

植物学报 ›› 2023, Vol. 58 ›› Issue (3): 417-432.DOI: 10.11983/CBB22065 cstr: 32102.14.CBB22065
收稿日期:2022-04-08
接受日期:2022-09-19
出版日期:2023-05-01
发布日期:2023-05-17
通讯作者:
*E-mail: tianxm06@lzu.edu.cn
基金资助:
Zhenzhou Chu, Gulbar Yisilam, Zezhong Qu, Xinmin Tian( )
)
Received:2022-04-08
Accepted:2022-09-19
Online:2023-05-01
Published:2023-05-17
Contact:
*E-mail: tianxm06@lzu.edu.cn
摘要: 刺木蓼(Atraphaxis spinosa)、额河木蓼(A. jrtyschensis)和细枝木蓼(A. decipiens)是同域分布在我国新疆北部的3种木蓼属物种。通过二代高通量测序, 对叶绿体基因组进行组装和注释, 比较3个物种的叶绿体基因组差异并进行系统发育分析。结果表明, 木蓼属3个物种的叶绿体全基因组大小为164 106-164 216 bp, 与其它绿色植物类似, 由1个大单拷贝区(LSC)、1个小单拷贝区(SSC)及介于二者之间的2个反向重复区(IRa/IRb)组成。在刺木蓼、额河木蓼和细枝木蓼中检测到48-49个串联重复序列及59-63个简单重复序列(SSR)。3个物种的核苷酸多样性平均值为0.000 96, Ka/Ks平均值为0.030 3, 遗传距离平均值为0.001 0。通过对3个物种的叶绿体基因组进行比较, 发现编码区比非编码区更保守。系统发育分析结果显示3个物种的亲缘关系较近。该研究基于叶绿体基因组对木蓼属物种的亲缘关系进行分析, 揭示了3个同域分布的物种之间的亲缘关系以及木蓼属在蓼科中的系统位置。研究结果可为木蓼属的分类学、系统学和生物地理学研究提供参考。
褚振州, 古丽巴哈尔·依斯拉木, 屈泽众, 田新民. 同域分布的3种木蓼属植物叶绿体基因组比较. 植物学报, 2023, 58(3): 417-432.
Zhenzhou Chu, Gulbar Yisilam, Zezhong Qu, Xinmin Tian. Comparative Analyses on the Chloroplast Genome of Three Sympatric Atraphaxis Species. Chinese Bulletin of Botany, 2023, 58(3): 417-432.
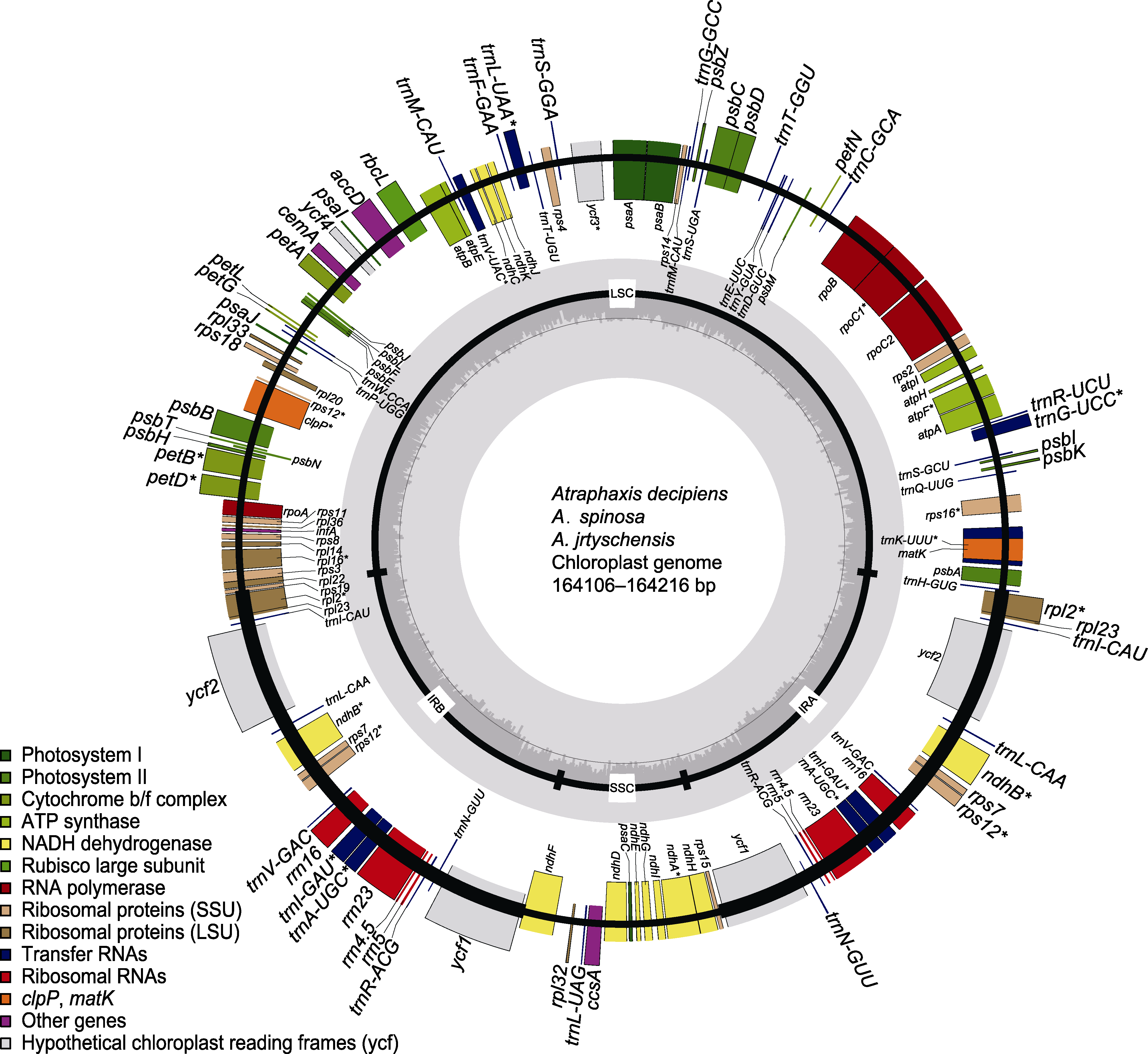
图1 木蓼属3个物种的叶绿体基因组 标注在大环外部的基因按照顺时针方向转录, 标注在大环内部的基因按照逆时针方向转录。不同基因功能标注不同颜色; 内环阴影部分代表3个物种叶绿体基因组的GC组成。LSC: 大单拷贝区; SSC: 小单拷贝区; IR: 反向重复区。* 含有内含子的基因
Figure 1 The chloroplast genome of the three Atraphaxis species Genes on the outside of the large circle are transcribed clockwise and those on the inside are transcribed counterclockwise. The genes are color-coded based on their function. The dashed area represents the GC composition of the chloroplast genome. LSC: Large single copy region; SSC: Small single copy region; IR: Inverted repeat region. * Genes including introns
