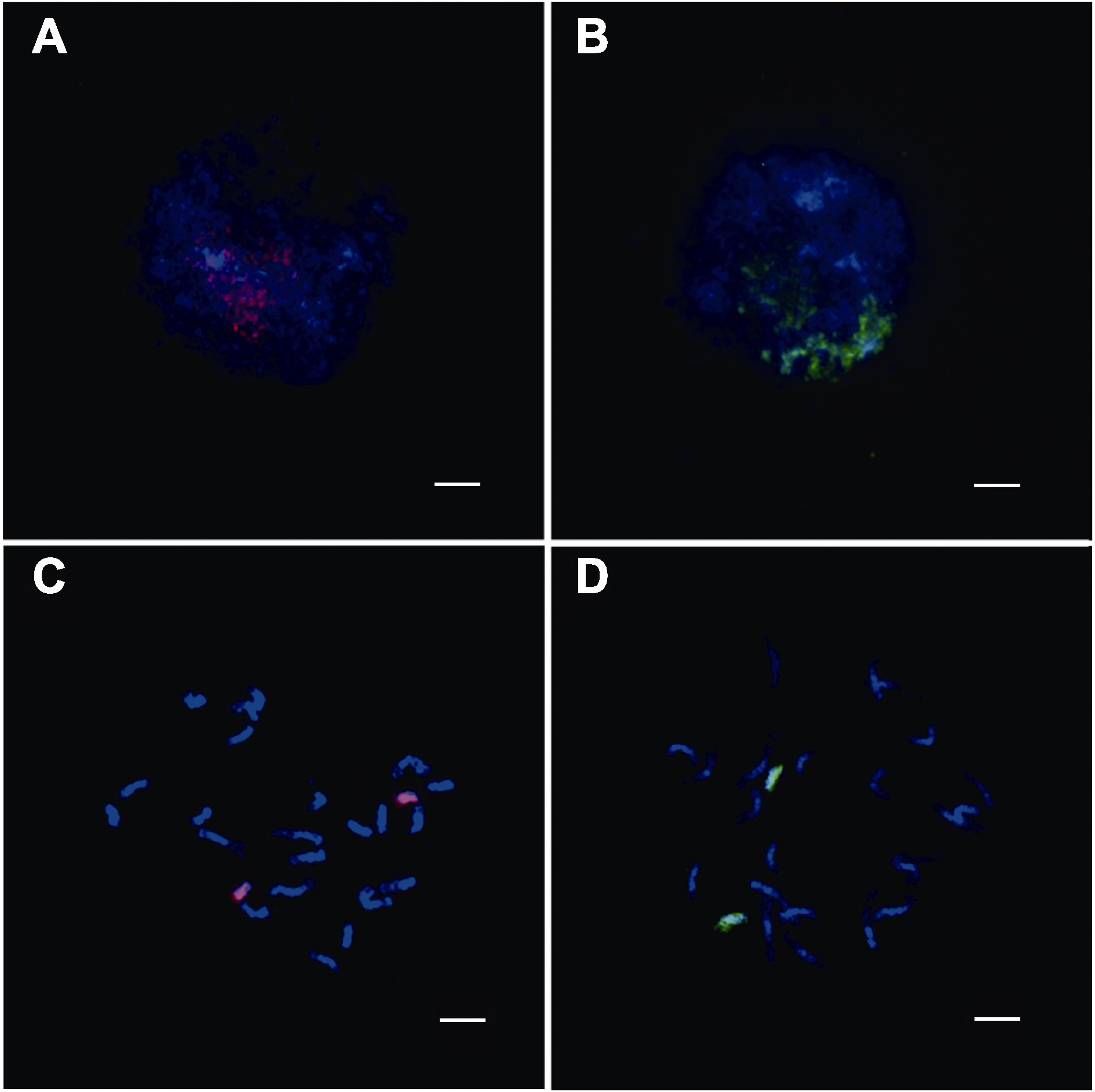
图3 双链寡核苷酸涂染荧光原位杂交技术靶向水稻日本晴有丝分裂时期第8号和10号染色体核型图 (A) 有丝分裂间期8号染色体的涂染核型图; (B) 有丝分裂间期10号染色体的涂染核型图; (C) 有丝分裂中期8号染色体的涂染核型图; (D) 有丝分裂中期10号染色体的涂染核型图。红色: TAMRA标记的8号全染色体的oligos探针信号; 绿色: FAM标记的10号全染色体的oligos探针信号。染色体由DAPI复染。Bars= 5 μm
Figure 3 Double-stranded labelled oligo-FISH based on chromosome painting targets the mitosis of chromosome 8 and 10 analysis of Nipponbare (A) Oligo-painting analysis on mitotic interphase chromoso-me 8 of Nipponbare; (B) Oligo-painting analysis on mitotic interphase chromosome 10 of Nipponbare; (C) Oligo-painting analysis on mitotic prometaphase chromosome 8 of Nippon-bare; (D) Oligo-painting analysis on mitotic prometaphase chromosome 10 of Nipponbare. Red: TAMRA-labelled probe signals of whole chromosome 8; Green: FAM-labelled probe signals of whole chromosome 10. Chromosomes were coun-terstained with DAPI. Bars=5 μm
| [1] |
Anderson LK, Stack SM, Mitchell JB (1982). An investiga-tion of the basis of a current hypothesis for the lack of G- banding in plant chromosomes. Exp Cell Res 138, 433-436.
PMID |
| [2] | Ban C, Chung S, Park DS, Shim YB (2004). Detection of protein-DNA interaction with a DNA probe: distinction between single-strand and double-strand DNA-protein interaction. Nucleic Acids Res 32, e110. |
| [3] |
Bi YF, Zhao QZ, Yan WK, Li MX, Liu YX, Cheng CY, Zhang L, Yu XQ, Li J, Qian CT, Wu YF, Chen JF, Lou QF (2020). Flexible chromosome painting based on multiplex PCR of oligo-nucleotides and its application for comparative chro-mosome analyses in Cucumis. Plant J 102, 178-186.
