

植物学报 ›› 2019, Vol. 54 ›› Issue (4): 441-454.DOI: 10.11983/CBB18191 cstr: 32102.14.CBB18191
赵月梅1,杨振艳2,赵永平1,李筱玲1,赵志新1,赵桂仿3,*( )
)
收稿日期:2018-09-06
接受日期:2019-04-23
出版日期:2019-07-01
发布日期:2020-01-08
通讯作者:
赵桂仿
基金资助:
Yuemei Zhao1,Zhenyan Yang2,Yongping Zhao1,Xiaoling Li1,Zhixin Zhao1,Guifang Zhao3,*( )
)
Received:2018-09-06
Accepted:2019-04-23
Online:2019-07-01
Published:2020-01-08
Contact:
Guifang Zhao
摘要: 木犀科11属19个种叶绿体基因组的一般特征和变异特征的比较分析显示, 结果表明, 该科叶绿体基因组大小为154-165 kb, 其差异主要是大单拷贝(LSC)长度的差异所致。Jasminum属3个物种的叶绿体基因组长度与其余物种有较大差异, 该属clpP基因内含子和accD基因丢失。共线性分析表明, Jasminum属3个物种多个基因出现基因重排现象, 倒位可能是重排的主要原因。Jasminum属在IRb/SSC和SSC/IRa边界的基因均与其它物种不同; 重复序列与SSR数量检测结果表明, Jasminum属与其余物种在数量及重复长度上差异较大。基于CDS数据构建的系统发育树表明, Abeliophyllum distichum和Forsythia suspensa为木犀科中较早分化的类群。
赵月梅,杨振艳,赵永平,李筱玲,赵志新,赵桂仿. 木犀科植物叶绿体基因组结构特征和系统发育关系. 植物学报, 2019, 54(4): 441-454.
Yuemei Zhao,Zhenyan Yang,Yongping Zhao,Xiaoling Li,Zhixin Zhao,Guifang Zhao. Chloroplast Genome Structural Characteristics and Phylogenetic Relationships of Oleaceae. Chinese Bulletin of Botany, 2019, 54(4): 441-454.
| Species | petD | rps12 | clpP | rpoC1 | rps16 |
|---|---|---|---|---|---|
| 1 | 483/1196 | 372/908 | 591/2047 | 2064/2821 | 267/1132 |
| 2 | 483/1218 | 372/908 | 591/2043 | 2073/2834 | 267/1147 |
| 3 | 483/1230 | 375/911 | 591/2045 | 2052/2820 | 267/1134 |
| 4 | 483/1261 | 381/917 | 591/2043 | 2073/2827 | 267/1153 |
| 5 | 483/1134 | 372/908 | 591/2045 | 2052/2811 | 255/1147 |
| 6 | 483/1148 | 372/908 | 591/2053 | 2073/2830 | 267/1136 |