| Atraphaxis spinosa | A. jrtyschensis | A. decipiens | |
|---|---|---|---|
| Gene size (bp) | 164106 | 164216 | 164187 |
| LSC (bp) | 88812 | 88900 | 88870 |
| SSC (bp) | 13456 | 13485 | 13485 |
| IR (bp) | 30919 | 30916 | 30916 |
| No. of genes | 133 | 133 | 133 |
| No. of PCGs | 88 | 88 | 88 |
| No. of tRNA | 37 | 37 | 37 |
| No. of rRNA | 8 | 8 | 8 |
| No. of duplicated genes | 20 | 20 | 20 |
| GC content (%) | 37.4 | 37.5 | 37.5 |
表1 木蓼属3个物种的叶绿体基因组基本信息
Table 1 Basic information on the chloroplast genome of the three Atraphaxis species
| Atraphaxis spinosa | A. jrtyschensis | A. decipiens | |
|---|---|---|---|
| Gene size (bp) | 164106 | 164216 | 164187 |
| LSC (bp) | 88812 | 88900 | 88870 |
| SSC (bp) | 13456 | 13485 | 13485 |
| IR (bp) | 30919 | 30916 | 30916 |
| No. of genes | 133 | 133 | 133 |
| No. of PCGs | 88 | 88 | 88 |
| No. of tRNA | 37 | 37 | 37 |
| No. of rRNA | 8 | 8 | 8 |
| No. of duplicated genes | 20 | 20 | 20 |
| GC content (%) | 37.4 | 37.5 | 37.5 |
| Gene category | Group of genes | Name of genes |
|---|---|---|
| Self-replication | Ribosomal RNAs | rrn4.5#, rrn5#, rrn16#, rrn23# |
| Transfer RNAs | trnV-GAC#, trnl-GAU#*, trnA-UGC#*, trnR-ACG#, trnN- GUU#, trnL-CAA#, trnl-CAU#, trnH-GUG, trnK-UUU*, trnQ-UUG, trnS-GUC, trnG-UCC*, trnR-UCU, trnC-GCA, trnD-GUC, trnY-GUA, trnE-UUC, trnT-GGU, trnS-UGA, trnG-GCC, trnfM-CAU, trnS-GGA, trnT-UGU, trnL-UAA*, trnF-GAA, trnM-CAU, trnV-UAC*, trnW-CCA, trnP-UGG | |
| Gene for photosynthesis | Small subunit of ribosomal proteins (SSU) | rps19, rps7#, rps12#*, rps15, rps2, rps14, rps4, rps18, rps3, rps11, rps16* |
| Large subunit of ribosomal proteins (LSU) | rpl2#*, rpl23#, rpl32, rpl33, rpl20, rpl14, rpl16*, rpl36, rpl22 | |