DOI URL |
| [4] |
Braz GT, do Vale Martins L, Zhang T, Albert PS, Birchler JA, Jiang JM, (2020). A universal chromosome identifica-tion system for maize and wild Zea species. Chromosome Res 28, 183-194.
DOI |
| [5] |
Braz GT, He L, Zhao HN, Zhang T, Semrau K, Rouillard JM, Torres GA, Jiang JM (2018). Comparative oligo-FISH mapping: an efficient and powerful methodology to reveal Karyo-typic and chromosomal evolution. Genetics 208, 513-523.
DOI URL |
| [6] |
do Vale Martins L, Yu F, Zhao HN, Dennison T, Lauter N, Wang HY, Deng ZH, Thompson A, Semrau K, Rouillard JM, Birchler JA, Jiang JM (2019). Meiotic crossovers characterized by haplotype-specific chromosome painting in maize. Nat Commun 10, 4604.
DOI PMID |
| [7] |
Fuchs J, Houben A, Brandes A, Schubert I (1996). Chro-mosome ‘painting’ in plants—a feasible technique? Chromosoma 104, 315-320.
PMID |
| [8] |
Hou LL, Xu M, Zhang T, Xu ZH, Wang WY, Zhang JX, Yu MM, Ji W, Zhu CW, Gong ZY, Gu MH, Jiang JM, Yu HX (2018). Chromosome painting and its applications in culti-vated and wild rice. BMC Plant Biol 18, 110.
DOI |
| [9] |
Huang W, Du Y, Zhao X, Jin WW (2016). B chromosome contains active genes and impacts the transcription of A chromosomes in maize (Zea mays L.). BMC Plant Biol 16, 88.
DOI PMID |
| [10] |
Jiang J, Gill BS, Wang GL, Ronald PC, Ward DC (1995). Metaphase and interphase fluorescence in situ hybridiza-tion mapping of the rice genome with bacterial artificial chromosomes. Proc Natl Acad Sci USA 92, 4487-4491.
