

植物学报 ›› 2023, Vol. 58 ›› Issue (2): 248-260.DOI: 10.11983/CBB22026 cstr: 32102.14.CBB22026
包金波, 丁志杰, 苗浩宇, 李雪丽, 任书贤, 焦若岩, 李浩, 邓茜茜, 李英姿, 田新民( )
)
收稿日期:2022-02-10
接受日期:2022-05-10
出版日期:2023-03-01
发布日期:2023-03-15
通讯作者:
*E-mail: 基金资助:Jinbo Bao, Zhijie Ding, Haoyu Miao, Xueli Li, Shuxian Ren, Ruoyan Jiao, Hao Li;Qianqian Deng, Yingzi Li, Xinmin Tian
Received:2022-02-10
Accepted:2022-05-10
Online:2023-03-01
Published:2023-03-15
Contact:
*E-mail: 摘要: 石栗(Aleurites moluccana)是大戟科石栗属的常绿阔叶乔木, 具有能源、药用和观赏价值。为填补石栗叶绿体基因组研究的空白, 通过二代高通量全基因组测序, 组装和注释了石栗叶绿体基因组, 并进行基因组特征和系统发育分析。结果显示, 石栗叶绿体基因组为典型的四段式结构, 总长度为163 298 bp, LSC、SSC及IR的长度分别为91 301、18 501和26 748 bp。石栗叶绿体基因组共有131个基因, 包括8个rRNA基因, 37个tRNA基因, 86个蛋白质编码基因。研究发现145个SSR位点, 检测到重复单元有单核苷酸、二核苷酸、三核苷酸和四核苷酸, 数目分别为80、53、10和2个。共线性分析结果表明, 石栗叶绿体基因组存在基因倒位和重排现象。利用最大似然法和贝叶斯法构建了系统发育树, 显示石栗与油桐(Vernicia fordii)和东京桐(Deutzianthus tonkinensis)亲缘关系较近, 并形成姐妹群。利用化石时间进行定年分析, 表明石栗属、油桐属和东京桐属的分化时间为25.94 Ma (95% HPD: 24.71-63.32 Ma)。该研究丰富了石栗基因组信息, 可为石栗种质资源的开发利用提供基础遗传数据, 同时为石栗属物种鉴定及系统发育研究提供参考。
包金波, 丁志杰, 苗浩宇, 李雪丽, 任书贤, 焦若岩, 李浩, 邓茜茜, 李英姿, 田新民. 石栗叶绿体基因组研究. 植物学报, 2023, 58(2): 248-260.
Jinbo Bao, Zhijie Ding, Haoyu Miao, Xueli Li, Shuxian Ren, Ruoyan Jiao, Hao Li, Qianqian Deng, Yingzi Li, Xinmin Tian. Analysis of Chloroplast Genomes of Aleurites moluccana. Chinese Bulletin of Botany, 2023, 58(2): 248-260.

图1 石栗自然种群 (A) 花序; (B) 枝条; (C) 幼枝和幼叶; (D) 果实
Figure 1 Natural population of Aleurites moluccana (A) Inflorescence; (B) Branches; (C) Young branches and leaves; (D) Fruit
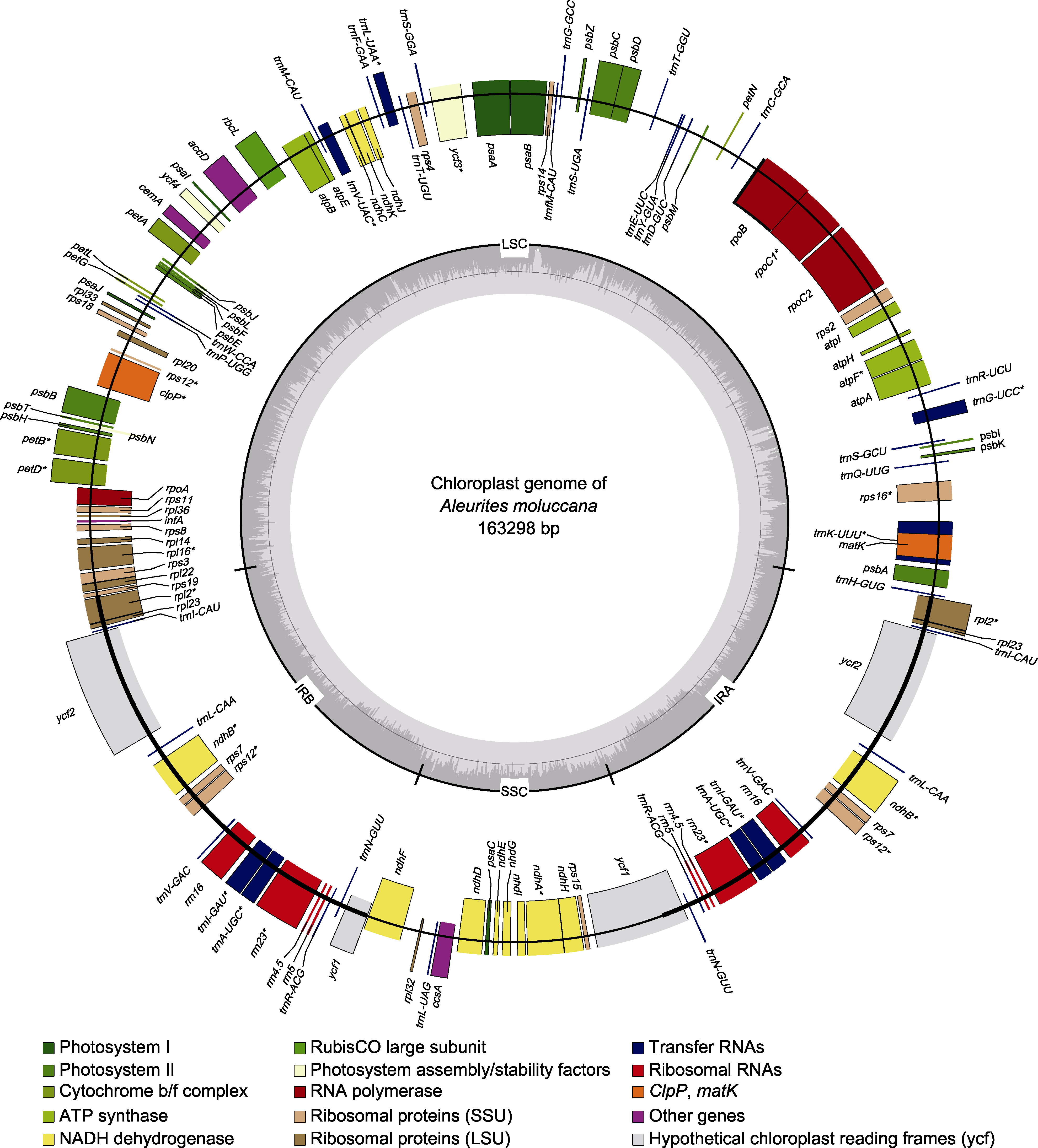
图2 石栗叶绿体基因组图谱 标注在大环外部的基因按照顺时针方向转录, 标注在大环内部的基因按照逆时针方向转录。不同颜色代表基因功能不同。内环阴影部分代表石栗叶绿体基因组的GC组成。LSC: 大单拷贝区; SSC: 小单拷贝区; IRA: 反向重复区A; IRB: 反向重复区B。* 含有内含子的基因。
