

植物学报 ›› 2025, Vol. 60 ›› Issue (4): 573-585.DOI: 10.11983/CBB24146 cstr: 32102.14.CBB24146
王传永1, 庄典2, 宋正达1, 翟恒华1, 李乃伟1,*( ), 张凡1,*(
), 张凡1,*( )
)
收稿日期:2024-09-24
接受日期:2025-02-09
出版日期:2025-07-10
发布日期:2025-02-08
通讯作者:
*E-mail: linaiwei@jib.ac.cn;mumizhongfeng@126.com
基金资助:
Chuanyong Wang1, Dian Zhuang2, Zhengda Song1, Henghua Zhai1, Naiwei Li1,*( ), Fan Zhang1,*(
), Fan Zhang1,*( )
)
Received:2024-09-24
Accepted:2025-02-09
Online:2025-07-10
Published:2025-02-08
Contact:
*E-mail: linaiwei@jib.ac.cn;mumizhongfeng@126.com
摘要: 黑涩楠(Aronia melanocarpa)因其观赏价值和经济价值而闻名, 但其与其它蔷薇科植物的系统进化关系仍不明确。该研究对黑涩楠叶绿体(cp)基因组进行测序, 并与13个蔷薇科物种的叶绿体基因组进行比较分析。结果表明, 黑涩楠的cp基因组大小为159 772 bp, 呈典型的四分结构; 其中大单拷贝区(LSC)长度为87 810 bp, 小单拷贝区(SSC)长度为19 200 bp, 中间含有2个26 381 bp的反向重复区(IRa和IRb)。共注释到132个基因, 包括87个蛋白质编码基因、37个tRNA和8个rRNA。还检测到76个简单重复序列(SSR)和50个长重复序列。系统进化分析表明, 黑涩楠与红涩楠(A. arbutifolia)的亲缘关系最近, 与榅桲(Cydonia oblonga)是姊妹支系。该研究提供的基因组信息将为后续的系统进化和种群遗传分析以及分子育种提供理论支持。
王传永, 庄典, 宋正达, 翟恒华, 李乃伟, 张凡. 黑涩楠叶绿体全基因组的结构和比较分析及系统进化推断. 植物学报, 2025, 60(4): 573-585.
Chuanyong Wang, Dian Zhuang, Zhengda Song, Henghua Zhai, Naiwei Li, Fan Zhang. Structural and Comparative Analysis of the Complete Chloroplast Genome of the Aronia melanocarpa and Its Phylogenetic Inference. Chinese Bulletin of Botany, 2025, 60(4): 573-585.
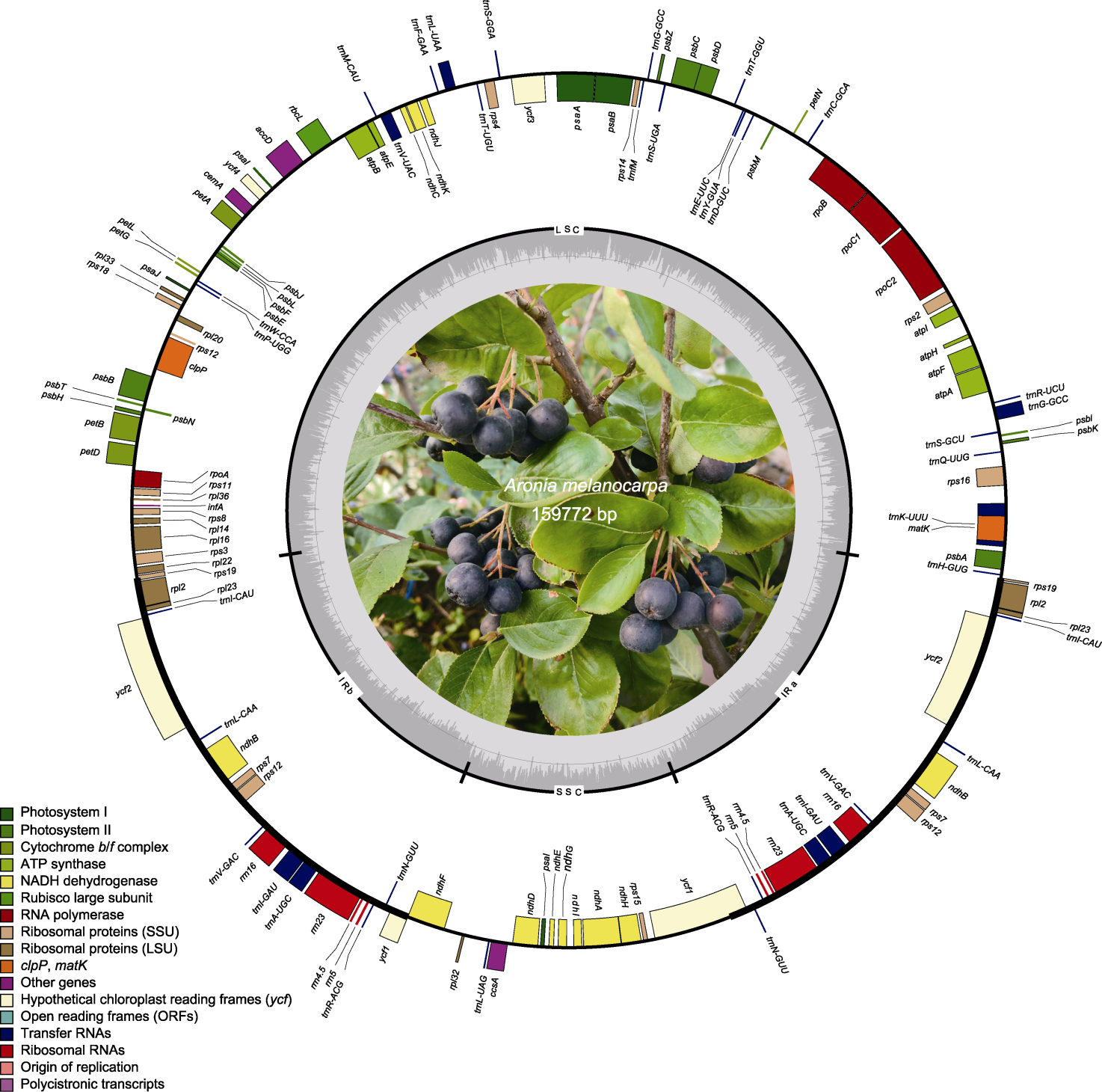
图1 黑涩楠叶绿体基因组图谱 圆圈内、外的基因分别按顺时针和逆时针方向转录。不同功能基因组用不同的颜色标记。内圈的GC和AT含量分别用深灰和浅灰色表示。
Figure 1 Map of the chloroplast genome of Aronia melanocarpa Genes inside and outside of the circle are transcribed in the clockwise and counterclockwise directions respectively. Different functional gene groups are color-coded accordingly. GC and AT content are represented on the inner circle by darker and lighter gray, respectively.
| Genome features | A. melanocarpa | Genome features | A. melanocarpa |
|---|---|---|---|
| Genome size (bp)/GC content (%) | 159772/36.6 | Number of unique genes | 110 |
| LSC size (bp)/GC content (%) | 87810/34.3 | Protein-coding genes | 87 |
| SSC size (bp)/GC content (%) | 19200/30.4 | tRNAs | 37 |
| IR size (bp)/GC content (%) | 52762/42.7 | rRNAs | 8 |
| Total gene number | 132 | Genes duplicated in the IRs | 22 |
表1 黑涩楠叶绿体基因组特征
Table 1 The Aronia melanocarpa chloroplast genome features
| Genome features | A. melanocarpa | Genome features | A. melanocarpa |