| 7 | 483/1217 | 372/908 | 591/2039 | 2073/2834 | 267/1142 |
| 8 | 483/1196 | 372/908 | 588/2045 | 2064/2822 | 267/1131 |
| 9 | 483/1196 | 381/917 | 660 | 2076/2844 | 267/1162 |
| 10 | 483/1203 | 387/923 | 786 | 2052/2808 | 267/1154 |
| 11 | 483/1213 | 387/923 | 786 | 2052/2807 | 267/1161 |
| 12 | 483/1215 | 381/917 | 591/2041 | 2073/2833 | 267/1143 |
| 13 | 483/1217 | 372/908 | 591/2043 | 2073/2830 | 267/1143 |
| 14 | 483/1215 | 372/908 | 591/2046 | 2073/2832 | 267/1141 |
| 15 | 483/1213 | 371/907 | 591/2043 | 2073/2831 | 267/1142 |
| 16 | 483/1213 | 372/908 | 591/2044 | 2073/2831 | 267/1142 |
| 17 | 483/1215 | 373/909 | 591/2041 | 2073/2833 | 267/1141 |
| 18 | 483/1203 | 372/908 | 591/2047 | 2073/2834 | 267/1147 |
| 19 | 483/1199 | 372/913 | 591/2041 | 2073/2784 | 237/1115 |
表2 叶绿体基因组中含内含子蛋白编码基因的编码区长度和基因全长
Table 2 Length of coding region and complete gene of intron-contained protein-coding genes of chloroplast genomes
| Species | petD | rps12 | clpP | rpoC1 | rps16 |
|---|---|---|---|---|---|
| 1 | 483/1196 | 372/908 | 591/2047 | 2064/2821 | 267/1132 |
| 2 | 483/1218 | 372/908 | 591/2043 | 2073/2834 | 267/1147 |
| 3 | 483/1230 | 375/911 | 591/2045 | 2052/2820 | 267/1134 |
| 4 | 483/1261 | 381/917 | 591/2043 | 2073/2827 | 267/1153 |
| 5 | 483/1134 | 372/908 | 591/2045 | 2052/2811 | 255/1147 |
| 6 | 483/1148 | 372/908 | 591/2053 | 2073/2830 | 267/1136 |
| 7 | 483/1217 | 372/908 | 591/2039 | 2073/2834 | 267/1142 |
| 8 | 483/1196 | 372/908 | 588/2045 | 2064/2822 | 267/1131 |
| 9 | 483/1196 | 381/917 | 660 | 2076/2844 | 267/1162 |