Figure 2 The chloroplast genome map of Aleurites moluccana Genes on the outside of the large circle are transcribed clockwise and those on the inside are transcribed counterclockwise. The genes are color-coded based on their function. The dashed area on the inside represents the GC composition of the A. moluccana chloroplast genome. LSC: Large single-copy region; SSC: Small single-copy region; IRA: Inverted repeat region A; IRB: Inverted repeat region B. * for genes containing introns.
| Categories of genes | Group of genes | Name of genes |
|---|---|---|
| Genes for photosynthesis | Subunits of photosystem I | psaA, psaB, psaC, psaI, psaJ |
| Subunits of photosystem II | psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbI, psbJ, psbK, psbL, psbM, psbN, psbT, psbZ | |
| Subunits of ATP synthase | atpA, atpB, atpE, atpF, atpH, atpI | |
| Subunits of cytochrome | petA, petB, petD, petG, petL, petN | |
| ATP-dependent protease subunits P gene | clpP | |
| Large subunits of Rubisco | rbcL | |
| Subunits of NADH dehydrogenase | ndhA, ndhB, ndhC, ndhD, ndhE, ndhF, ndhG, ndhH, ndhI, ndhJ, ndhK | |
| Self replication | Small subunit of ribosome | rps2, rps3, rps4, rps7, rps8, rps11, rps12, rps14, rps15, rps16, rps18, rps19 |
| Large subunit of ribosome | rpl2, rpl14, rpl16, rpl20, rpl2, rpl23, rpl32, rpl33, rpl36, rpl22 | |
| DNA dependent RNA polymerase | rpoA, rpoB, rpoC1, rpoC2 | |
| Ribosomal RNA genes | rrn5, rrn4.5, rrn16, rrn23 | |
| Transfer RNA genes | trnN-GUU, trnR-ACG, trnH-GUG, trnL-CAU, trnA-UGC, trnL-GAU, trnV-GAC, trnL- CAA, trnL-CAU, trnP-UGG, trnW-CCA, trnM-CAU, trnV-UAC, trnF-GAA, trnL-UAA, trnT-UGU, trnS-GGA, trnfM-CAU, trnG-GCC, trnS-UGA, trnT-GGU, trnE-UUC, trnY-GUA, trnD-GUC, trnC-GCA, trnR-UCU, trnS-GCU, trnQ-UUG, trnK-UUU, trnL-CAA, trnV-GAC, trnL-GAU, trnR-UGC, trnL-UAG, trnR-ACG, trnN-GUU | |
| Other genes | Maturase | matK |
| Envelop membrane protein | cemA | |
| Translation initiation factor IF-1 | infA | |
| C-type cytochrome synthesis gene | ccsA | |