|---|---|---|---|
| Genome size (bp)/GC content (%) | 159772/36.6 | Number of unique genes | 110 |
| LSC size (bp)/GC content (%) | 87810/34.3 | Protein-coding genes | 87 |
| SSC size (bp)/GC content (%) | 19200/30.4 | tRNAs | 37 |
| IR size (bp)/GC content (%) | 52762/42.7 | rRNAs | 8 |
| Total gene number | 132 | Genes duplicated in the IRs | 22 |
| Category | Gene group | Name of gene | Number |
|---|---|---|---|
| Self-replication | Proteins of the large ribosomal subunit | rpl2ab, rpl14, rpl16b, rpl20, rpl22, rpl23a, rpl32, rpl33, rpl36 | 11 |
| Proteins of the small ribosomal subunit | rps2, rps3, rps4, rps7a, rps8, rps11, rps12ac, rps14, rps15, rps16b, rps18, rps19ab | 15 | |
| Subunits of RNA polymerase | rpoA, rpoB, rpoC1b, rpoC2 | 4 | |
| rRNAs | rrn23Sa, rrn16Sa, rrn5Sa, rrn4.5Sa | 8 | |
| tRNAs | trnH-GUG, trnK-UUUb, trnQ-UUG, trnS-GCU, trnG- GCCab, trnR-UCU, trnC-GCA, trnD-GUC, trnY-GUA, trnE- UUC, trnT-GGU, trnS-UGA, trnfM, trnS-GGA, trnS-GGA, trnT-UGU, trnL-UAAb, trnF-GAA, trnV-UACb, trnM-CAU, trnW-CCA, trnP-UGG, trnI-CAUa, trnL-CAAa, trnV-GACa, trnI-GAUab, trnA-UGCab, trnR-ACGa, trnN-GUUa, trnL-UAG | 37 | |
| Photosynthesis | Subunits of photosystem I | psaA, psaB, psaIa, psaJ | 5 |
| Subunits of photosystem II | psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbIa, psbJ, psbK, psbL, psbM, psbN, psbT, psbZ | 15 | |
| Subunits of NADH dehydrogenase | ndhAb, ndhBab, ndhC, ndhD, ndhE, ndhF, ndhG, ndhH, ndhI, ndhJ, ndhK | 12 | |
| Subunits of cytochrome b/f complex | petA, petBb, petDb, petG, petL, petN | 6 | |
| Subunits of ATP synthase | atpA, atpB, atpE, atpFb, atpH, atpI | 6 | |
| Large subunit of Rubisco | rbcL | 1 | |
| Biosynthesis | Maturase | matK | 1 |
| Protease | clpPc | 1 | |
| Envelope membrane protein | cemA | 1 | |
| Acetyl-CoA carboxylase | accD | 1 | |
| C-type cytochrome synthesis gene | ccsA | 1 | |
| Translation initiation factor | infA | 1 | |
| Unknown function | Conserved hypothetical chloroplast reading frames | ycf1a, ycf2a, ycf3c, ycf4 | 6 |
表2 本研究测序的黑涩楠叶绿体基因组中的基因
Table 2 Genes in the chloroplast genome of Aronia melanocarpa sequenced in this study
| Category | Gene group | Name of gene | Number |
|---|---|---|---|