| 10 | 483/1203 | 387/923 | 786 | 2052/2808 | 267/1154 |
| 11 | 483/1213 | 387/923 | 786 | 2052/2807 | 267/1161 |
| 12 | 483/1215 | 381/917 | 591/2041 | 2073/2833 | 267/1143 |
| 13 | 483/1217 | 372/908 | 591/2043 | 2073/2830 | 267/1143 |
| 14 | 483/1215 | 372/908 | 591/2046 | 2073/2832 | 267/1141 |
| 15 | 483/1213 | 371/907 | 591/2043 | 2073/2831 | 267/1142 |
| 16 | 483/1213 | 372/908 | 591/2044 | 2073/2831 | 267/1142 |
| 17 | 483/1215 | 373/909 | 591/2041 | 2073/2833 | 267/1141 |
| 18 | 483/1203 | 372/908 | 591/2047 | 2073/2834 | 267/1147 |
| 19 | 483/1199 | 372/913 | 591/2041 | 2073/2784 | 237/1115 |
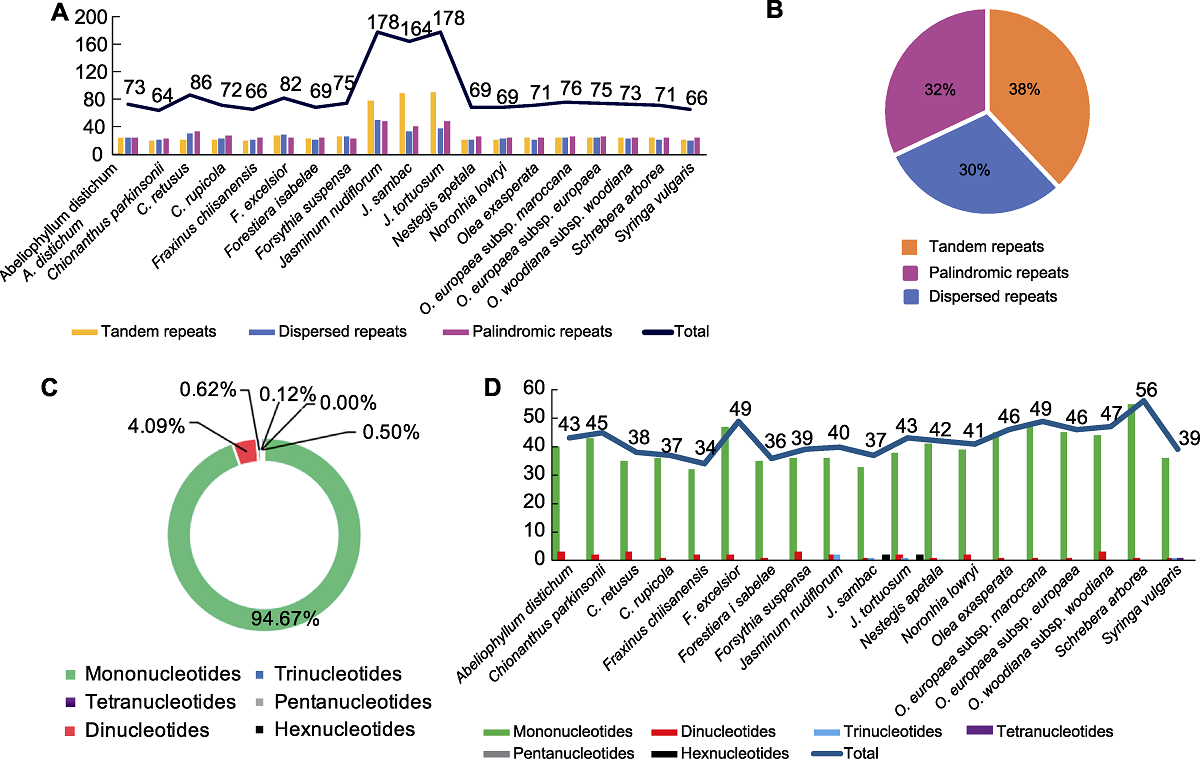
图1 19种木犀科植物叶绿体基因组中重复序列类型和比例 (A) 3种重复类型的数目; (B) 3种重复类型的比例; (C) SSR类型的比例; (D) SSR的数目和类型
Figure 1 The type and percentage of repeated sequences in the chloroplast genomes of 19 species in Oleaceae (A) Number of three type repeats; (B) Percentage of three type repeats; (C) Percentage of SSR types; (D) Number and types of SSRs
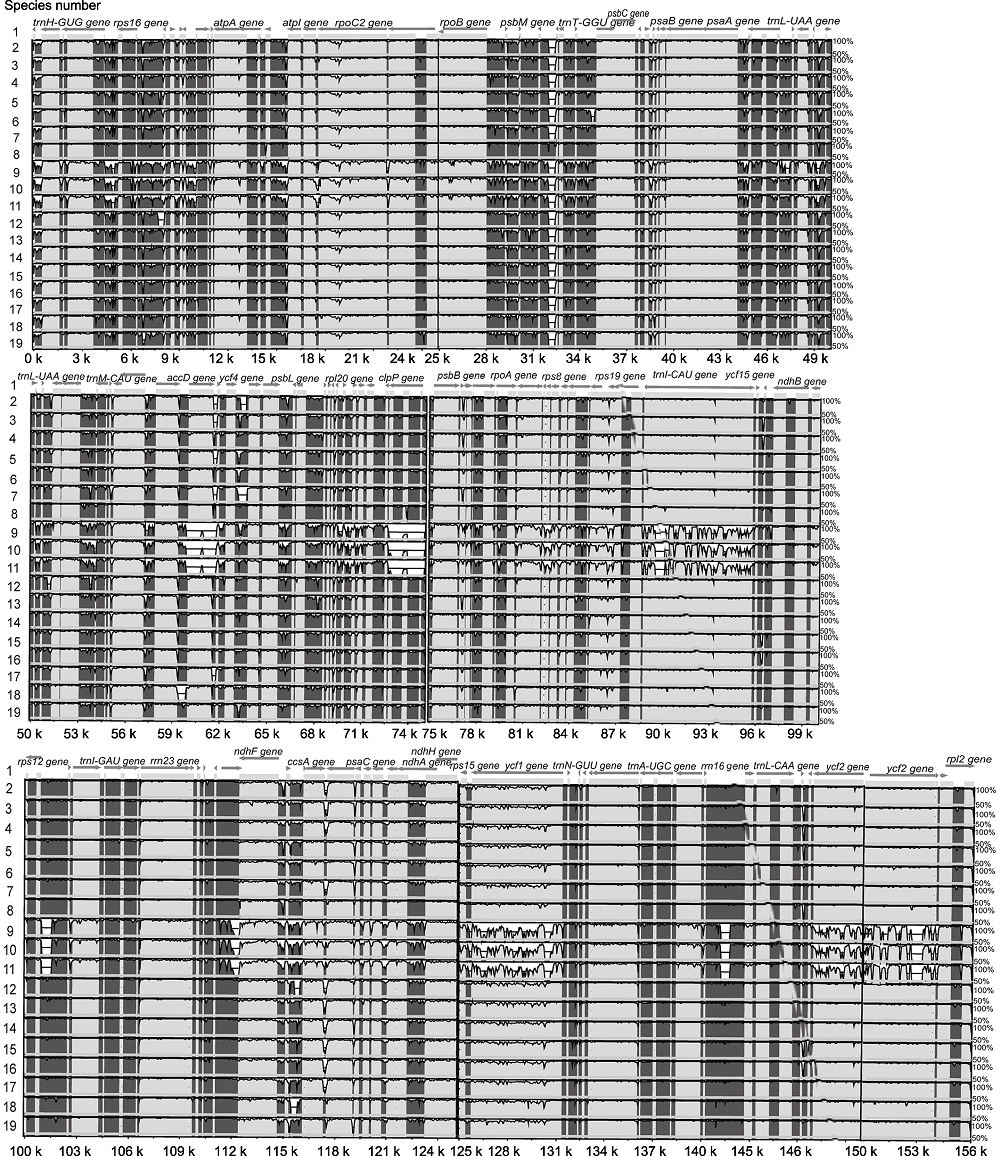
图3 木犀科19个物种叶绿体基因组的比对分析 物种编号1-19同表1。以Abeliophyllum distichum叶绿体基因组序列为参考序列(x轴), 各物种叶绿体基因组与参考序列一致度范围为50%-100%(y轴)。箭头表示基因及转录方向。
Figure 3 Graphic view of the alignment of chloroplast genomes from 19 species in Oleaceae The species number 1-19 same as Table 1. Sequence identity varying between 50% and 100% are drawn on the y axis of the plot, the x axis corresponds to the coordinates on the Abeliophyllum distichum chloroplast genome. Arrows indicate the annotated genes and their transcriptional direction.