| Unknown function | Conserved open reading frames | ycf1, ycf2, ycf3, ycf4, ycf15 |
表1 石栗叶绿体基因组功能基因注释
Table 1 Annotation of functional genes in the chloroplast genome of Aleurites moluccana
| Categories of genes | Group of genes | Name of genes |
|---|---|---|
| Genes for photosynthesis | Subunits of photosystem I | psaA, psaB, psaC, psaI, psaJ |
| Subunits of photosystem II | psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbI, psbJ, psbK, psbL, psbM, psbN, psbT, psbZ | |
| Subunits of ATP synthase | atpA, atpB, atpE, atpF, atpH, atpI | |
| Subunits of cytochrome | petA, petB, petD, petG, petL, petN | |
| ATP-dependent protease subunits P gene | clpP | |
| Large subunits of Rubisco | rbcL | |
| Subunits of NADH dehydrogenase | ndhA, ndhB, ndhC, ndhD, ndhE, ndhF, ndhG, ndhH, ndhI, ndhJ, ndhK | |
| Self replication | Small subunit of ribosome | rps2, rps3, rps4, rps7, rps8, rps11, rps12, rps14, rps15, rps16, rps18, rps19 |
| Large subunit of ribosome | rpl2, rpl14, rpl16, rpl20, rpl2, rpl23, rpl32, rpl33, rpl36, rpl22 | |
| DNA dependent RNA polymerase | rpoA, rpoB, rpoC1, rpoC2 | |
| Ribosomal RNA genes | rrn5, rrn4.5, rrn16, rrn23 | |
| Transfer RNA genes | trnN-GUU, trnR-ACG, trnH-GUG, trnL-CAU, trnA-UGC, trnL-GAU, trnV-GAC, trnL- CAA, trnL-CAU, trnP-UGG, trnW-CCA, trnM-CAU, trnV-UAC, trnF-GAA, trnL-UAA, trnT-UGU, trnS-GGA, trnfM-CAU, trnG-GCC, trnS-UGA, trnT-GGU, trnE-UUC, trnY-GUA, trnD-GUC, trnC-GCA, trnR-UCU, trnS-GCU, trnQ-UUG, trnK-UUU, trnL-CAA, trnV-GAC, trnL-GAU, trnR-UGC, trnL-UAG, trnR-ACG, trnN-GUU | |
| Other genes | Maturase | matK |
| Envelop membrane protein | cemA | |
| Translation initiation factor IF-1 | infA | |
| C-type cytochrome synthesis gene | ccsA | |
| Unknown function | Conserved open reading frames | ycf1, ycf2, ycf3, ycf4, ycf15 |