| Self-replication | Proteins of the large ribosomal subunit | rpl2ab, rpl14, rpl16b, rpl20, rpl22, rpl23a, rpl32, rpl33, rpl36 | 11 |
| Proteins of the small ribosomal subunit | rps2, rps3, rps4, rps7a, rps8, rps11, rps12ac, rps14, rps15, rps16b, rps18, rps19ab | 15 | |
| Subunits of RNA polymerase | rpoA, rpoB, rpoC1b, rpoC2 | 4 | |
| rRNAs | rrn23Sa, rrn16Sa, rrn5Sa, rrn4.5Sa | 8 | |
| tRNAs | trnH-GUG, trnK-UUUb, trnQ-UUG, trnS-GCU, trnG- GCCab, trnR-UCU, trnC-GCA, trnD-GUC, trnY-GUA, trnE- UUC, trnT-GGU, trnS-UGA, trnfM, trnS-GGA, trnS-GGA, trnT-UGU, trnL-UAAb, trnF-GAA, trnV-UACb, trnM-CAU, trnW-CCA, trnP-UGG, trnI-CAUa, trnL-CAAa, trnV-GACa, trnI-GAUab, trnA-UGCab, trnR-ACGa, trnN-GUUa, trnL-UAG | 37 | |
| Photosynthesis | Subunits of photosystem I | psaA, psaB, psaIa, psaJ | 5 |
| Subunits of photosystem II | psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbIa, psbJ, psbK, psbL, psbM, psbN, psbT, psbZ | 15 | |
| Subunits of NADH dehydrogenase | ndhAb, ndhBab, ndhC, ndhD, ndhE, ndhF, ndhG, ndhH, ndhI, ndhJ, ndhK | 12 | |
| Subunits of cytochrome b/f complex | petA, petBb, petDb, petG, petL, petN | 6 | |
| Subunits of ATP synthase | atpA, atpB, atpE, atpFb, atpH, atpI | 6 | |
| Large subunit of Rubisco | rbcL | 1 | |
| Biosynthesis | Maturase | matK | 1 |
| Protease | clpPc | 1 | |
| Envelope membrane protein | cemA | 1 | |
| Acetyl-CoA carboxylase | accD | 1 | |
| C-type cytochrome synthesis gene | ccsA | 1 | |
| Translation initiation factor | infA | 1 | |
| Unknown function | Conserved hypothetical chloroplast reading frames | ycf1a, ycf2a, ycf3c, ycf4 | 6 |
| Nucleotide(s) | Number of repeats | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 6 | 7 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | Total | |
| A | - | - | 5 | 5 | 4 | 2 | 2 | 4 | 2 | - | - | - | 1 | 25 |
| C | - | - | 2 | 1 | - | 1 | - | - | - | - | - | - | - | 4 |
| G | - | - | 1 | 1 | - | - | - | - | - | - | - | - | - | 2 |
| T | - | - | 13 | 9 | 6 | 2 | 1 | 4 | 1 | 2 | 2 | 1 | - | 41 |
| AT | 2 | - | - | - | - | - | - | - | - | - | - | - | 2 | |
| TA | 1 | 1 | - | - | - | - | - | - | - | - | - | - | - | 2 |
表3 黑涩楠叶绿体基因组中简单重复序列(SSR)的鉴定频率
Table 3 The frequency of identified of simple sequence repeats (SSRs) in the Aronia melanocarpa chloroplast genome