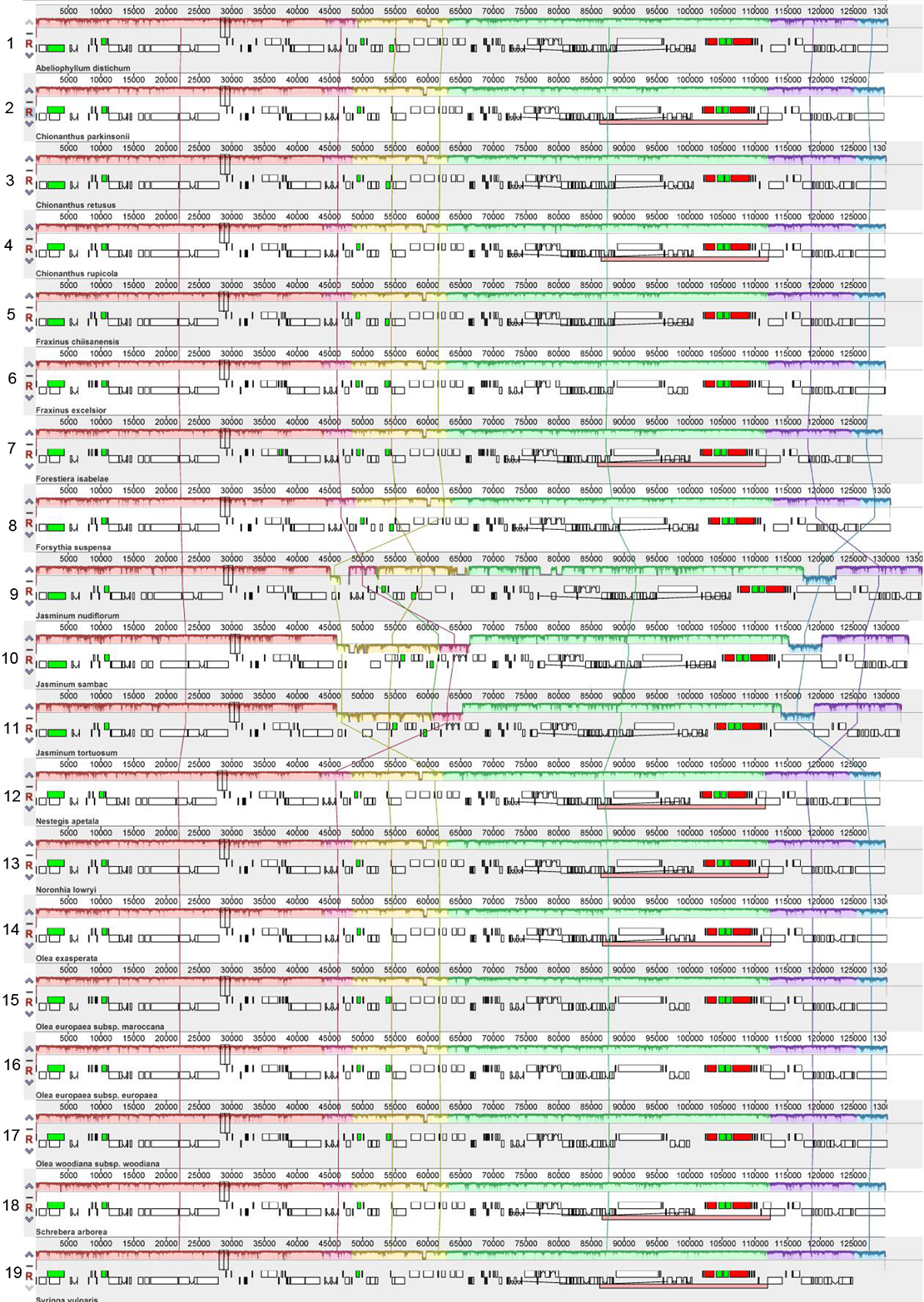
图4 木犀科19种植物的叶绿体基因组共线性分析 物种编号1-19同表1。
Figure 4 Synteny analyses of chloroplast genomes from 19 species in Oleaceae The species number 1-19 same as Table 1.