| RNA polymerase | rpoC2, rpoC1*, rpoB, rpoA | |
| Photosystem I | psaC, psaA, psaB, psaJ, psaI | |
| Photosystem II | psbK, psbI, psbM, psbD, psbC, psbZ, psbJ, psbL, psbF, psbE, psbB, psbT, psbH, psbA, psbN | |
| Other genes | Subunits of cytochrome b/f complex | petN, petA, petL, petG, petB*, petD* |
| Protease | clpP** | |
| Subunits of ATP synthase | atpA, atpF*, atpH, atpI, atpE, atpB | |
| Large subunit of Rubisco | rbcL | |
| Subunits of NADH-dehydrogenase | ndhB#*, ndhF, ndhD, ndhC, ndhE, ndhG, ndhI, ndhA*, ndhH, ndhJ, ndhK, ndhC | |
| Maturase | matK | |
| Envelop membrane protein | cemA | |
| Subunit of acetyl-CoA-carboxylase | accD | |
| C-type cytochrome synthesis gene | ccsA | |
| Translational initiation factor | infA | |
| Gene of unknown function | Hypothetical chloroplast reading frames (ycf) | ycf1#, ycf2#, ycf3**, ycf4, ycf15 |
表2 木蓼属3个物种叶绿体基因组的基因注释信息
Table 2 Gene annotation in the chloroplast genome of the three Atraphaxis species
| Gene category | Group of genes | Name of genes |
|---|---|---|
| Self-replication | Ribosomal RNAs | rrn4.5#, rrn5#, rrn16#, rrn23# |
| Transfer RNAs | trnV-GAC#, trnl-GAU#*, trnA-UGC#*, trnR-ACG#, trnN- GUU#, trnL-CAA#, trnl-CAU#, trnH-GUG, trnK-UUU*, trnQ-UUG, trnS-GUC, trnG-UCC*, trnR-UCU, trnC-GCA, trnD-GUC, trnY-GUA, trnE-UUC, trnT-GGU, trnS-UGA, trnG-GCC, trnfM-CAU, trnS-GGA, trnT-UGU, trnL-UAA*, trnF-GAA, trnM-CAU, trnV-UAC*, trnW-CCA, trnP-UGG | |
| Gene for photosynthesis | Small subunit of ribosomal proteins (SSU) | rps19, rps7#, rps12#*, rps15, rps2, rps14, rps4, rps18, rps3, rps11, rps16* |
| Large subunit of ribosomal proteins (LSU) | rpl2#*, rpl23#, rpl32, rpl33, rpl20, rpl14, rpl16*, rpl36, rpl22 | |