| SSR repeat type (number of copies) | SSR repeat sequence | Number of copies | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16-30 | Total | ||
| Mononucleotide (80) | A/T | 24 | 17 | 7 | 14 | 3 | 3 | 68 | ||||||
| C/G | 7 | 3 | 2 | 12 | ||||||||||
| Dinucleotide (53) | AT/TA/AC/CA/AG/GA | 18 | 7 | 6 | 4 | 6 | 2 | 2 | 45 | |||||
| CG/GC | 4 | 2 | 1 | 1 | 8 | |||||||||
| Trinucleotide (10) | CGG/AAG | 2 | 2 | 1 | 5 | |||||||||
| TGT/TTA | 1 | 2 | 3 | |||||||||||
| ATT/AAT | 1 | 1 | 2 | |||||||||||
| Tetranucleotide (2) | TACA/TGGT | 1 | 1 | 2 | ||||||||||
表2 石栗叶绿体基因组简单重复序列(SSR)信息
Table 2 Information of simple sequence repeat (SSR) identified in the chloroplast genome of Aleurites moluccana
| SSR repeat type (number of copies) | SSR repeat sequence | Number of copies | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16-30 | Total | ||
| Mononucleotide (80) | A/T | 24 | 17 | 7 | 14 | 3 | 3 | 68 | ||||||
| C/G | 7 | 3 | 2 | 12 | ||||||||||
| Dinucleotide (53) | AT/TA/AC/CA/AG/GA | 18 | 7 | 6 | 4 | 6 | 2 | 2 | 45 | |||||
| CG/GC | 4 | 2 | 1 | 1 | 8 | |||||||||
| Trinucleotide (10) | CGG/AAG | 2 | 2 | 1 | 5 | |||||||||
| TGT/TTA | 1 | 2 | 3 | |||||||||||
| ATT/AAT | 1 | 1 | 2 | |||||||||||
| Tetranucleotide (2) | TACA/TGGT | 1 | 1 | 2 | ||||||||||
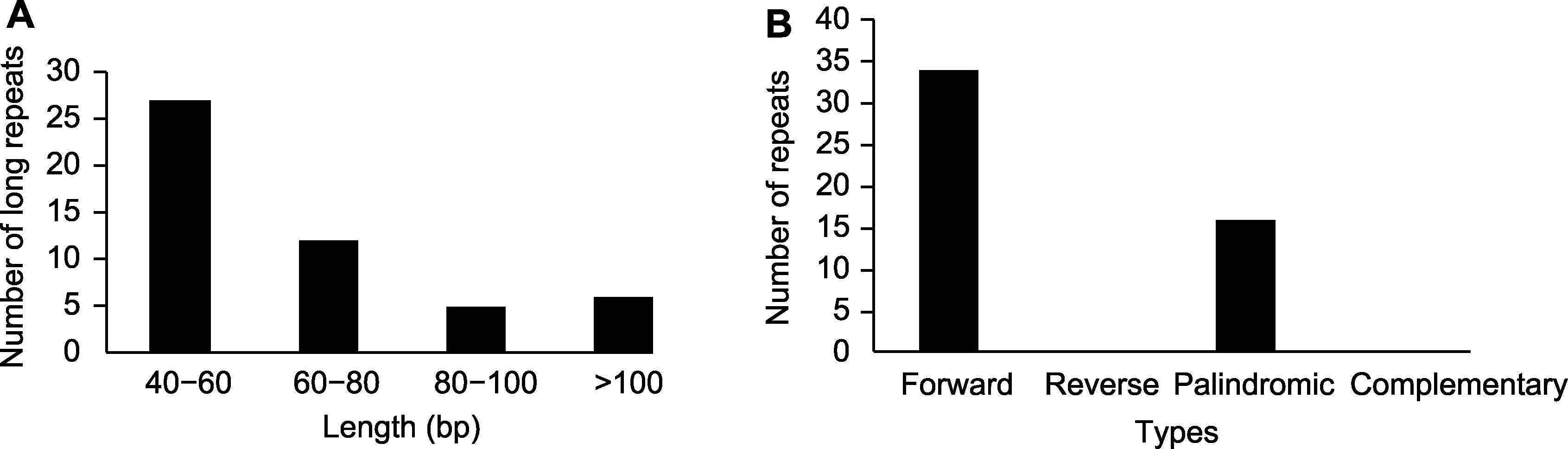
图3 石栗叶绿体基因组重复序列长度及类型 (A) 重复序列长度; (B) 重复序列类型
Figure 3 The length and type of repeat sequences in the chloroplast genome of Aleurites moluccana (A) The length of repeat sequences; (B) The type of repeat sequences
| Amino acid | Codon | No. of codon | RSCU | Amino acid | Codon | No. of codon | RSCU |
|---|---|---|---|---|---|---|---|
| Ala | GCU | 431 | 1.20 | Cys | UGU | 698 | 1.20 |
| GCC | 306 | 0.86 | UGC | 468 | 0.80 | ||
| GCA | 427 | 1.19 | Glu | GAA | 1416 | 1.40 | |
| GCG | 267 | 0.75 | GAG | 611 | 0.60 | ||
| Asp | GAU | 1073 | 1.41 | Gly | GGU | 534 | 0.93 |
| GAC | 448 | 0.59 | GGC | 382 | 0.67 | ||