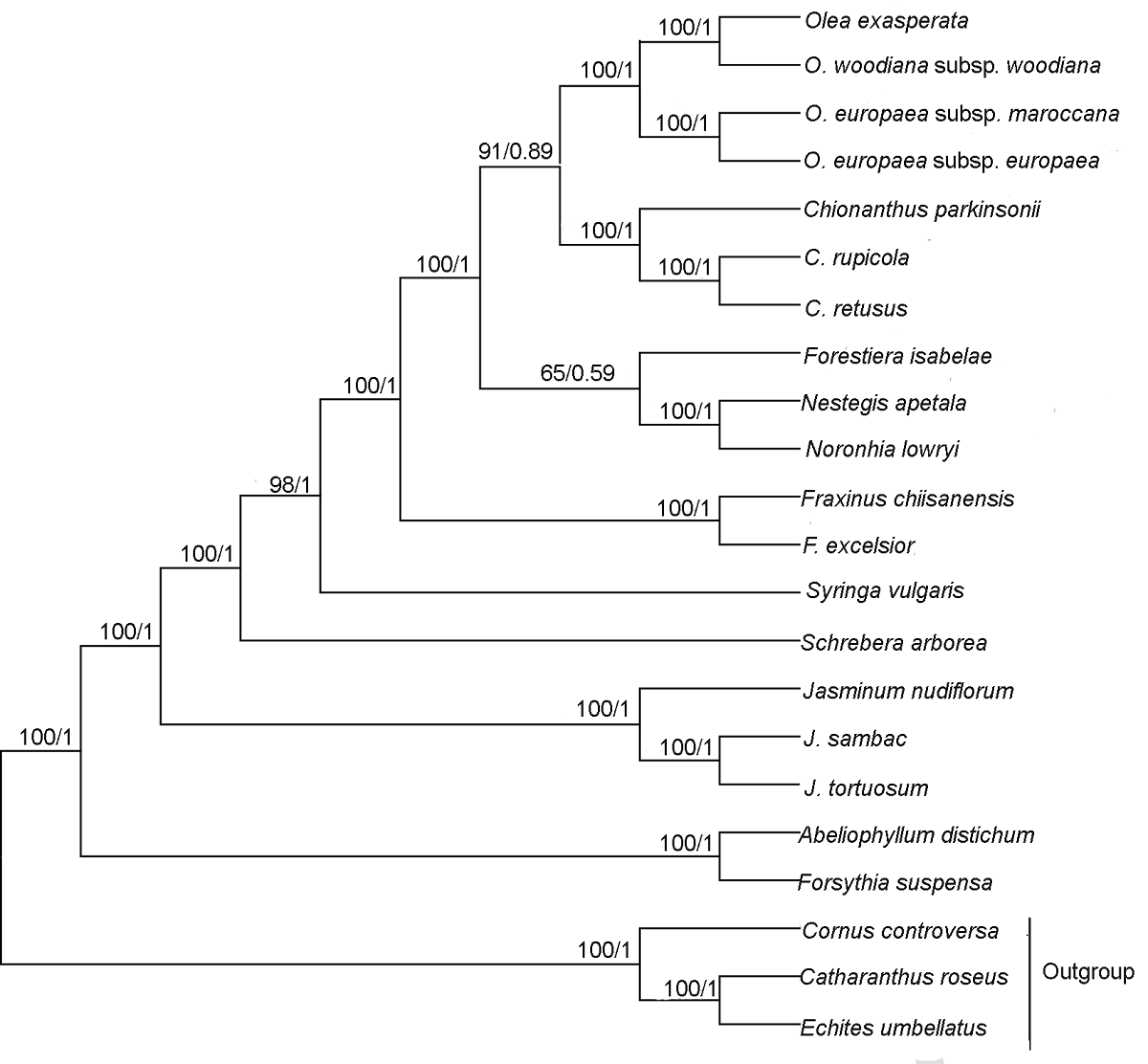
图5 基于木犀科19个物种共有蛋白编码基因构建的系统发育树 每个节点附近的数字分别表示ML树的自展支持率和BI树的后验概率。以Cornus controversa、Echites umbellatus和Catharanthus roseus为外类群。ML: 最大似然法; BI: 贝叶斯法
Figure 5 Phylogenetic relationship of 19 species in Oleaceae inferred from ML and BI analyses based on shared protein-coding genes The numbers near each node are bootstrap support values in ML and posterior probability in BI. The outgroups are Cornus controversa, Echites umbellatus and Catharanthus roseus. ML: Maximum likelihood; BI: Bayesian inference
| [1] | 李巧丽, 延娜, 宋琼, 郭军战 ( 2018). 鲁桑叶绿体基因组序列及特征分析. 植物学报 53, 94-103. |
| [2] | 唐萍, 阮秋燕, 彭程 ( 2011). 禾本科植物叶绿体基因组结构的系统进化研究. 中国农学通报 27(30), 17. |
| [3] | Baali-Cherif D, Besnard G ( 2005). High genetic diversity and clonal growth in relict populations of Olea europaea subsp. laperrinei ( Oleaceae) from Hoggar, Algeria. Ann Bot 96, 823-830. |
| [4] | Benson G ( 1999). Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res 27, 573-580. |
| [5] | Besnard G, de Casas RR, Christin PA, Vargas P ( 2009). Phylogenetics of Olea( Oleaceae) based on plastid and nuclear ribosomal DNA sequences: tertiary climatic shifts and lineage differentiation times. Ann Bot 104, 143-160. |
| [6] | Besnard G, Hernández P, Khadari B, Dorado G, Savolainen V ( 2011). Genomic profiling of plastid DNA variation in the mediterranean olive tree. BMC Plant Biol 11, 80. |
| [7] | Besnard G, Khadari B, Baradat P, Bervillé A ( 2002). Olea europaea( Oleaceae) phylogeography based on chloroplast DNA polymorphism. Theor Appl Genet 104, 1353-1361. |
| [8] | Blazier JC, Ruhlman TA, Weng ML, Rehman SK, Sabir JSM, Jansen RK ( 2016). Divergence of RNA polymerase α subunits in angiosperm plastid genomes is mediated by genomic rearrangement. Sci Rep 6, 24595. |
| [9] | Chumley TW, Palmer JD, Mower JP, Fourcade HM, Calie PJ, Boore JL, Jansen RK ( 2006). The complete chloroplast genome sequence of Pelargonium x hortorum: organization and evolution of the largest and most highly rearranged chloroplast genome of land plants. Mol Biol Evol 23, 2175-2190. |
| [10] | Darling ACE, Mau B, Blattner FR, Perna ANT ( 2004). Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res 14, 1394-1403. |
| [11] | Dong WP, Xu C, Cheng T, Lin K, Zhou SL ( 2013). Sequencing angiosperm plastid genomes made easy: a complete set of universal primers and a case study on the phylogeny of Saxifragales. Genome Biol Evol 5, 989-997. |
| [12] | Flora of China Editorial Committee ( 1995). Flora of China, Vol. 16. Beijing & St. Louis: Science Press & Missouri Botanical Garden Press. pp. 143-188. |
| [13] | Flora of China Editorial Committee ( 1996). Flora of China, Vol. 15. Beijing & St. Louis: Science Press & Missouri Botanical Garden Press. pp. 272-319. |