| Nucleotide(s) | Number of repeats | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 6 | 7 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | Total | |
| A | - | - | 5 | 5 | 4 | 2 | 2 | 4 | 2 | - | - | - | 1 | 25 |
| C | - | - | 2 | 1 | - | 1 | - | - | - | - | - | - | - | 4 |
| G | - | - | 1 | 1 | - | - | - | - | - | - | - | - | - | 2 |
| T | - | - | 13 | 9 | 6 | 2 | 1 | 4 | 1 | 2 | 2 | 1 | - | 41 |
| AT | 2 | - | - | - | - | - | - | - | - | - | - | - | 2 | |
| TA | 1 | 1 | - | - | - | - | - | - | - | - | - | - | - | 2 |
| Amino acids | Codon | No. | RSCU | Amino acids | Codon | No. | RSCU |
|---|---|---|---|---|---|---|---|
| Phe | UUU | 975 | 1.3 | Ala | GCU | 645 | 1.84 |
| Phe | UUC | 526 | 0.7 | Ala | GCC | 217 | 0.62 |
| Leu | UUA | 912 | 1.95 | Ala | GCA | 390 | 1.11 |
| Leu | UUG | 565 | 1.21 | Ala | GCG | 148 | 0.42 |
| Leu | CUU | 593 | 1.27 | TER | UAA | 51 | 1.76 |
| Leu | CUC | 186 | 0.4 | TER | UAG | 21 | 0.72 |
| Leu | CUA | 363 | 0.78 | TER | UGA | 15 | 0.52 |
| Leu | CUG | 181 | 0.39 | His | CAU | 493 | 1.55 |
| Ile | AUU | 1123 | 1.47 | His | CAC | 145 | 0.45 |
| Ile | AUC | 440 | 0.58 | Gln | CAA | 727 | 1.54 |
| Ile | AUA | 730 | 0.96 | Gln | CAG | 217 | 0.46 |
| Met | AUG | 627 | 1 | Asn | AAU | 987 | 1.53 |
| Val | GUU | 524 | 1.44 | Asn | AAC | 305 | 0.47 |
| Val | GUC | 167 | 0.46 | Lys | AAA | 1068 | 1.49 |
| Val | GUA | 552 | 1.52 | Lys | AAG | 364 | 0.51 |
| Val | GUG | 208 | 0.57 | Asp | GAU | 889 | 1.62 |
| Ser | UCU | 573 | 1.69 | Asp | GAC | 208 | 0.38 |
| Ser | UCC | 330 | 0.97 | Glu | GAA | 1035 | 1.48 |
| Ser | UCA | 408 | 1.2 | Glu | GAG | 363 | 0.52 |
| Ser | UCG | 190 | 0.56 | Cys | UGU | 226 | 1.49 |
| Ser | AGU | 411 | 1.21 | Cys | UGC | 77 | 0.51 |
| Ser | AGC | 128 | 0.38 | Trp | UGG | 458 | 1 |
| Pro | CCU | 421 | 1.56 | Arg | CGU | 340 | 1.27 |
| Pro | CCC | 201 | 0.74 | Arg | CGC | 112 | 0.42 |
| Pro | CCA | 310 | 1.15 | Arg | CGA | 370 | 1.38 |
| Pro | CCG | 149 | 0.55 | Arg | CGG | 121 | 0.45 |
| Thr | ACU | 551 | 1.6 | Arg | AGA | 493 | 1.84 |
| Thr | ACC | 251 | 0.73 | Arg | AGG | 173 | 0.65 |
| Thr | ACA | 423 | 1.23 | Gly | GGU | 589 | 1.31 |
| Thr | ACG | 153 | 0.44 | Gly | GGC | 183 | 0.41 |
| Tyr | UAU | 801 | 1.61 | Gly | GGA | 725 | 1.62 |
| Tyr | UAC | 194 | 0.39 | Gly | GGG | 295 | 0.66 |
表4 黑涩楠叶绿体基因组中的密码子用法