| Phe | UUU | 2457 | 1.26 | GGA | 803 | 1.40 | |
| UUC | 1452 | 0.74 | GGG | 572 | 1.00 | ||
| His | CAU | 874 | 1.36 | Ile | AUU | 1927 | 1.19 |
| CAC | 412 | 0.64 | AUC | 1184 | 0.73 | ||
| Lys | AAA | 2578 | 1.38 | AUA | 1728 | 1.07 | |
| AAG | 1162 | 0.62 | Leu | UUA | 1299 | 1.46 | |
| Trp | UGG | 748 | 1.00 | UUG | 1125 | 1.26 | |
| Asn | AAU | 2122 | 1.43 | CUU | 1021 | 1.15 | |
| AAC | 843 | 0.57 | CUC | 577 | 0.65 | ||
| Arg | CGU | 341 | 0.58 | CUA | 844 | 0.95 | |
| CGC | 246 | 0.42 | CUG | 442 | 0.50 | ||
| CGA | 590 | 1.00 | Pro | CCU | 568 | 1.02 | |
| CGG | 412 | 0.70 | CCC | 556 | 1.00 | ||
| AGA | 1253 | 2.12 | CCA | 699 | 1.25 | ||
| AGG | 702 | 1.19 | CCG | 408 | 0.73 | ||
| Thr | ACU | 683 | 1.18 | Met | AUG | 887 | 1.00 |
| ACC | 550 | 0.95 | Gln | CAA | 1041 | 1.38 | |
| ACA | 696 | 1.20 | CAG | 463 | 0.62 | ||
| ACG | 383 | 0.66 | Ser | UCU | 1070 | 1.36 | |
| Val | GUU | 737 | 1.34 | UCC | 840 | 1.07 | |
| GUC | 402 | 0.73 | UCA | 1002 | 1.27 | ||
| GUA | 667 | 1.21 | UCG | 565 | 0.72 | ||
| GUG | 393 | 0.71 | AGU | 720 | 0.92 | ||
| Tyr | UAU | 1842 | 1.43 | AGC | 521 | 0.66 | |
| UAC | 737 | 0.57 |
表3 石栗叶绿体基因组密码子使用偏好性
Table 3 Codon usage bias in the chloroplast genome of Aleurites moluccana
| Amino acid | Codon | No. of codon | RSCU | Amino acid | Codon | No. of codon | RSCU |
|---|---|---|---|---|---|---|---|
| Ala | GCU | 431 | 1.20 | Cys | UGU | 698 | 1.20 |
| GCC | 306 | 0.86 | UGC | 468 | 0.80 | ||
| GCA | 427 | 1.19 | Glu | GAA | 1416 | 1.40 | |
| GCG | 267 | 0.75 | GAG | 611 | 0.60 | ||
| Asp | GAU | 1073 | 1.41 | Gly | GGU | 534 | 0.93 |
| GAC | 448 | 0.59 | GGC | 382 | 0.67 | ||
| Phe | UUU | 2457 | 1.26 | GGA | 803 | 1.40 | |
| UUC | 1452 | 0.74 | GGG | 572 | 1.00 | ||
| His | CAU | 874 | 1.36 | Ile | AUU | 1927 | 1.19 |
| CAC | 412 | 0.64 | AUC | 1184 | 0.73 | ||