DOI PMID |
| [11] |
Jiang JM (2019). Fluorescence in situ hybridization in plants: recent developments and future applications. Chromosome Res 27, 153-165.
DOI |
| [12] |
Jiang JM, Gill BS (2006). Current status and the future of fluorescence in situ hybridization (FISH) in plant genome research. Genome 49, 1057-1068.
DOI URL |
| [13] |
Landegent JE, Jansen in de Wal N, Dirks RW, van der Ploeg M (1987). Use of whole cosmid cloned genomic sequences for chromosomal localization by non-radioactive in situ hybridization. Hum Genet 77, 366-370.
PMID |
| [14] |
Langer-Safer PR, Levine M, Ward DC (1982). Immunological method for mapping genes on drosophila polytene chromosomes. Proc Natl Acad Sci USA 79, 4381-4385.
DOI PMID |
| [15] |
Li ZA, Bi YF, Wang X, Wang YZ, Yang SQ, Zhang ZT, Chen JF, Lou QF (2018). Chromosome identification in Cucumis anguria revealed by cross-species single-copy gene FISH. Genome 61, 397-404.
DOI URL |
| [16] |
Liu XY, Sun S, Wu Y, Zhou Y, Gu SW, Yu HX, Yi CD, Gu MH, Jiang JM, Liu B, Zhang T, Gong ZY (2020). Dual- color oligo-FISH can reveal chromosomal variations and evolution in Oryza species. Plant J 101, 112-121.
DOI URL |
| [17] |
Lou QF, Zhang YX, He YH, Li J, Jia L, Cheng CY, Guan W, Yang SQ, Chen JF (2014). Single-copy gene-based chromosome painting in cucumber and its application for chromosome rearrangement analysis in Cucumis. Plant J 78, 169-179.
DOI URL |
| [18] |
Mukai Y, Nakahara Y, Yamamoto M (1993). Simultaneous discrimination of the three genomes in hexaploid wheat by multicolor fluorescence in situ hybridization using total ge-nomic and highly repeated DNA probes. Genome 36, 489-494.
DOI PMID |
| [19] | Ouyang S, Zhu W, Hamilton J, Lin HN, Campbell M, Childs K, Thibaud-Nissen F, Malek RL, Lee Y, Zheng L, Orvis J, Haas B, Wortman J, Buell CR (2007). The TIGR rice genome annotation resource: improvements and new features. Nucleic Acids Res 35, D883-D887. |
| [20] |
Šimonikova D, Němečková A, Čížková J, Brown A, Swennen R, Doležel J, Hřibová E (2020). Chromosome painting in cultivated bananas and their wild relatives (Musa spp.) reveals differences in chromosome structure. Int J Mol Sci 21, 7915.
DOI URL |
| [21] |
Tang XM, Bao WD, Zhang WL, Cheng ZK (2007). Identifica-tion of chromosomes from multiple rice genomes using a universal molecular cytogenetic marker system. J Integr Plant Biol 49, 953-960.
DOI URL |
| [22] | Trent JM, Thompson FH (1987). Methods for chromosome banding of human and experimental tumors in vitro. Methods Enzymol 151, 267-279. |
| [23] |
Xin HY, Zhang T, Wu YF, Zhang WL, Zhang PD, Xi ML, Jiang JM (2020). An extraordinarily stable karyotype of the woody Populus species revealed by chromosome painting. Plant J 101, 253-264.
DOI URL |
| [24] |
Zhang T, Liu GQ, Zhao HN, Braz GT, Jiang JM (2021). Chorus2: design of genome-scale oligonucleotide-based probes for fluorescence in situ hybridization. Plant Biotechnol J 19, 1967-1978.
DOI PMID |
| [25] |
Zhao QZ, Meng Y, Wang PQ, Qin XD, Cheng CY, Zhou JG, Yu XQ, Li J, Lou QF, Jahn M, Chen JF (2021). Reconst-ruction of ancestral karyotype illuminates chromosome evolution in the genus Cucumis. Plant J 107, 1243-1259.