| [14] | Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I ( 2004). Vista: computational tools for comparative genomics. Nucleic Acids Res 32, W273-W279. |
| [15] | Gao L, Su YJ, Wang T ( 2010). Plastid genome sequencing, comparative genomics, and phylogenomics: current status and prospects. J Syst Evol 48, 77-93. |
| [16] | Gray BN, Ahner BA, Hanson MR ( 2009). Extensive homologous recombination between introduced and native regulatory plastid DNA elements in transplastomic plants. Transgenic Res 18, 559-572. |
| [17] | Greiner S, Wang X, Rauwolf U, Silber MV, Mayer K, Meurer J, Haberer G, Herrmann RG ( 2008). The complete nucleotide sequences of the five genetically distinct plastid genomes of Oenothera, subsection Oenothera. I. Sequence evaluation and plastome evolution. Nucleic Acids Res 36, 2366-2378. |
| [18] | Guisinger MM, Kuehl JV, Boore JL, Jansen RK ( 2011). Extreme reconfiguration of plastid genomes in the angiosperm family Geraniaceae: rearrangements, repeats, and codon usage. Mol Biol Evol 28, 583-600. |
| [19] | Hansen DR, Dastidar SG, Cai ZQ, Penaflor C, Kuehl JV, Boore JL, Jansen RK ( 2007). Phylogenetic and evolutionary implications of complete chloroplast genome sequences of four early-diverging angiosperms: Buxus( Buxaceae), Chloranthus 45, 547-563. |
| [20] | He YX, Liu LX, Yang SH, Dong MF, Yuan WJ, Shang FD ( 2017). Characterization of the complete chloroplast genome of chinese fringetree (Chionanthus retusus). Conserv Genet Resour 9, 431-434. |
| [21] | Hirao T, Watanabe A, Kurita M, Kondo T, Takata K ( 2008). Complete nucleotide sequence of the Cryptomeria japonica D. Don. chloroplast genome and comparative chloroplast genomics: diversified genomic structure of coniferous species. BMC Plant Biol 8, 70. |
| [22] | Hiratsuka J, Shimada H, Whittier R, Ishibashi T, Sakamoto M, Mori M, Kondo C, Honji Y, Sun CR, Meng BY, Li YQ, Kanno A, Nishizawa Y, Hirai A, Shinozaki K, Sugiura M ( 1989). The complete sequence of the rice (Oryza sativa) chloroplast genome: intermolecular recombination between distinct tRNA genes accounts for a major plastid DNA inversion during the evolution of the cereals. Mol Gen Genet 217, 185-194. |
| [23] | Hu YC, Zhang Q, Rao GY , Sodmergen ( 2008). Occurrence of plastids in the sperm cells of Caprifoliaceae: biparental plastid inheritance in angiosperms is unilaterally derived from maternal inheritance. Plant Cell Physiol 49, 958-968. |
| [24] | Huang H, Shi C, Liu Y, Mao SY, Gao LZ ( 2014). Thirteen Camellia chloroplast genome sequences determined by high-throughput sequencing: genome structure and phylogenetic relationships. BMC Evol Biol 14, 151. |