| RNA polymerase | rpoC2, rpoC1*, rpoB, rpoA | |
| Photosystem I | psaC, psaA, psaB, psaJ, psaI | |
| Photosystem II | psbK, psbI, psbM, psbD, psbC, psbZ, psbJ, psbL, psbF, psbE, psbB, psbT, psbH, psbA, psbN | |
| Other genes | Subunits of cytochrome b/f complex | petN, petA, petL, petG, petB*, petD* |
| Protease | clpP** | |
| Subunits of ATP synthase | atpA, atpF*, atpH, atpI, atpE, atpB | |
| Large subunit of Rubisco | rbcL | |
| Subunits of NADH-dehydrogenase | ndhB#*, ndhF, ndhD, ndhC, ndhE, ndhG, ndhI, ndhA*, ndhH, ndhJ, ndhK, ndhC | |
| Maturase | matK | |
| Envelop membrane protein | cemA | |
| Subunit of acetyl-CoA-carboxylase | accD | |
| C-type cytochrome synthesis gene | ccsA | |
| Translational initiation factor | infA | |
| Gene of unknown function | Hypothetical chloroplast reading frames (ycf) | ycf1#, ycf2#, ycf3**, ycf4, ycf15 |
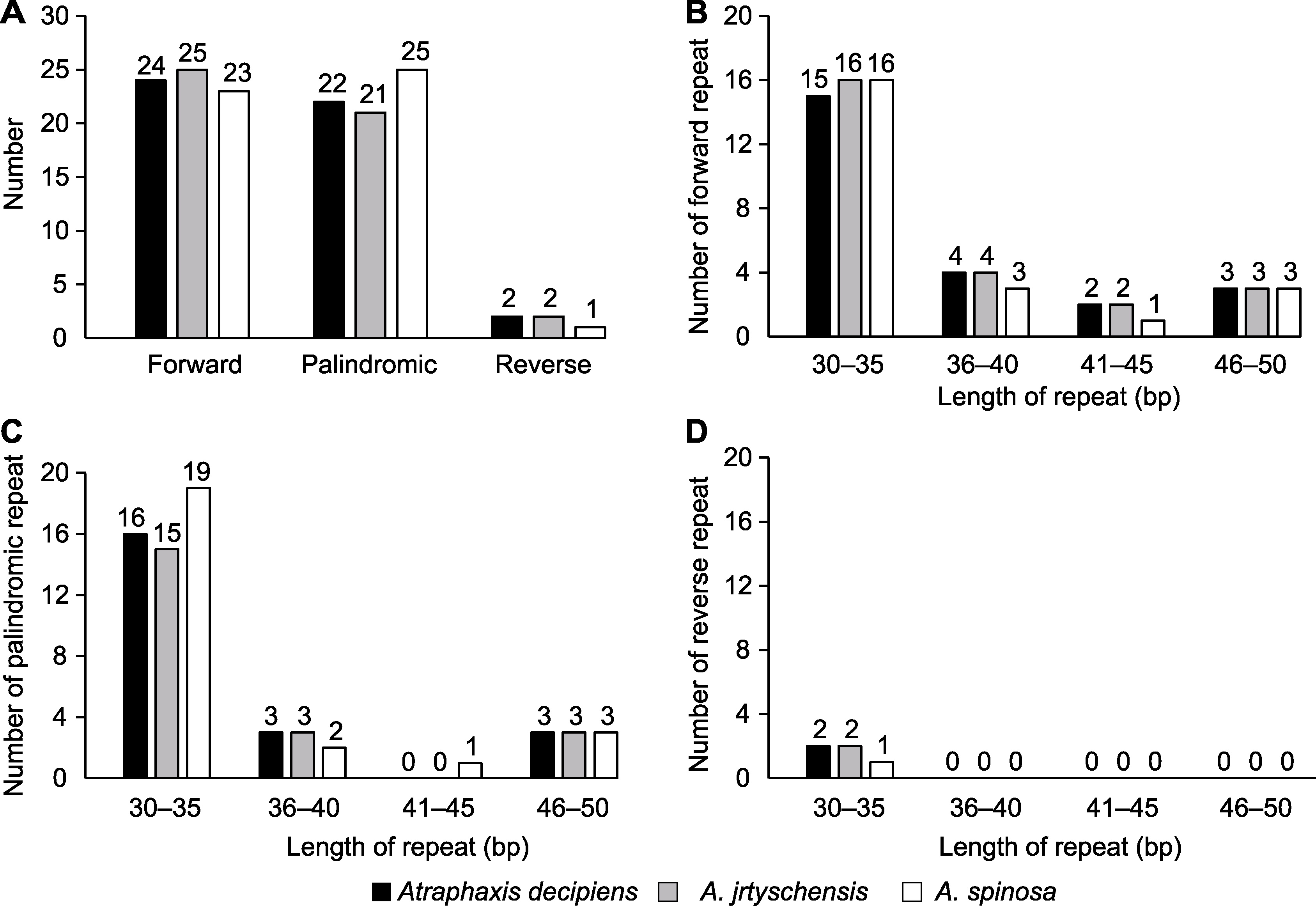
图2 木蓼属3个物种叶绿体基因组中的重复序列分析 (A) 正向重复、反向重复和回文重复的总数; (B) 正向重复序列的数量; (C) 回文重复序列的数量; (D) 反向重复序列的数量
Figure 2 Analysis of the repeat sequences in the chloroplast genome of the three Atraphaxis species (A) Total number of the forward repeats, reverse repeats and palindromic repeats; (B) Number of the forward repeats; (C) Number of the palindromic repeats; (D) Number of the reverse repeats
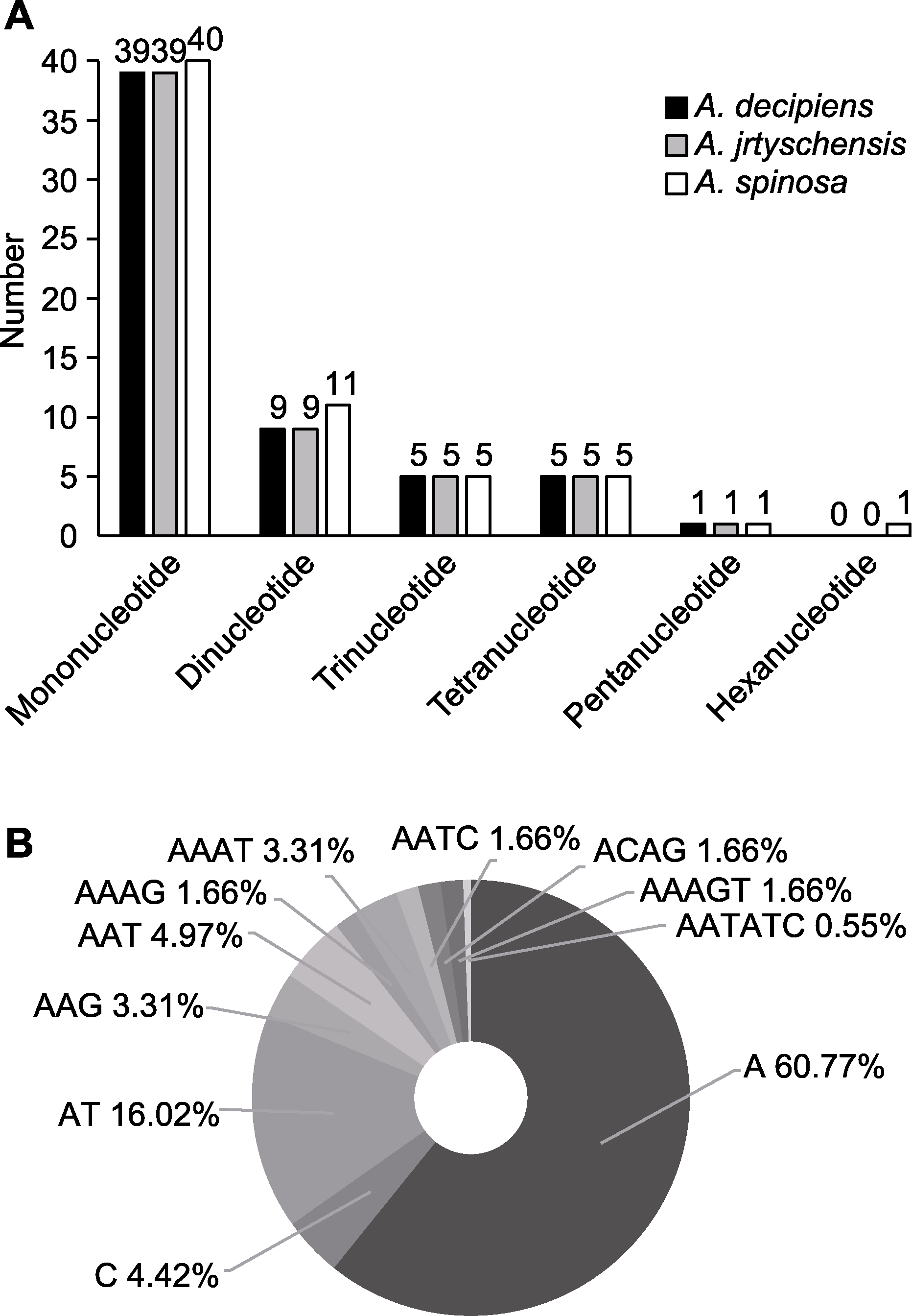
图3 木蓼属3个物种叶绿体基因组中的简单重复序列(SSRs) (A) 不同类型SSR的分布; (B) 不同类型SSR的频率
Figure 3 Simple sequence repeats (SSRs) in the chloroplast genome of the three Atraphaxis species (A) Distribution of different SSR types; (B) Frequency of different type of SSRs
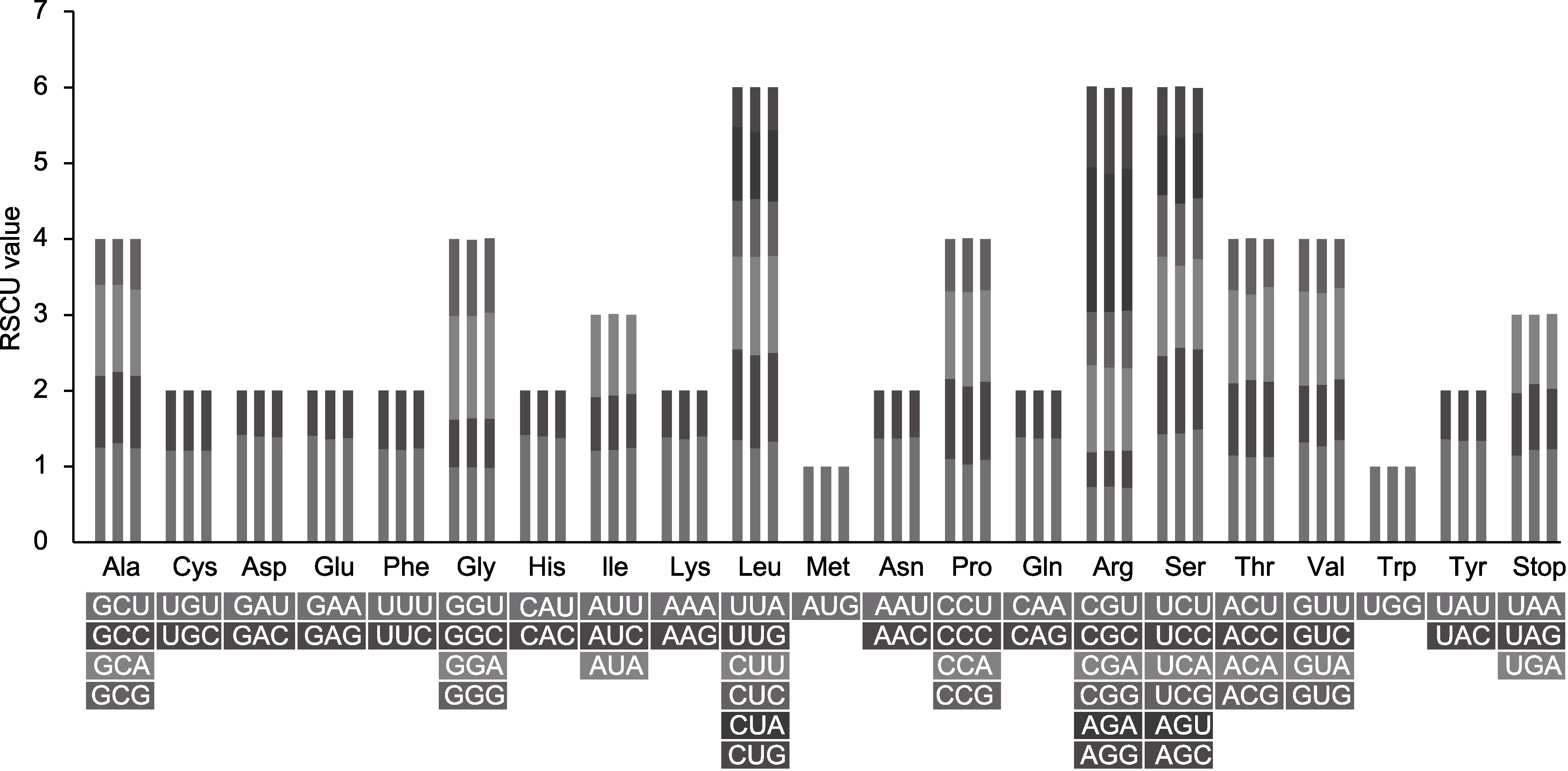
图4 木蓼属3个物种的叶绿体基因组密码子使用偏好性 每个氨基酸的3个柱形从左到右依次为刺木蓼(Atraphaxis spinosa)、额河木蓼(A. jrtyschensis)和细枝木蓼(A. decipiens)。RSCU: 相对同义密码子使用度