Table 4 Codon usage in the Aronia melanocarpa chloroplast genome
| Amino acids | Codon | No. | RSCU | Amino acids | Codon | No. | RSCU |
|---|---|---|---|---|---|---|---|
| Phe | UUU | 975 | 1.3 | Ala | GCU | 645 | 1.84 |
| Phe | UUC | 526 | 0.7 | Ala | GCC | 217 | 0.62 |
| Leu | UUA | 912 | 1.95 | Ala | GCA | 390 | 1.11 |
| Leu | UUG | 565 | 1.21 | Ala | GCG | 148 | 0.42 |
| Leu | CUU | 593 | 1.27 | TER | UAA | 51 | 1.76 |
| Leu | CUC | 186 | 0.4 | TER | UAG | 21 | 0.72 |
| Leu | CUA | 363 | 0.78 | TER | UGA | 15 | 0.52 |
| Leu | CUG | 181 | 0.39 | His | CAU | 493 | 1.55 |
| Ile | AUU | 1123 | 1.47 | His | CAC | 145 | 0.45 |
| Ile | AUC | 440 | 0.58 | Gln | CAA | 727 | 1.54 |
| Ile | AUA | 730 | 0.96 | Gln | CAG | 217 | 0.46 |
| Met | AUG | 627 | 1 | Asn | AAU | 987 | 1.53 |
| Val | GUU | 524 | 1.44 | Asn | AAC | 305 | 0.47 |
| Val | GUC | 167 | 0.46 | Lys | AAA | 1068 | 1.49 |
| Val | GUA | 552 | 1.52 | Lys | AAG | 364 | 0.51 |
| Val | GUG | 208 | 0.57 | Asp | GAU | 889 | 1.62 |
| Ser | UCU | 573 | 1.69 | Asp | GAC | 208 | 0.38 |
| Ser | UCC | 330 | 0.97 | Glu | GAA | 1035 | 1.48 |
| Ser | UCA | 408 | 1.2 | Glu | GAG | 363 | 0.52 |
| Ser | UCG | 190 | 0.56 | Cys | UGU | 226 | 1.49 |
| Ser | AGU | 411 | 1.21 | Cys | UGC | 77 | 0.51 |
| Ser | AGC | 128 | 0.38 | Trp | UGG | 458 | 1 |
| Pro | CCU | 421 | 1.56 | Arg | CGU | 340 | 1.27 |
| Pro | CCC | 201 | 0.74 | Arg | CGC | 112 | 0.42 |
| Pro | CCA | 310 | 1.15 | Arg | CGA | 370 | 1.38 |
| Pro | CCG | 149 | 0.55 | Arg | CGG | 121 | 0.45 |
| Thr | ACU | 551 | 1.6 | Arg | AGA | 493 | 1.84 |
| Thr | ACC | 251 | 0.73 | Arg | AGG | 173 | 0.65 |
| Thr | ACA | 423 | 1.23 | Gly | GGU | 589 | 1.31 |
| Thr | ACG | 153 | 0.44 | Gly | GGC | 183 | 0.41 |
| Tyr | UAU | 801 | 1.61 | Gly | GGA | 725 | 1.62 |
| Tyr | UAC | 194 | 0.39 | Gly | GGG | 295 | 0.66 |

图2 14种蔷薇科植物叶绿体基因组比较 图中纵坐标和横坐标分别代表50%-100%的相同率和序列长度。注释基因显示在顶部。
Figure 2 A comparison of the chloroplast genomes among 14 Rosaceae species The vertical scale and horizontal axes in the figure represent the percentage of identity ranging from 50% to 100% and the sequence length, respectively. Annotated genes are displayed on the top.

图3 蔷薇科14个物种的LSC、SSC和IR区域的边界距离比较 LSC、SSC和IR同表1。