| [25] | Jeandroz S, Roy A, Bousquet J ( 1997). Phylogeny and phylogeography of the circumpolar genus Fraxinus( Oleaceae) based on internal transcribed spacer sequences of nuclear ribosomal DNA. Mol Phylogenet Evol 7, 241-251. |
| [26] | Katayama H, Ogihara Y ( 1996). Phylogenetic affinities of the grasses to other monocots as revealed by molecular analysis of chloroplast DNA. Curr Genet 29, 572-581. |
| [27] | Khakhlova O, Bock R ( 2006). Elimination of deleterious mutations in plastid genomes by gene conversion. Plant J 46, 85-94. |
| [28] | Kim C, Kim HJ, Do HDK, Jung J, Kim JH ( 2018). Characterization of the complete chloroplast genome of Fraxinus chiisanensis( Oleaceae), an endemic to Korea. Conserv Genet Resour 11, 63-66. |
| [29] | Kim HW, Lee HL, Lee DK, Kim KJ ( 2016). Complete plastid genome sequences of Abeliophyllum distichum Nakai (Oleaceae), a Korea endemic genus. Mitochondr DNA Part B 1, 596-598. |
| [30] | Knox EB, Downie SR, Palmer JD ( 1993). Chloroplast genome rearrangements and the evolution of giant lobelias from herbaceous ancestors. Mol Biol Evol 10, 414-430. |
| [31] | Kurtz S, Choudhuri JV, Ohlebusch E, Schleiermacher C, Stoye J, Giegerich R ( 2001). Reputer: the manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res 29, 4633-4642. |
| [32] | Lee HL, Jansen RK, Chumley TW, Kim KJ ( 2007). Gene relocations within chloroplast genomes of Jasminum and Menodora( Oleaceae) are due to multiple, overlapping inversions. Mol Biol Evol 24, 1161-1180. |
| [33] | Li JH, Alexander JH, Zhang DL ( 2002). Paraphyletic Syringa( Oleaceae): evidence from sequences of nuclear ribosomal DNA ITS and ETS regions. Syst Bot 27, 592-597. |
| [34] | Maier RM, Neckermann K, Igloi GL, Kösel H ( 1995). Complete sequence of the maize chloroplast genome: gene content, hotspots of divergence and fine tuning of genetic information by transcript editing. J Mol Biol 251, 614-628. |
| [35] | Ogihara Y, Terachi T, Sasakuma T ( 1988). Intramolecular recombination of chloroplast genome mediated by short direct-repeat sequences in wheat species. Proc Natl Acad Sci USA 85, 8573-8577. |
| [36] | Palmer JD ( 1985). Comparative organization of chloroplast genomes. Annu Rev Genet 19, 325-354. |
| [37] | Posada D ( 2008). jModeltest: phylogenetic model averaging. Mol Biol Evol 25, 1253-1256. |
| [38] | Ronquist F, Huelsenbeck JP ( 2003). Mrbayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572-1574. |
| [39] | Stamatakis A, Hoover P, Rougemont J ( 2008). A rapid bootstrap algorithm for the raxml web servers. Syst Biol 57, 758-771. |
| [40] | Thiel T, Michalek W, Varshney R, Graner A ( 2003). Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley (Hordeum vulgare L.). Theor Appl Genet 106, 411-422. |
| [41] | Van de Paer C, Bouchez O, Besnard G ( 2018). Prospects on the evolutionary mitogenomics of plants: a case study on the olive family (Oleaceae). Mol Ecol Res 18, 407-423. |