Figure 4 The codon usage bias of the chloroplast genomes in the three Atraphaxis species Columns above each amino acid from left to right are A. spinosa, A. jrtyschensis and A. decipiens, respectively. RSCU: Relative synonymous codon usage
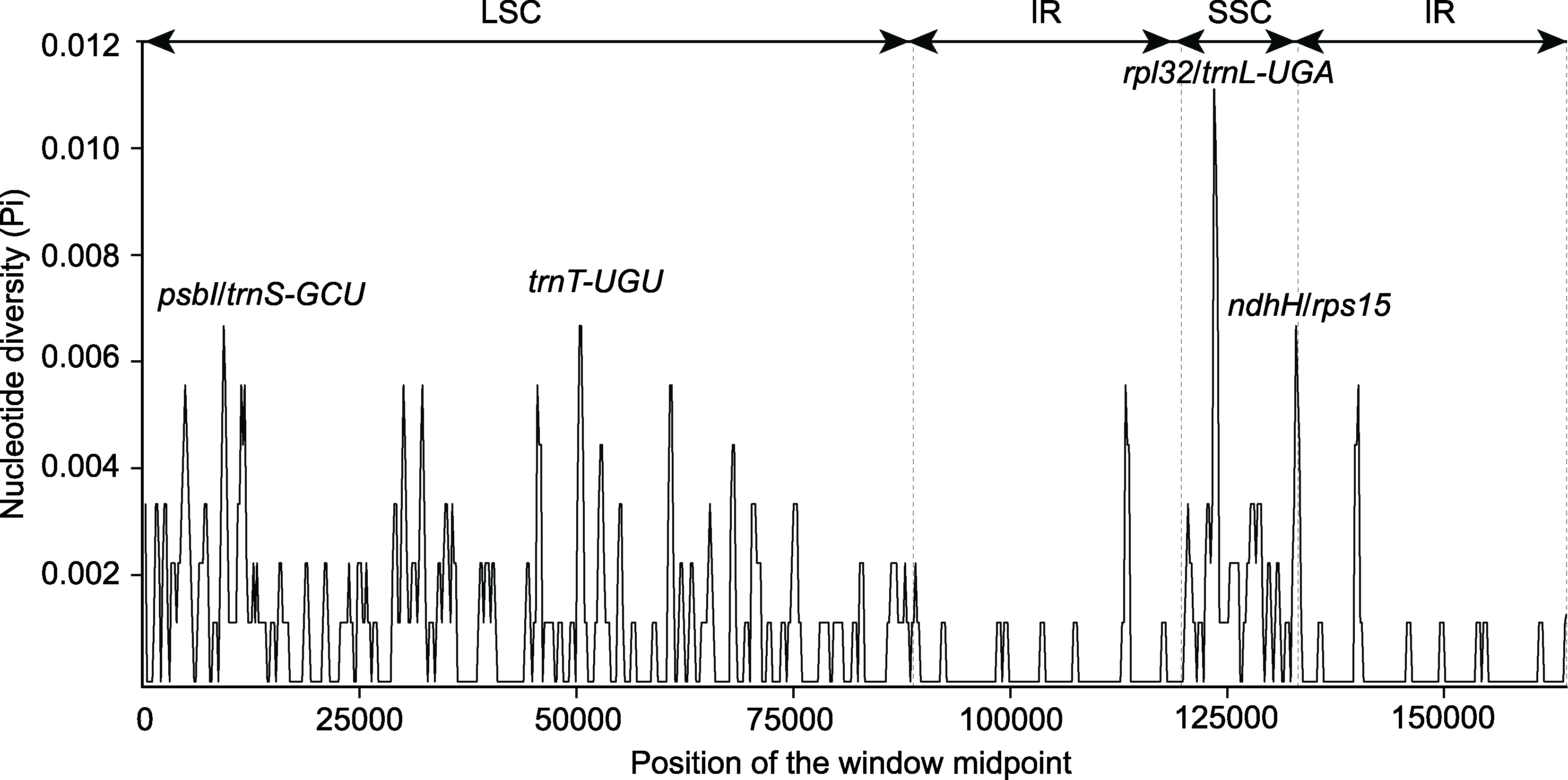
图5 木蓼属3个物种的叶绿体基因组核苷酸多样性 LSC、SSC和IR同图1。
Figure 5 The nucleotide diversity of the chloroplast genomes of the three Atraphaxis species LSC, SSC and IR are the same as shown in Figure 1.
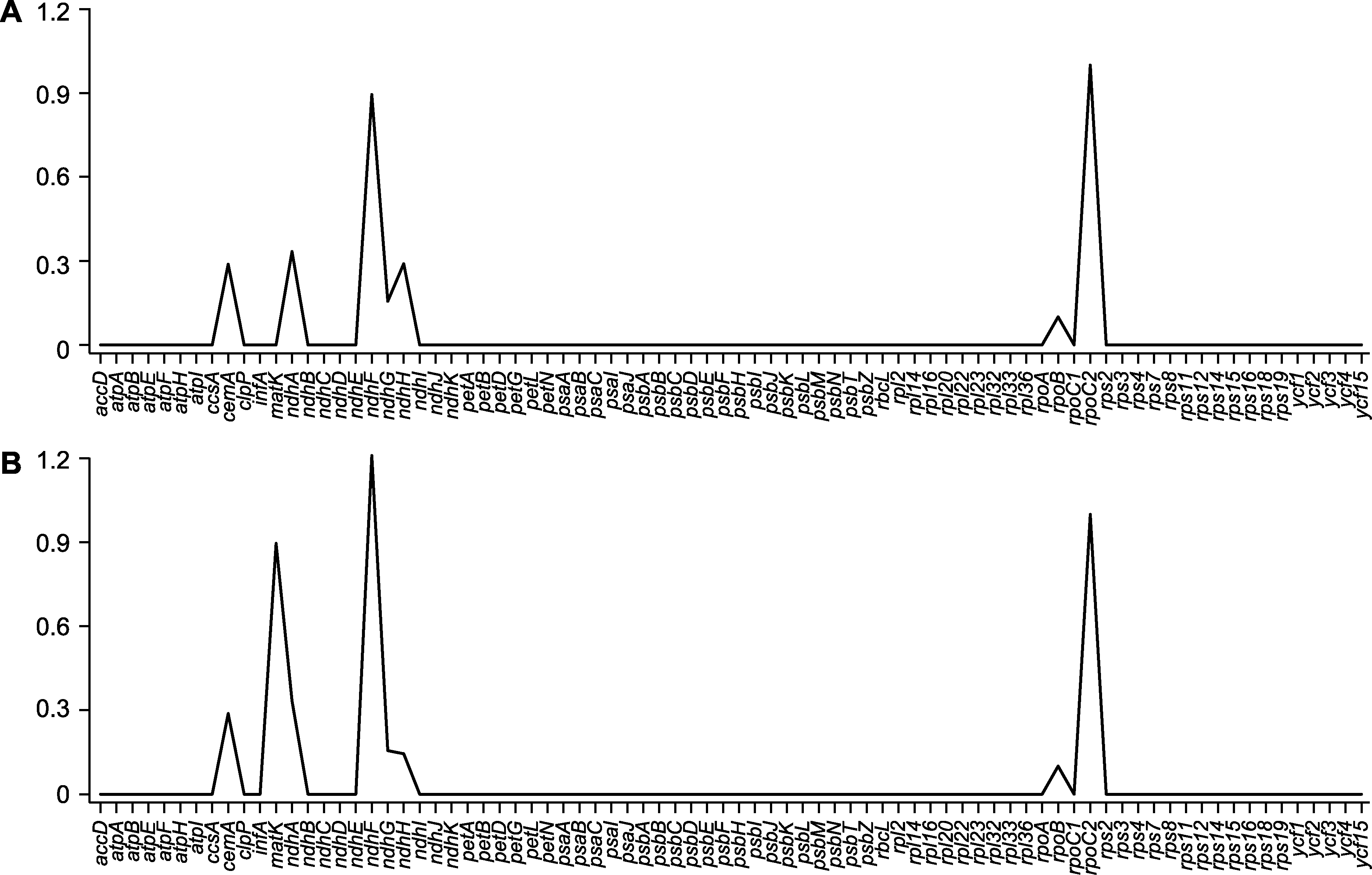
图6 木蓼属3个物种中80个蛋白质编码基因的非同义替换/同义替换(Ka/Ks)比值 (A) 刺木蓼vs额河木蓼; (B) 刺木蓼vs细枝木蓼
Figure 6 The non-synonymous-to-synonymous substitution (Ka/Ks) ratios of 80 protein-coding genes among the three Atraphaxis species (A) A. spinosa vs A. jrtyschensis; (B) A. spinosa vs A. decipiens
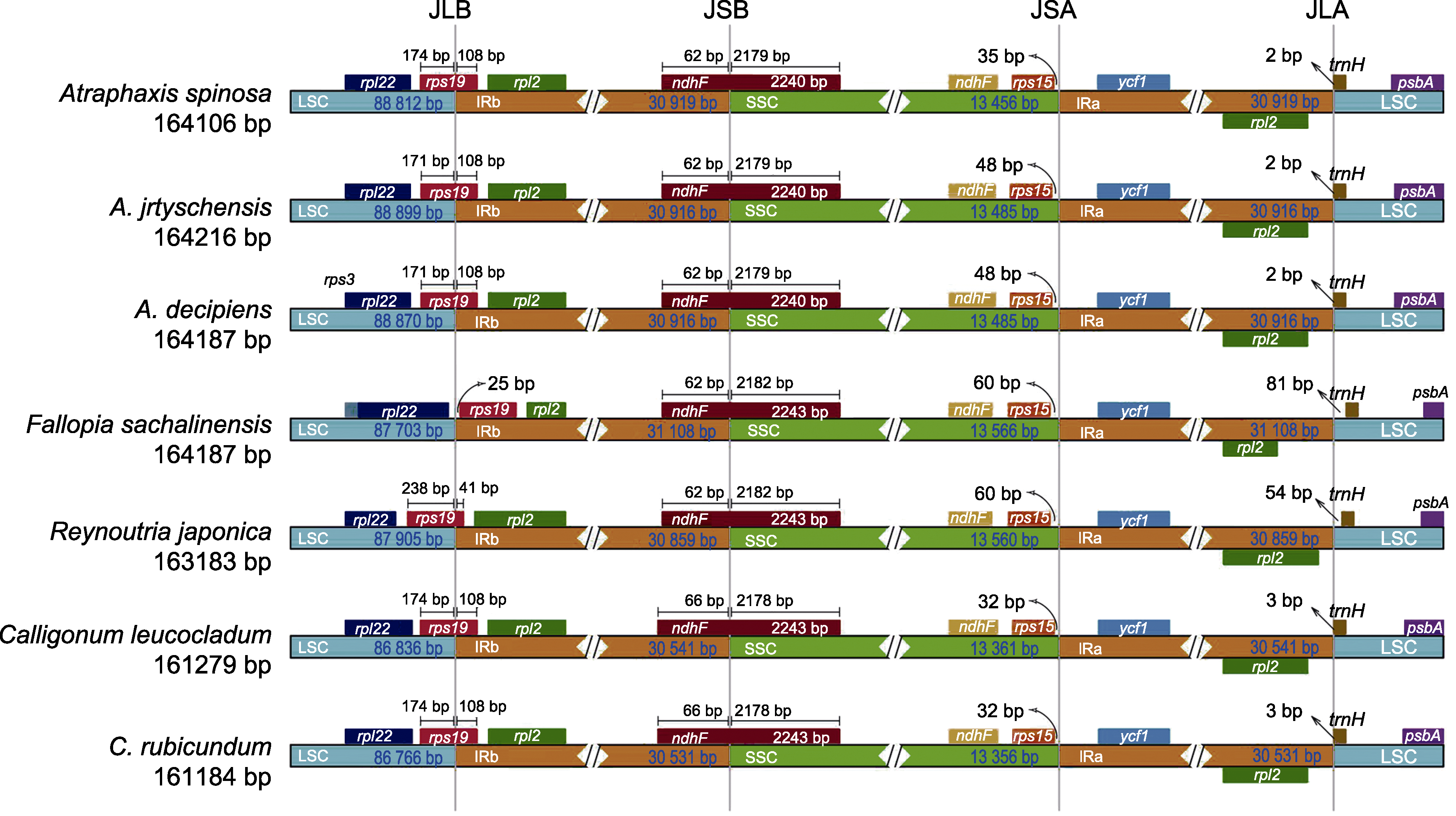
图7 蓼科7个物种之间的LSC、SSC和IR区域的边界位置比较 LSC、SSC和IR同图1。JLB: LSC与IRb的边界; JSB: IRb与SSC的边界; JSA: SSC与IRa的边界; JLA: IRa与LSC的边界。
Figure 7 Comparison of the border positions of LSC, SSC, and IR regions among the seven species of the Polygonaceae LSC, SSC and IR are the same as shown in Figure 1. JLB: Junction line between LSC and IRb; JSB: Junction line between IRb and SSC; JSA: Junction line between SSC and IRa; JLA: Junction line between IRa and LSC.
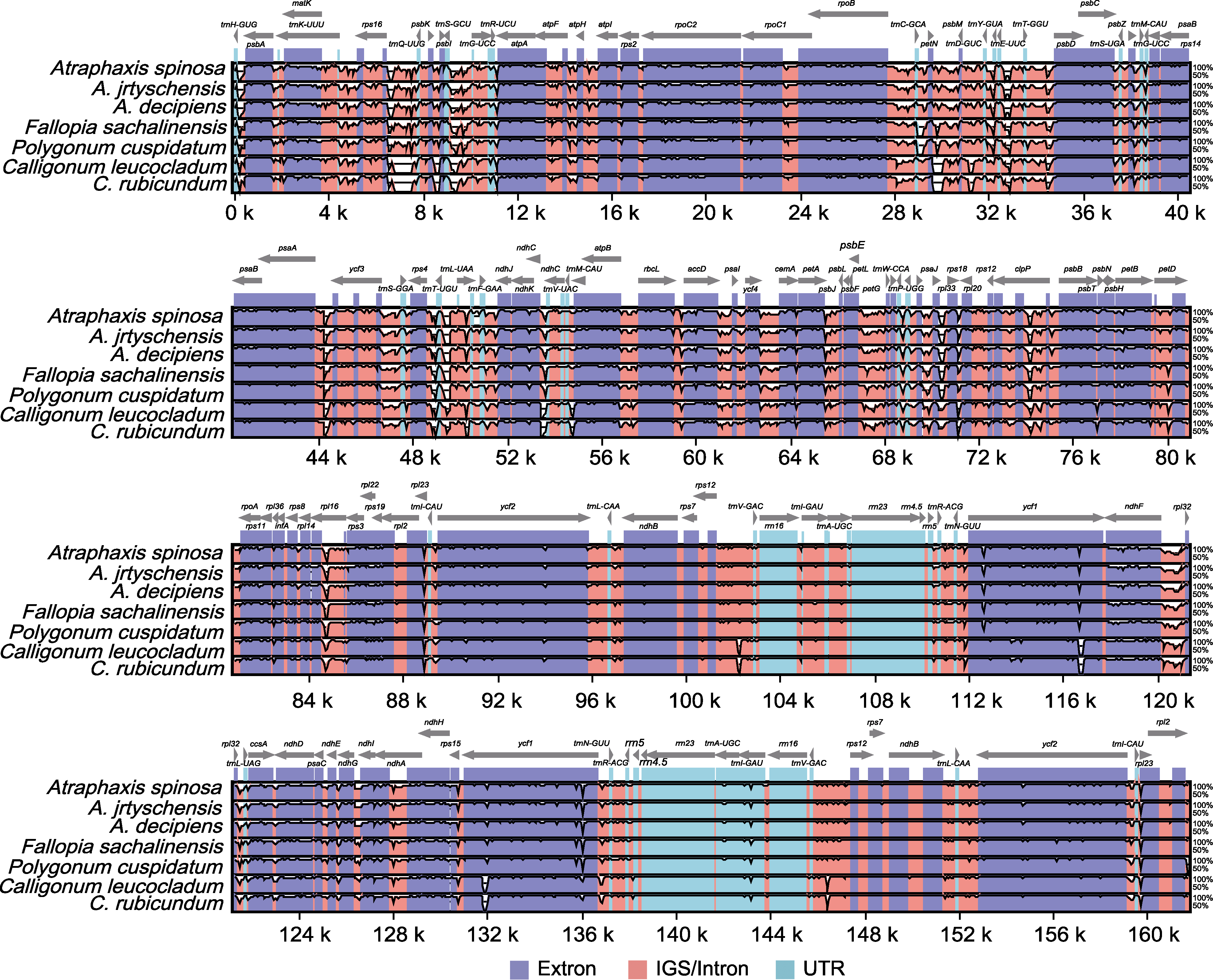
图8 7种蓼科植物叶绿体基因组mVISTA序列分析 灰线表示基因方向及在LSC、SSC和IR区域的位置。序列一致性在y轴上显示为50%-100%的百分比。基因组不同区域用不同颜色区分。
Figure 8 mVISTA sequence analysis on the chloroplast genome among seven species of the Polygonaceae The gray lines show the gene orientation and the position in the LSC, SSC, and IR regions. Sequence identity is shown as a percentage between 50%-100% on y-axis. Genome regions are distinguished by different colors.