| Lys | AAA | 2578 | 1.38 | AUA | 1728 | 1.07 | |
| AAG | 1162 | 0.62 | Leu | UUA | 1299 | 1.46 | |
| Trp | UGG | 748 | 1.00 | UUG | 1125 | 1.26 | |
| Asn | AAU | 2122 | 1.43 | CUU | 1021 | 1.15 | |
| AAC | 843 | 0.57 | CUC | 577 | 0.65 | ||
| Arg | CGU | 341 | 0.58 | CUA | 844 | 0.95 | |
| CGC | 246 | 0.42 | CUG | 442 | 0.50 | ||
| CGA | 590 | 1.00 | Pro | CCU | 568 | 1.02 | |
| CGG | 412 | 0.70 | CCC | 556 | 1.00 | ||
| AGA | 1253 | 2.12 | CCA | 699 | 1.25 | ||
| AGG | 702 | 1.19 | CCG | 408 | 0.73 | ||
| Thr | ACU | 683 | 1.18 | Met | AUG | 887 | 1.00 |
| ACC | 550 | 0.95 | Gln | CAA | 1041 | 1.38 | |
| ACA | 696 | 1.20 | CAG | 463 | 0.62 | ||
| ACG | 383 | 0.66 | Ser | UCU | 1070 | 1.36 | |
| Val | GUU | 737 | 1.34 | UCC | 840 | 1.07 | |
| GUC | 402 | 0.73 | UCA | 1002 | 1.27 | ||
| GUA | 667 | 1.21 | UCG | 565 | 0.72 | ||
| GUG | 393 | 0.71 | AGU | 720 | 0.92 | ||
| Tyr | UAU | 1842 | 1.43 | AGC | 521 | 0.66 | |
| UAC | 737 | 0.57 |
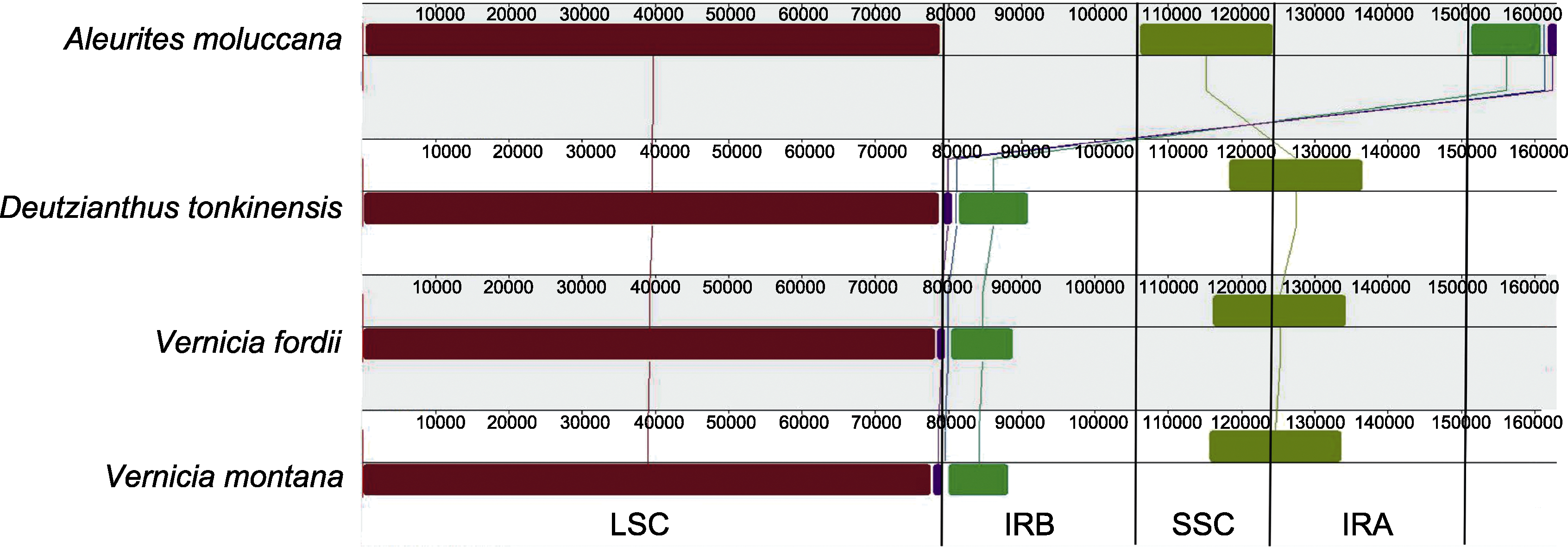
图5 石栗叶绿体基因组共线性分析 LSC、SSC、IRA和IRB同图2。
Figure 5 Collinear analysis of Aleurites moluccana chloroplast genome LSC, SSC, IRA, and IRB are the same as in Figure 2.
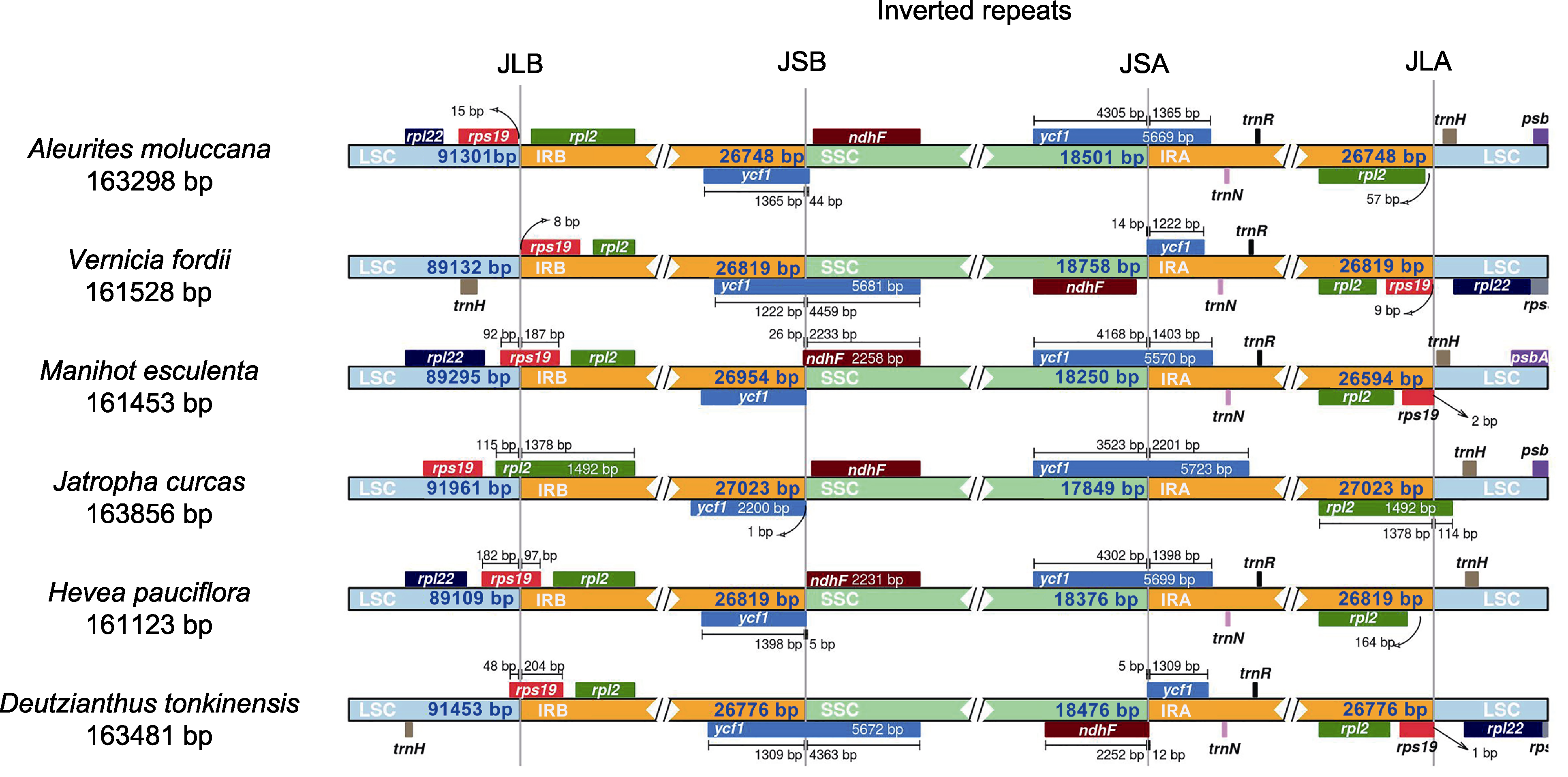
图6 石栗叶绿体基因组反向重复区的收缩与扩张 LSC、SSC、IRA和IRB同图2。JLB: LSC与IRB的边界; JSB: SSC与IRB的边界; JSA: SSC与IRA的边界; JLA: LSC与IRA的边界
Figure 6 Contraction and expansion of inverted repeat region in the chloroplast genome of Aleurites moluccana LSC, SSC, IRA, and IRB are the same as in Figure 2. JLB: Boundary between LSC and IRB; JSB: Boundary between SSC and IRB; JSA: Boundary between SSC and IRA; JLA: Boundary between LSC and IRA
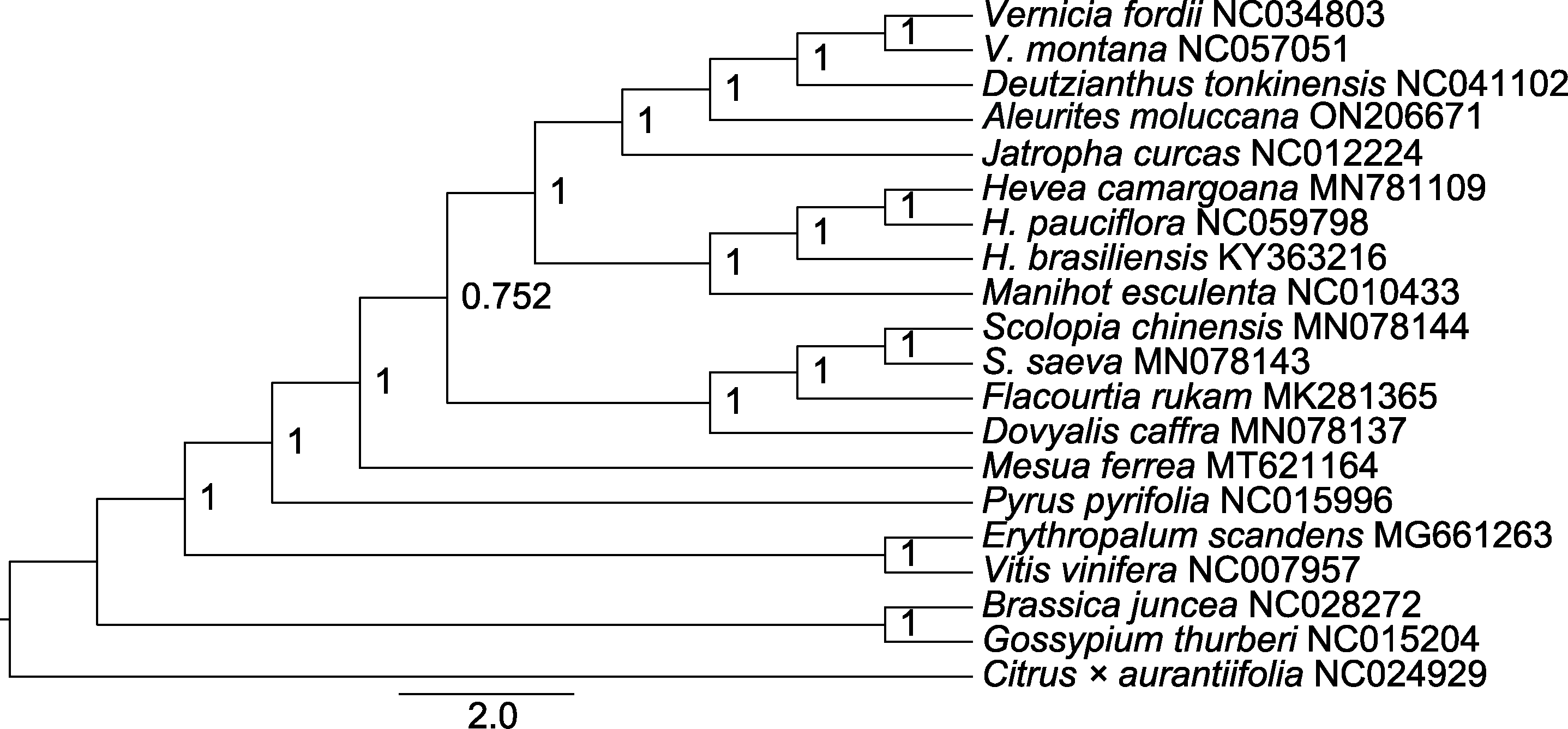
图7 基于蛋白编码序列构建的石栗与其它19个物种贝叶斯系统发育树 分支上的数值为后验概率。
Figure 7 Bayesian phylogenetic tree of Aleurites moluccana and other 19 species based on protein coding sequence The values on the branch are posteriori probability.