Figure 3 Comparisons of the boundary distances for LSC, SSC, and IR regions among 14 species from the Rosaceae LSC, SSC, and IR are the same as shown in Table 1.
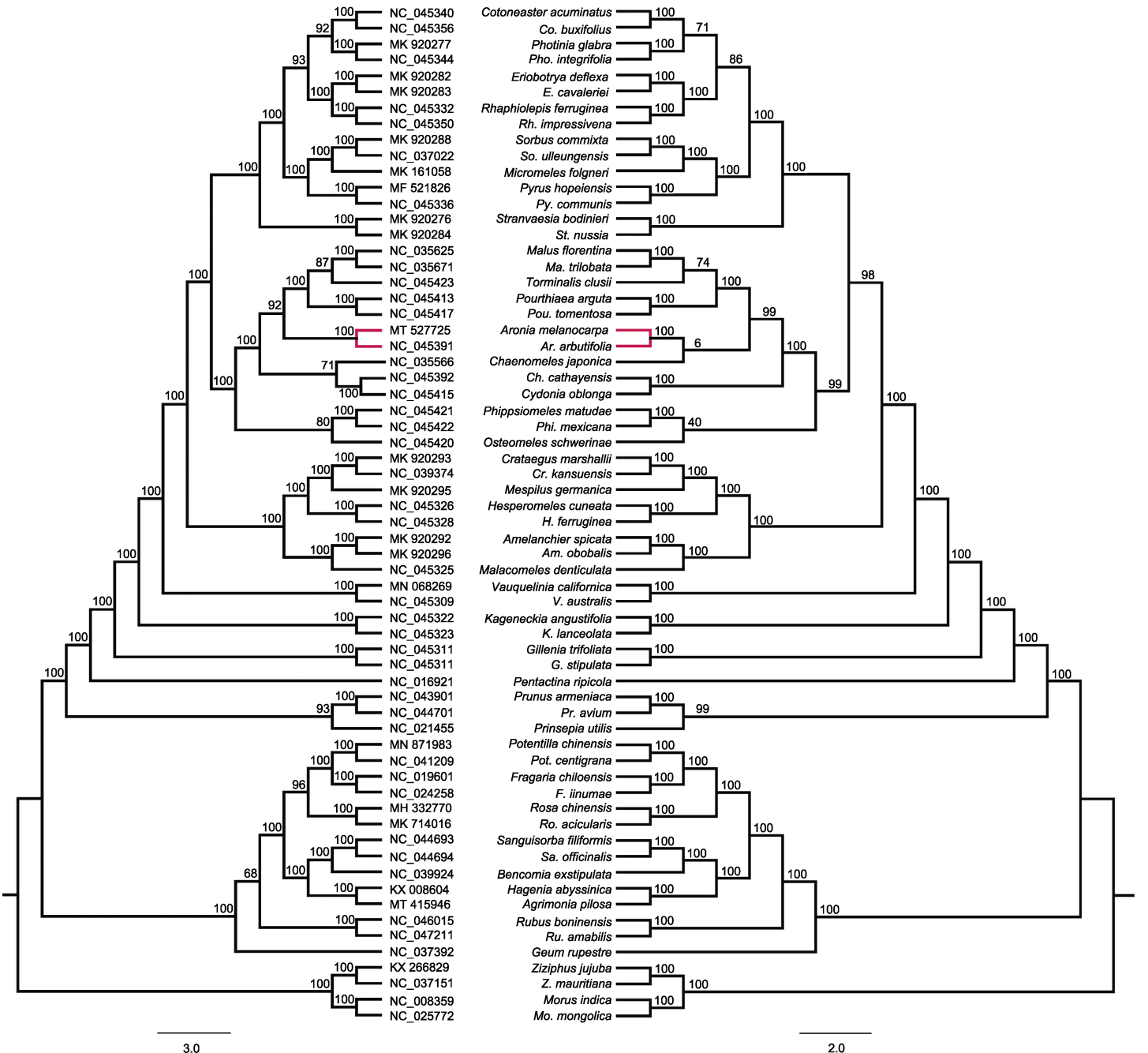
图4 利用完整的叶绿体基因组对60个蔷薇科物种进行系统进化分析 左侧为利用iQTree构建的系统发育树, 节点上方的数字为自展支持率; 右侧为利用FastTree构建的系统发育树, 节点上方的数字为SH-like local支持率。
Figure 4 Phylogenetic analyses on 60 Rosaceae species using their complete chloroplast genomes On the left, phylogenetic tree was constructed by iQTree, numbers above the nodes are bootstrap values; on the right, phylogenetic tree was constructed by FastTree, numbers above the nodes are SH-like local values.
| [1] |
Amiryousefi A, Hyvönen J, Poczai P (2018). IRscope: an online program to visualize the junction sites of chloroplast genomes. Bioinformatics 34, 3030-3031.
DOI PMID |
| [2] |
Benson G (1999). Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res 27, 573-580.
DOI PMID |
| [3] | Brand M (2010). Aronia: native shrubs with untapped potential. Arnoldia 67, 14-25. |
| [4] | Campbell CS, Evans RC, Morgan DR, Dickinson TA, Arsenault MP (2007). Phylogeny of subtribe Pyrinae (formerly the Maloideae, Rosaceae): limited resolution of a complex evolutionary history. Plant Syst Evol 266, 119-145. |
| [5] | Chiapella JO, Barfuss MHJ, Xue ZQ, Greimler J (2019). The plastid genome of Deschampsia cespitosa (Poaceae). Molecules 24, 216. |
| [6] | Chu ZZ, Gulbar Yisilam, Qu ZZ, Tian XM (2023). Comparative analyses on the chloroplast genome of three sympatric Atraphaxis species. Chin Bull Bot 58, 417-432. (in Chinese) |
|
褚振州, 古丽巴哈尔·依斯拉木, 屈泽众, 田新民 (2023). 同域分布的3种木蓼属植物叶绿体基因组比较. 植物学报 58, 417-432.
DOI |
|
| [7] | Connolly BA (2014). Collection, Description, Taxonomic Relationships, Fruit Biochemistry, and Utilization of Aronia melanocarpa, A. arbutifolia, A. prunifolia, and A. mitschurinii. PhD dissertation. Connecticut: University of Connecticut. pp. 1-1. |
| [8] |
Daniell H, Lin CS, Yu M, Chang WJ (2016). Chloroplast genomes: diversity, evolution, and applications in genetic engineering. Genome Biol 17, 134.
DOI PMID |
| [9] | Dierckxsens N, Mardulyn P, Smits G (2017). NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res 45, e18. |
| [10] |
Domarew CA, Holt RR, Goodman-Snitkoff G (2002). A study of Russian phytomedicine and commonly used herbal remedies. J Herb Pharmacother 2, 31-48.