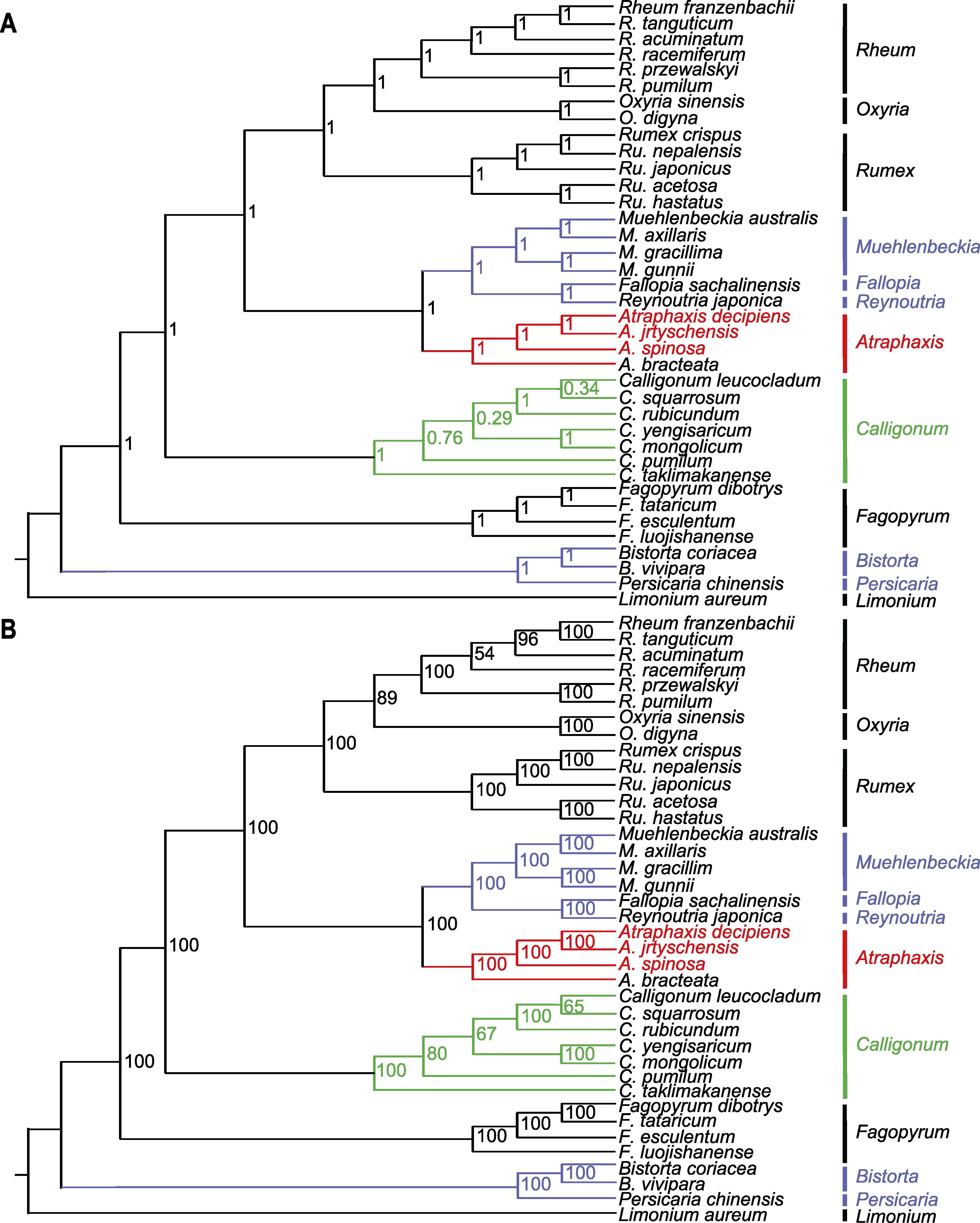
图9 基于叶绿体基因组中蛋白编码序列构建的37个蓼科物种的系统发育树 (A) 基于贝叶斯法(BI)构建的系统发育树, 分支上的数值为后验概率; (B) 基于最大似然法(ML)构建的系统发育树, 分支上的数值为自举值。以黄花补血草为外类群。红色代表本研究的3个物种。紫色代表蓼族(Trib. Polygoneae)的物种, 绿色代表木蓼族(Trib. Atraphaxideae)的物种。
Figure 9 Phylogenetic tree of 37 species in the Polygonaceae based on the protein coding sequences in their chloroplast genomes (A) The Bayesian inference (BI) tree with posterior probabilities values on the branches; (B) Maximum likelihood (ML) tree with bootstrap values on the branches. Limonium aureum is used as outgroup. The red color represents the three species in this study. The purple color represents the species of Trib. Polygoneae, and the green color represents the species of Trib. Atraphaxideae.
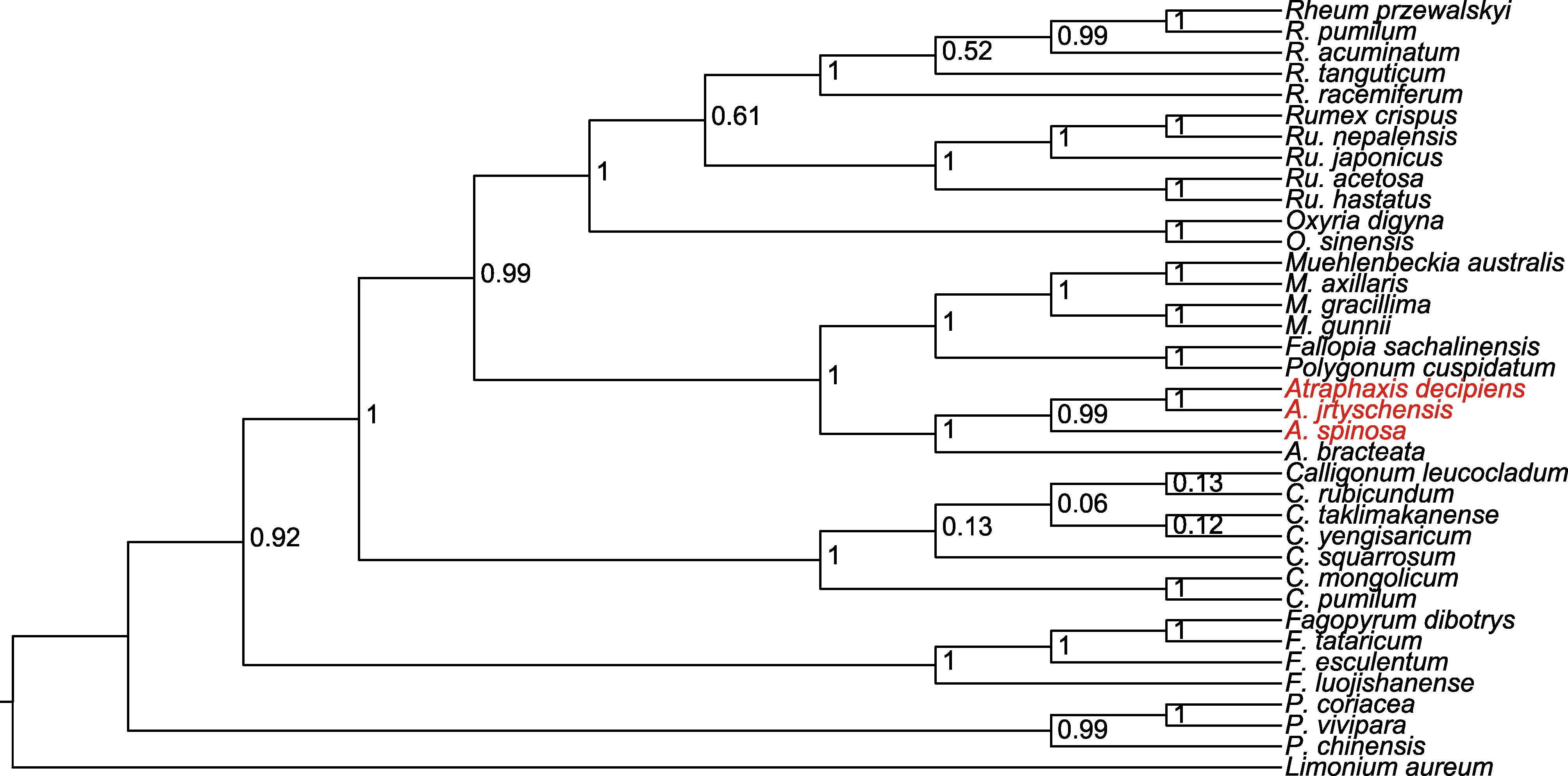
图10 基于贝叶斯法(BI), 利用ycf1、ndhA、atpA和atpI高变异位点联合构建的36个蓼科物种的系统发育树 以黄花补血草作为外类群。
Figure 10 Phylogenetic tree of 36 species in the Polygonaceae based on Bayesian inference (BI) with a combination of high variance loci of ycf1, ndhA, atpA and atpI Limonium aureum is used as outgroup.