| [42] | Wallander E ( 2008). Systematics of Fraxinus( Oleaceae) and evolution of dioecy. Plant Syst Evol 273, 25-49. |
| [43] | Wallander E, Albert VA ( 2000). Phylogeny and classification of Oleaceae based on rps16 and trnL-F sequence data. Am J Bot 87, 1827-1841. |
| [44] | Wang WB, Yu H, Wang JH, Lei WJ, Gao JH, Qiu XP, Wang JS ( 2017). The complete chloroplast genome sequences of the medicinal plant Forsythia suspensa( Oleaceae). Int J Mol Sci 18, 2288. |
| [45] | Weng ML, Blazier JC, Govindu M, Jansen RK ( 2014). Reconstruction of the ancestral plastid genome in Geraniaceae reveals a correlation between genome rearrangements, repeats, and nucleotide substitution rates. Mol Biol Evol 31, 645-659. |
| [46] | Zhang TW, Fang YJ, Wang XM, Deng X, Zhang XW, Hu SN, Yu J ( 2012). The complete chloroplast and mitochondrial genome sequences of Boea hygrometrica: insights into the evolution of plant organellar genomes. PLoS One 7, e30531. |
| [47] | Zhang X, Zhou T, Yang J, Sun JJ, Ju MM, Zhao YM, Zhao GF ( 2018). Comparative analyses of chloroplast genomes of Cucurbitaceae species: lights into selective pressures and phylogenetic relationships. Molecules 23, 2165. |
| [1] | 王传永, 庄典, 宋正达, 翟恒华, 李乃伟, 张凡. 黑果腺肋花楸叶绿体全基因组的结构和比较分析及系统进化推断[J]. 植物学报, 2025, 60(4): 1-0. |
| [2] | 孙亚君. 何谓高等或低等生物——澄清《物种起源》所蕴含的生物等级性的涵义及其成立性[J]. 生物多样性, 2025, 33(1): 24394-. |
| [3] | 艾妍雨, 胡海霞, 沈婷, 莫雨轩, 杞金华, 宋亮. 附生维管植物多样性及其与宿主特征的相关性: 以哀牢山中山湿性常绿阔叶林为例[J]. 生物多样性, 2024, 32(5): 24072-. |
| [4] | 吕燕文, 王子韵, 肖钰, 何梓晗, 吴超, 胡新生. 谱系分选理论与检测方法的研究进展[J]. 生物多样性, 2024, 32(4): 23400-. |
| [5] | 曹可欣, 王敬雯, 郑国, 武鹏峰, 李英滨, 崔淑艳. 降水格局改变及氮沉降对北方典型草原土壤线虫多样性的影响[J]. 生物多样性, 2024, 32(3): 23491-. |
| [6] | 王斌, 钟艺倩, 杨美雪, 吴淼锐, 王艳萍, 陆芳, 陶旺兰, 李健星, 赵弘明, 刘晟源, 向悟生, 李先琨. 喀斯特季节性雨林优势树种叶片非结构性碳水化合物空间变异及生态驱动因素[J]. 生物多样性, 2024, 32(12): 24325-. |
| [7] | 杨向林, 赵彩云, 李俊生, 种方方, 李文金. 植物入侵导致群落谱系结构更加聚集: 以广西国家级自然保护区草本植物为例[J]. 生物多样性, 2024, 32(11): 24175-. |
| [8] | 何林君, 杨文静, 石宇豪, 阿说克者莫, 范钰, 王国严, 李景吉, 石松林, 易桂花, 彭培好. 火烧干扰下植物群落系统发育和功能多样性对紫茎泽兰入侵的影响[J]. 生物多样性, 2024, 32(11): 24269-. |
| [9] | 王振宇, 黄志群. 亚热带27种木本植物叶片性状对植食作用的影响: 验证生长-防御权衡假说[J]. 植物生态学报, 2024, 48(11): 1501-1509. |
| [10] | 李庆多, 栗冬梅. 全球蝙蝠巴尔通体流行状况分析[J]. 生物多样性, 2023, 31(9): 23166-. |
| [11] | 褚振州, 古丽巴哈尔·依斯拉木, 屈泽众, 田新民. 同域分布的3种木蓼属植物叶绿体基因组比较[J]. 植物学报, 2023, 58(3): 417-432. |
| [12] | 宋会银, 胡征宇, 刘国祥. 绿藻门小球藻科的分类学研究进展[J]. 生物多样性, 2023, 31(2): 22083-. |
| [13] | 李治中, 彭帅, 王青锋, 李伟, 梁士楚, 陈进明. 中国海菜花属植物隐种多样性[J]. 生物多样性, 2023, 31(2): 22394-. |
| [14] | 包金波, 丁志杰, 苗浩宇, 李雪丽, 任书贤, 焦若岩, 李浩, 邓茜茜, 李英姿, 田新民. 石栗叶绿体基因组研究[J]. 植物学报, 2023, 58(2): 248-260. |
| [15] | 朱瑞良, 马晓英, 曹畅, 曹子寅. 中国苔藓植物多样性研究进展[J]. 生物多样性, 2022, 30(7): 22378-. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||