PMID |
| [11] | Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I (2004). VISTA: computational tools for comparative genomics. Nucleic Acids Res 32, W273-W279. |
| [12] | Fu PC, Zhang YZ, Geng HM, Chen SL (2016). The complete chloroplast genome sequence of Gentiana lawrencei var. farreri (Gentianaceae) and comparative analysis with its congeneric species. PeerJ 4, e2540. |
| [13] | Guo HJ, Liu JS, Luo L, Wei XP, Zhang J, Qi YD, Zhang BG, Liu HT, Xiao PG (2017). Complete chloroplast genome sequences of Schisandra chinensis: genome structure, comparative analysis, and phylogenetic relationship of basal angiosperms. Sci China Life Sci 60, 1286-1290. |
| [14] | Guo W, Yu YJ, Shen RJ, Liao WB, Chin SW, Potter D (2011). A phylogeny of Photinia sensu lato (Rosaceae) and related genera based on nrITS and cpDNA analysis. Plant Syst Evol 291, 91-102. |
| [15] | Hardin JW (1973). The enigmatic chokeberries (Aronia, Rosaceae). Bull Torrey Bot Club 100, 178-184. |
| [16] | He Y, Xiao HT, Deng C, Xiong L, Yang J, Peng C (2016). The complete chloroplast genome sequences of the medicinal plant Pogostemon cablin. Int J Mol Sci 17, 820. |
| [17] |
Hildebrand M, Hallick RB, Passavant CW, Bourque DP (1988). Trans-splicing in chloroplasts: the rps 12 loci of Nicotiana tabacum. Proc Natl Acad Sci USA 85, 372-376.
DOI PMID |
| [18] |
Katoh K, Rozewicki J, Yamada KD (2019). MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform 20, 1160-1166.
DOI PMID |
| [19] |
Kulling SE, Rawel HM (2008). Chokeberry (Aronia melanocarpa)—a review on the characteristic components and potential health effects. Planta Med 74, 1625-1634.
DOI PMID |
| [20] |
Kurtz S, Choudhuri JV, Ohlebusch E, Schleiermacher C, Stoye J, Giegerich R (2001). REPuter: the manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res 29, 4633-4642.
DOI PMID |
| [21] | Li QY, Guo W, Liao WB, Macklin JA, Li JH (2012). Generic limits of Pyrinae: insights from nuclear ribosomal DNA sequences. Bot Stud 53, 151-164. |
| [22] | Li YF, Sylvester SP, Li M, Zhang C, Li X, Duan YF, Wang XR (2019). The complete plastid genome of Magnolia zenii and genetic comparison to Magnoliaceae species. Molecules 24, 261. |
| [23] | Lim JD, Cha HS, Choung MG, Choi RN, Choi DJ, Youn AR (2014). Antioxidant activities of acidic ethanol extract and the anthocyanin rich fraction from Aronia melanocarpa. Korean J Food Cookery Sci 30, 573-578. |
| [24] | Liu BB, Liu GN, Hong DY, Wen J (2020). Eriobotrya belongs to Rhaphiolepis (Maleae, Rosaceae): evidence from chloroplast genome and nuclear ribosomal DNA data. Front Plant Sci 10, 1731. |
| [25] | Liu HY, Yu Y, Deng YQ, Li J, Huang ZX, Zhou SD (2018). The chloroplast genome of Lilium henrici: genome structure and comparative analysis. Molecules 23, 1276. |
| [26] |
Liu QP, Xue QZ (2005). Comparative studies on codon usage pattern of chloroplasts and their host nuclear genes in four plant species. J Genet 84, 55-62.
DOI PMID |
| [27] | Liu YC, Lin BY, Lin JY, Wu WL, Chang CC (2016). Evaluation of chloroplast DNA markers for intraspecific identification of Phalaenopsis equestris cultivars. Sci Hortic 203, 86-94. |
| [28] |
Lohse M, Drechsel O, Bock R (2007). OrganellarGenomeDRAW (OGDRAW): a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr Genet 52, 267-274.
DOI PMID |
| [29] |
Mayor C, Brudno M, Schwartz JR, Poliakov A, Rubin EM, Frazer KA, Pachter LS, Dubchak I (2000). VISTA: visualizing global DNA sequence alignments of arbitrary length. Bioinformatics 16, 1046-1047.
DOI PMID |
| [30] |
Morton BR (2003). The role of context-dependent mutations in generating compositional and codon usage bias in grass chloroplast DNA. J Mol Evol 56, 616-629.
DOI PMID |
| [31] |
Mudunuri SB, Nagarajaram HA (2007). IMEx: imperfect microsatellite extractor. Bioinformatics 23, 1181-1187.
DOI PMID |
| [32] | Palmer JD (1987). Chloroplast DNA evolution and biosystematic uses of chloroplast DNA variation. Am Nat 130, S6-S29. |
| [33] | Price MN, Dehal PS, Arkin AP (2010). FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One 5, e9490. |
| [34] | Ravi V, Khurana JP, Tyagi AK, Khurana P (2008). An update on chloroplast genomes. Plant Syst Evol 271, 101-122. |
| [35] | Robertson KR, Phipps JB, Rohrer JR, Smith PG (1991). A synopsis of genera in Maloideae (Rosaceae). Syst Bot 16, 376-394. |
| [36] | Seidemann J (1993). Chockberries a fruit little known till now. Deutsch Lebensm Rundsch (Ger) 89, 149-151. |
| [37] |
Sharp PM, Li WH (1987). The codon adaptation index—a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res 15, 1281-1295.