| [1] | 艾对元 (2008). 基因组中重复序列的意义. 生命的化学 28, 343-345. |
| [2] | 李安仁, 高作经, 毛祖美, 刘玉兰 (1998). 中国植物志, 第25卷第1分册. 北京: 科学出版社. pp. 133-141. |
| [3] |
李巧丽, 延娜, 宋琼, 郭军战 (2018). 鲁桑叶绿体基因组序列及特征分析. 植物学报 53, 94-103.
DOI |
| [4] | 马克平 (1993). 试论生物多样性的概念. 生物多样性 1, 20-22. |
| [5] | 王成龙 (2018). 野生荞麦叶绿体基因组比较分析及荞麦属植物系统进化研究. 博士论文. 雅安: 四川农业大学. pp. 52-53. |
| [6] | 王继玥, 白禹, 石登红, 刘燕 (2021). 桑科植物叶绿体基因组研究进展. 北方园艺 (8), 124-130. |
| [7] | 徐惠梅, 杨惠芳, 靳春霞 (2008). 沙木蓼和杨柴治沙造林试验. 现代农业科技 (19), 39, 41. |
| [8] | 杨昌友 (1984). 新疆木蓼属新种. 植物研究 4(2), 150-151. |
| [9] | 杨梦婷, 黄洲, 干建平, 徐君驰, 庞基良 (2019). SSR分子标记的研究进展. 杭州师范大学学报(自然科学版) 18, 429-436. |
| [10] |
Amiryousefi A, Hyvönen J, Poczai P (2018). IRscope: an online program to visualize the junction sites of chloroplast genomes. Bioinformatics 34, 3030-3031.
DOI PMID |
| [11] | Bao BJ, Li AJ (1993). A study of the genus Atraphaxis in China and the system of Atraphaxideae (Polygonaceae). J Syst Evol 31, 127-139. |
| [12] | Behura SK, Severson DW (2013). Codon usage bias: causative factors, quantification methods and genome- wide patterns: with emphasis on insect genomes. Biol Rev 88, 49-61. |
| [13] |
Burke JM, Sanchez A, Kron K, Luckow M (2010). Placing the woody tropical genera of Polygonaceae: a hypothesis of character evolution and phylogeny. Am J Bot 97, 1377-1390.
DOI PMID |
| [14] |
Cosner ME, Raubeson LA, Jansen RK (2004). Chloroplast DNA rearrangements in Campanulaceae: phylogenetic utility of highly rearranged genomes. BMC Evolut Biol 4, 27.
DOI URL |
| [15] | Curci PL, De Paola D, Danzi D, Vendramin GG, Sonnante G (2015). Complete chloroplast genome of the multifunctional crop globe artichoke and comparison with other Asteraceae. PLoS One 10, e0120589. |
| [16] |
Daniell H, Lin CS, Yu M, Chang WJ (2016). Chloroplast genomes: diversity, evolution, and applications in genetic engineering. Genome Biol 17, 134.
DOI PMID |
| [17] | Dierckxsens N, Mardulyn P, Smits G (2017). NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res 45, e18. |
| [18] | Doyle JJ, Doyle JL (1987). A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19, 11-15. |
| [19] | Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I (2004). VISTA: computational tools for comparative genomics. Nucleic Acids Res 32, W273-W279. |
| [20] |
Gandhi SG, Awasthi P, Bedi YS (2010). Analysis of SSR dynamics in chloroplast genomes of Brassicaceae family. Bioinformation 5, 16-20.
DOI PMID |
| [21] |
Gao FL, Chen CJ, Arab DA, Du ZG, He YH, Ho SYW (2019). EasyCodeML: a visual tool for analysis of selection using CodeML. Ecol Evol 9, 3891-3898.
DOI |
| [22] |
Gao XY, Zhang X, Meng HH, Li J, Zhang D, Liu CN (2018). Comparative chloroplast genomes of Paris sect. Marmorata: insights into repeat regions and evolutionary implications. BMC Genomics 19, 878.
DOI |
| [23] | Greiner S, Lehwark P, Bock R (2019). OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res 47, W59-W64. |
| [24] |
Hershberg R, Petrov DA (2008). Selection on codon bias. Annu Rev Genet 42, 287-299.
DOI PMID |
| [25] |
Jansen RK, Wojciechowski MF, Sanniyasi E, Lee SB, Daniell S (2008). Complete plastid genome sequence of the chickpea (Cicer arietinum) and the phylogenetic distribution of rps12and clpP intron losses among legumes (Leguminosae). Mol Phylogenet Evol 48, 1204-1217.
DOI URL |
| [26] | Jia J, Xue QZ (2009). Codon usage biases of transposable elements and host nuclear genes in Arabidopsis thaliana and Oryza sativa. Genom Proteomics Bioinf 7, 175-184. |
| [27] |
Kalyaanamoorthy S, Minh BQ, Wong TKF, Von Haesele A, Jermiin LS (2017). ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods 14, 587-589.
DOI PMID |
| [28] |
Katoh K, Rozewicki J, Yamada KD (2019). MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform 20, 1160-1166.
DOI PMID |
| [29] |
Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A (2012). Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647-1649.
DOI PMID |
| [30] | Khan A, Asaf S, Khan AL, Al-Harrasi A, Al-Sudairy O, AbdulKareem NM, Khan A, Shehzad T, Alsaady N, Al-Lawati A, Al-Rawahi A, Shinwari ZK (2019). First complete chloroplast genomics and comparative phylogenetic analysis of Commiphora gileadensis and C. foliacea: myrrh producing trees. PLoS One 14, e0208511. |
| [31] |
Kurtz S, Choudhuri JV, Ohlebusch E, Schleiermacher C, Stoye J, Giegerich R (2001). REPuter: the manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res 29, 4633-4642.
DOI PMID |
| [32] |
Li L, Hu YF, He M, Zhang B, Wu W, Cai PM, Huo D, Hong YC (2021). Comparative chloroplast genomes: insights into the evolution of the chloroplast genome of Camellia sinensis and the phylogeny of Camellia. BMC Genomics 22, 138.
DOI PMID |
| [33] |
Li X, Li YF, Zang MY, Li MZ, Fang YM (2018). Complete chloroplast genome sequence and phylogenetic analysis of Quercus acutissima. Int J Mol Sci 19, 2443.
DOI URL |
| [34] |
Librado P, Rozas J (2009). DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451-1452.
DOI PMID |
| [35] | Lyu XL, Liu Y (2020). Nonoptimal codon usage is critical for protein structure and function of the master general amino acid control regulator CPC-1. mBio 11, e02605-20. |
| [36] |
Marais G, Mouchiroud D, Duret L (2001). Does recombination improve selection on codon usage? Lessons from nematode and fly complete genomes. Proc Natl Acad Sci USA 98, 5688-5692.