DOI URL |
| [1] | 叶灿, 姚林波, 金莹, 高蓉, 谭琪, 李旭映, 张艳军, 陈析丰, 马伯军, 章薇, 张可伟. 水稻水杨酸代谢突变体高通量筛选方法的建立与应用[J]. 植物学报, 2025, 60(4): 1-0. |
| [2] | 赵凌, 管菊, 梁文化, 张勇, 路凯, 赵春芳, 李余生, 张亚东. 基于高密度Bin图谱的水稻苗期耐热性QTL定位[J]. 植物学报, 2025, 60(3): 342-353. |
| [3] | 李新宇, 谷月, 徐非非, 包劲松. 水稻胚乳淀粉合成相关蛋白的翻译后修饰研究进展[J]. 植物学报, 2025, 60(2): 256-270. |
| [4] | 李建国, 张怡, 张文君. 水稻根系铁膜形成及对磷吸收的影响[J]. 植物学报, 2025, 60(1): 132-143. |
| [5] | 姚瑞枫, 谢道昕. 水稻独脚金内酯信号感知的激活和终止[J]. 植物学报, 2024, 59(6): 873-877. |
| [6] | 连锦瑾, 唐璐瑶, 张伊诺, 郑佳兴, 朱超宇, 叶语涵, 王跃星, 商文楠, 傅正浩, 徐昕璇, 吴日成, 路梅, 王长春, 饶玉春. 水稻抗氧化性状遗传位点挖掘及候选基因分析[J]. 植物学报, 2024, 59(5): 738-751. |
| [7] | 黄佳慧, 杨惠敏, 陈欣雨, 朱超宇, 江亚楠, 胡程翔, 连锦瑾, 芦涛, 路梅, 张维林, 饶玉春. 水稻突变体pe-1对弱光胁迫的响应机制[J]. 植物学报, 2024, 59(4): 574-584. |
| [8] | 周俭民. 收放自如的明星战车[J]. 植物学报, 2024, 59(3): 343-346. |
| [9] | 朱超宇, 胡程翔, 朱哲楠, 张芷宁, 汪理海, 陈钧, 李三峰, 连锦瑾, 唐璐瑶, 钟芊芊, 殷文晶, 王跃星, 饶玉春. 水稻穗部性状QTL定位及候选基因分析[J]. 植物学报, 2024, 59(2): 217-230. |
| [10] | 夏婧, 饶玉春, 曹丹芸, 王逸, 柳林昕, 徐雅婷, 牟望舒, 薛大伟. 水稻中乙烯生物合成关键酶OsACS和OsACO调控机制研究进展[J]. 植物学报, 2024, 59(2): 291-301. |
| [11] | 方妍力, 田传玉, 苏如意, 刘亚培, 王春连, 陈析丰, 郭威, 纪志远. 水稻抗细菌性条斑病基因挖掘与初定位[J]. 植物学报, 2024, 59(1): 1-9. |
| [12] | 朱宝, 赵江哲, 张可伟, 黄鹏. 水稻细胞分裂素氧化酶9参与调控水稻叶夹角发育[J]. 植物学报, 2024, 59(1): 10-21. |
| [13] | 贾绮玮, 钟芊芊, 顾育嘉, 陆天麒, 李玮, 杨帅, 朱超宇, 胡程翔, 李三峰, 王跃星, 饶玉春. 水稻茎秆细胞壁相关组分含量QTL定位及候选基因分析[J]. 植物学报, 2023, 58(6): 882-892. |
| [14] | 田传玉, 方妍力, 沈晴, 王宏杰, 陈析丰, 郭威, 赵开军, 王春连, 纪志远. 2019-2021年我国南方稻区白叶枯病菌的毒力与遗传多样性调查研究[J]. 植物学报, 2023, 58(5): 743-749. |
| [15] | 戴若惠, 钱心妤, 孙静蕾, 芦涛, 贾绮玮, 陆天麒, 路梅, 饶玉春. 水稻叶色调控机制及相关基因研究进展[J]. 植物学报, 2023, 58(5): 799-812. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||