DOI PMID |
| [38] | Shi LC, Chen HM, Jiang M, Wang LQ, Wu X, Huang LF, Liu C (2019). CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res 47, W65-W73. |
| [39] | Shipunov A, Gladkova S, Timoshina P, Lee HJ, Choi J, Despiegelaere S, Connolly B (2019). Mysterious chokeberries: new data on the diversity and phylogeny of Aronia Medik. (Rosaceae). Eur J Taxon 570, 1-14. |
| [40] | Shukla S, Mehta A (2015). Anticancer potential of medicinal plants and their phytochemicals: a review. Braz J Bot 38, 199-210. |
| [41] | Szopa A, Kokotkiewicz A, Kubica P, Banaszczak P, Wojtanowska-Krośniak A, Krośniak M, Marzec-Wróblewska U, Badura A, Zagrodzki P, Bucinski A, Luczkiewicz M, Ekiert H (2017). Comparative analysis of different groups of phenolic compounds in fruit and leaf extracts of Aronia sp.: A. melanocarpa, A. arbutifolia, and A. × prunifolia and their antioxidant activities. Eur Food Res Technol 243, 1645-1657. |
| [42] | Wang T, Zeng XY, Hu YQ, Li N (2023). Optimization of brewing technology and antioxidant activity of wild cherry berry wine. Food Ind 44(6), 26-30. (in Chinese) |
| 汪娣, 曾雪莹, 胡玉清, 李楠 (2023). 野樱莓果酒酿造工艺优化及抗氧化活性分析. 食品工业 44(6), 26-30. | |
| [43] |
Wyman SK, Jansen RK, Boore JL (2004). Automatic annotation of organellar genomes with DOGMA. Bioinformatics 20, 3252-3255.
DOI PMID |
| [44] | Xu C, Dong WP, Li WQ, Lu YZ, Xie XM, Jin XB, Shi JP, He KH, Suo ZL (2017). Comparative analysis of six Lagerstroemia complete chloroplast genomes. Front Plant Sci 8, 15. |
| [45] | Yang L, Ding X, Jiang ZP, Chen WW, Chen S, Ma N, Xu SY, Wang JY (2024). Optimization of fermentation technology of Aronia melanocarpa pomace enzyme and study on its antioxidant activity. Chin Cond 49(9), 29-37. (in Chinese) |
| 杨柳, 丁雪, 姜志鹏, 陈文文, 陈生, 马宁, 徐圣喻, 王静怡 (2024). 野樱莓果渣酵素发酵工艺优化及抗氧化活性研究. 中国调味品 49(9), 29-37. | |
| [46] | Zhao F, Drew BT, Chen YP, Hu GX, Li B, Xiang CL (2020). The chloroplast genome of Salvia: genomic characterization and phylogenetic analysis. Int J Mol Sci 181, 812-830. |
| [47] | Zhao JL, Qi B, Ding LJ, Tang XQ (2010). Based on RSCU and QRSCU research codon bias of F/10 and G/11 Xylanase. J Food Sci Biotechnol 29, 755-764. |
| [48] | Zhao YM, Yang ZY, Zhao YP, Li XL, Zhao ZX, Zhao GF (2019). Chloroplast genome structural characteristics and phylogenetic relationships of Oleaceae. Chin Bull Bot 54, 441-454. (in Chinese) |
|
赵月梅, 杨振艳, 赵永平, 李筱玲, 赵志新, 赵桂仿 (2019). 木犀科植物叶绿体基因组结构特征和系统发育关系. 植物学报 54, 441-454.
DOI |
| [1] | 褚振州, 古丽巴哈尔·依斯拉木, 屈泽众, 田新民. 同域分布的3种木蓼属植物叶绿体基因组比较[J]. 植物学报, 2023, 58(3): 417-432. |
| [2] | 包金波, 丁志杰, 苗浩宇, 李雪丽, 任书贤, 焦若岩, 李浩, 邓茜茜, 李英姿, 田新民. 石栗叶绿体基因组研究[J]. 植物学报, 2023, 58(2): 248-260. |
| [3] | 赵月梅,杨振艳,赵永平,李筱玲,赵志新,赵桂仿. 木犀科植物叶绿体基因组结构特征和系统发育关系[J]. 植物学报, 2019, 54(4): 441-454. |
| [4] | 李巧丽, 延娜, 宋琼, 郭军战. 鲁桑叶绿体基因组序列及特征分析[J]. 植物学报, 2018, 53(1): 94-103. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||