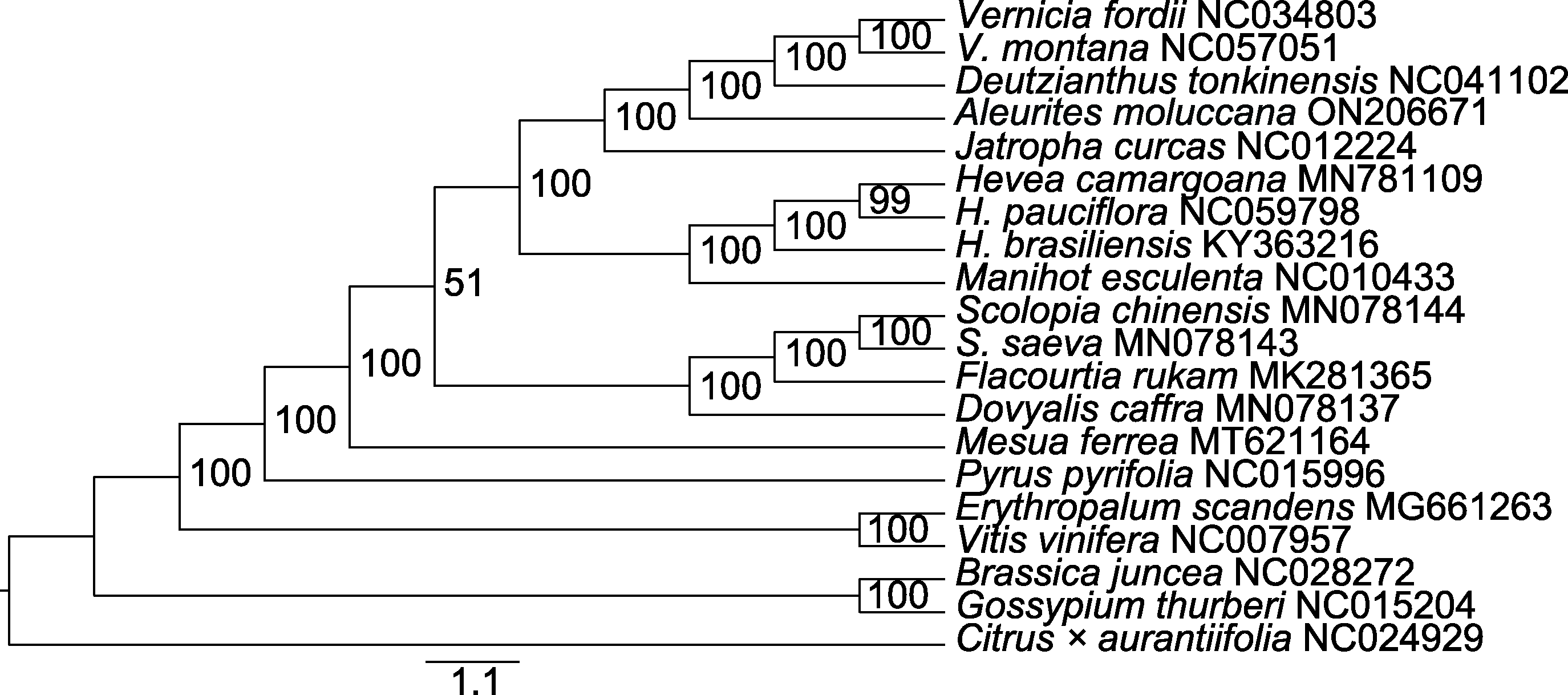
图8 基于蛋白编码序列构建的石栗与其它19个物种最大似然(ML)系统发育树 分支上的数值为后验概率。
Figure 8 Maximum likelihood (ML) phylogenetic tree of Aleurites moluccana and other 19 species based on protein coding sequence The values on the branch are posteriori probability.