DOI PMID |
| [37] | Qian J, Song JY, Gao HH, Zhu YJ, Xu J, Pang XH, Yao H, Sun C, Li XE, Li CY, Liu JY, Xu HB, Chen SL (2013). The complete chloroplast genome sequence of the medicinal plant Salvia miltiorrhiza. PLoS One 8, e57607. |
| [38] |
Saski C, Lee SB, Daniell H, Wood TC, Tomkins J, Kim HG, Jansen RK (2005). Complete chloroplast genome sequence of Glycine max and comparative analyses with other legume genomes. Plant Mol Biol 59, 309-322.
DOI URL |
| [39] |
Sun YX, Zhang ML (2012). Molecular phylogeny of tribe Atraphaxideae (Polygonaceae) evidenced from five cpDNA genes. J Arid Land 4, 180-190.
DOI |
| [40] |
Tamura K, Stecher G, Kumar S (2021). MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol 38, 3022-3027.
DOI PMID |
| [41] |
Thiel T, Michalek W, Varshney R, Graner A (2003). Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley (Hordeum vulgare L.). Theor Appl Genet 106, 411-422.
DOI PMID |
| [42] | Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S (2017). GeSeq-versatile and accurate annotation of organelle genomes. Nucleic Acids Res 45, W6-W11. |
| [43] |
Wang QY, Yuan ZY, Zhang Y, Mamtimin S, Tian XM (2018a). The complete chloroplast genome of Atraphaxis jrtyschensis (Polygonaceae), an endemic and endangered desert shrub to Xinjiang, China. Mitochondrial DNA Part B 3, 1104-1105.
DOI URL |
| [44] |
Wang WC, Chen SY, Zhang XZ (2018b). Whole-genome comparison reveals divergent IR borders and mutation hotspots in chloroplast genomes of herbaceous bamboos (Bambusoideae: Olyreae). Molecules 23, 1537.
DOI URL |
| [45] | Wang XH (2018). Phytochemical Investigations of Atraphaxis spinosa and Camphorosma lessingii Litv. Master's thesis. Tianjin: Tianjin University. pp. 41-66. |
| [46] |
Wanga VO, Dong X, Oulo MA, Mkala EM, Yang JX, Onjalalaina GE, Gichua MK, Kirika PM, Gituru RW, Hu GW, Wang QF (2021). Complete chloroplast genomes of Acanthochlamys bracteata (China) and Xerophyta (Africa) (Velloziaceae): comparative genomics and phylogenomic placement. Front Plant Sci 12, 691833.
DOI URL |
| [47] |
Wicke S, Schneeweiss GM, dePamphilis CW, Müller KF, Quandt D (2011). The evolution of the plastid chromosome in land plants: gene content, gene order, gene function. Plant Mol Biol 76, 273-297.
DOI PMID |
| [48] |
Wu S, Chen JY, Li Y, Liu A, Li A, Yin M, Shrestha N, Liu JQ, Ren GP (2021). Extensive genomic rearrangements mediated by repetitive sequences in plastomes of Medicago and its relatives. BMC Plant Biol 21, 421.
DOI |
| [49] |
Xie DF, Yu Y, Deng YQ, Li J, Liu HY, Zhou SD, He XJ (2018). Comparative analysis of the chloroplast genomes of the Chinese endemic genus Urophysa and their contribution to chloroplast phylogeny and adaptive evolution. Int J Mol Sci 19, 1847.
DOI URL |
| [50] | Xue JH, Wang S, Zhou SL (2012). Polymorphic chloroplast microsatellite loci in Nelumbo (Nelumbonaceae). Am J Bot 99, e240-e244. |
| [51] |
Yang BB, Li LD, Liu JQ, Zhang LS (2021). Plastome and phylogenetic relationship of the woody buckwheat Fagopyrum tibeticum in the Qinghai-Tibet Plateau. Plant Diversity 43, 198-205.
DOI URL |
| [52] | Yang Y, Dang YY, Li Q, Lu JJ, Li XW, Wang YT (2014). Complete chloroplast genome sequence of poisonous and medicinal plant Datura stramonium: organizations and implications for genetic engineering. PLoS One 9, e110656. |
| [53] |
Yang ZH (2007). PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol 24, 1586-1591.
DOI PMID |
| [54] | Yurtseva OV, Kuznetsova OI, Mavrodieva ME, Mavrodiev EV (2016). What is Atraphaxis L. (Polygonaceae, Polygoneae): cryptic taxa and resolved taxonomic complexity instead of the formal lumping and the lack of morphological synapomorphies. Peer J 4, e1977. |
| [55] |
Zhang D, Gao FL, Jakovlić I, Zou H, Zhang J, Li WX, Wang GT (2020). PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol Ecol Resour 20, 348-355.
DOI PMID |
| [56] | Zhou JH, Ding YZ, He Y, Chu YF, Zhao P, Ma LY, Wang XJ, Li XR, Liu YS (2014). The effect of multiple evolutionary selections on synonymous codon usage of genes in the Mycoplasma bovis genome. PLoS One 9, e108949. |
| [57] |
Zhou T, Chen C, Wei Y, Chang YX, Bai GQ, Li ZH, Kanwal N, Zhao GF (2016). Comparative transcriptome and chloroplast genome analyses of two related Dipteronia species. Front Plant Sci 7, 1512.
PMID |
| [1] | 王传永, 庄典, 宋正达, 翟恒华, 李乃伟, 张凡. 黑果腺肋花楸叶绿体全基因组的结构和比较分析及系统进化推断[J]. 植物学报, 2025, 60(4): 1-0. |
| [2] | 夏琳凤, 李瑞, 王海政, 冯大领, 王春阳. 轮藻门植物基因组学研究进展[J]. 植物学报, 2025, 60(2): 271-282. |
| [3] | 孙亚君. 何谓高等或低等生物——澄清《物种起源》所蕴含的生物等级性的涵义及其成立性[J]. 生物多样性, 2025, 33(1): 24394-. |
| [4] | 艾妍雨, 胡海霞, 沈婷, 莫雨轩, 杞金华, 宋亮. 附生维管植物多样性及其与宿主特征的相关性: 以哀牢山中山湿性常绿阔叶林为例[J]. 生物多样性, 2024, 32(5): 24072-. |
| [5] | 吕燕文, 王子韵, 肖钰, 何梓晗, 吴超, 胡新生. 谱系分选理论与检测方法的研究进展[J]. 生物多样性, 2024, 32(4): 23400-. |
| [6] | 曹可欣, 王敬雯, 郑国, 武鹏峰, 李英滨, 崔淑艳. 降水格局改变及氮沉降对北方典型草原土壤线虫多样性的影响[J]. 生物多样性, 2024, 32(3): 23491-. |
| [7] | 王斌, 钟艺倩, 杨美雪, 吴淼锐, 王艳萍, 陆芳, 陶旺兰, 李健星, 赵弘明, 刘晟源, 向悟生, 李先琨. 喀斯特季节性雨林优势树种叶片非结构性碳水化合物空间变异及生态驱动因素[J]. 生物多样性, 2024, 32(12): 24325-. |
| [8] | 杨向林, 赵彩云, 李俊生, 种方方, 李文金. 植物入侵导致群落谱系结构更加聚集: 以广西国家级自然保护区草本植物为例[J]. 生物多样性, 2024, 32(11): 24175-. |
| [9] | 何林君, 杨文静, 石宇豪, 阿说克者莫, 范钰, 王国严, 李景吉, 石松林, 易桂花, 彭培好. 火烧干扰下植物群落系统发育和功能多样性对紫茎泽兰入侵的影响[J]. 生物多样性, 2024, 32(11): 24269-. |
| [10] | 王振宇, 黄志群. 亚热带27种木本植物叶片性状对植食作用的影响: 验证生长-防御权衡假说[J]. 植物生态学报, 2024, 48(11): 1501-1509. |
| [11] | 李庆多, 栗冬梅. 全球蝙蝠巴尔通体流行状况分析[J]. 生物多样性, 2023, 31(9): 23166-. |
| [12] | 包金波, 丁志杰, 苗浩宇, 李雪丽, 任书贤, 焦若岩, 李浩, 邓茜茜, 李英姿, 田新民. 石栗叶绿体基因组研究[J]. 植物学报, 2023, 58(2): 248-260. |
| [13] | 宋会银, 胡征宇, 刘国祥. 绿藻门小球藻科的分类学研究进展[J]. 生物多样性, 2023, 31(2): 22083-. |
| [14] | 李治中, 彭帅, 王青锋, 李伟, 梁士楚, 陈进明. 中国海菜花属植物隐种多样性[J]. 生物多样性, 2023, 31(2): 22394-. |
| [15] | 朱瑞良, 马晓英, 曹畅, 曹子寅. 中国苔藓植物多样性研究进展[J]. 生物多样性, 2022, 30(7): 22378-. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||