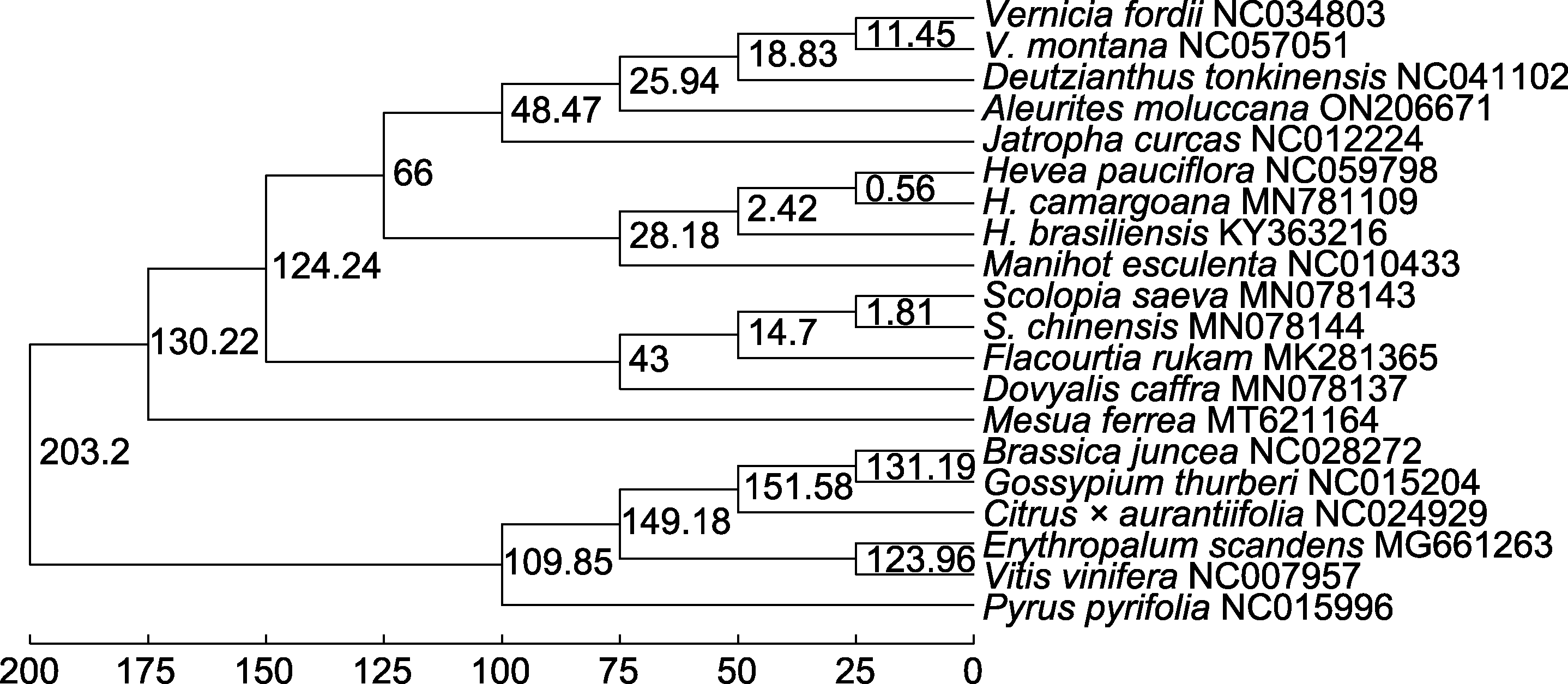
图9 基于宽松分子钟模型构建的石栗与其它19个物种系统发育时间树 分支上的数值为分化时间(单位: 百万年前)。
Figure 9 Phylogenetic dating tree of Aleurites moluccana and other 19 species based on relax molecular clock model The values on the branch are the differentiation time (unit: million years ago).
| [1] | 蔡金标, 丁建祖, 陈必勇 (1997). 中国油桐品种、类型的分类. 经济林研究 15(4), 47-50. |
| [2] | 曹晖, 肖艳华, 王绍云 (2007). 石栗属和油桐属的化学成分和生物活性. 凯里学院学报 25(006), 43-45. |
| [3] | 陈琴怡 (2017). 两种五加科植物的叶绿体全基因组研究及其系统发育分析. 硕士论文. 杭州: 浙江大学. pp. 22-34. |
| [4] |
李巧丽, 延娜, 宋琼, 郭军战 (2018). 鲁桑叶绿体基因组序列及特征分析. 植物学报 53, 94-103.
DOI |
| [5] | 梁文汇, 李开祥, 邓力, 曾祥艳, 邓福春 (2011). 广西生物柴油原料树种石栗的综合评价. 广西林业科学 40, 333-335. |
| [6] | 凌建群, 张新英, 陈耀堂 (1995). 油桐、千年桐和石栗的木材比较解剖. 北京大学学报(自然科学版) 31, 745-751. |
| [7] | 刘昌盛, 黄凤洪, 李重屹, 王明霞, 南占东, 韩伟, 廖李 (2008). 木本油料石栗的初步研究. 中国油料作物学报 30, 106-107, 111. |
| [8] | 罗群凤, 冯源恒, 贾婕, 陈虎, 杨章旗 (2018). 马尾松叶绿体基因组测序及特征分析. 广西林业科学 47, 7. |
| [9] | 苏梦云, 周国璋 (1988). 油桐属与石栗属叶绿体的核酸、蛋白质及超微结构的初步研究. 林业科学研究 1, 424-427. |
| [10] | 孙雨晴 (2018). 四种葱蒜类蔬菜叶绿体DNA提取优化及比较基因组学研究. 硕士论文. 长春: 吉林农业大学. pp. 36-38. |
| [11] | 王劲风, 方嘉兴, 刘兴温, 周国璋, 苏梦云, 成小飞 (1986). 油桐属种分类及其品种类型鉴别方法的探讨. 全国林木遗传育种第五次学术报告会论文汇编. pp. 119-121. |
| [12] | 王磊 (2013). 石栗种子内含物变化及accD基因的克隆研究. 硕士论文. 南宁: 广西大学. pp. 45-68. |
| [13] |
谢海坤, 焦健, 樊秀彩, 张颖, 姜建福, 孙海生, 刘崇怀 (2017). 基于高通量测序组装赤霞珠叶绿体基因组及其特征分析. 中国农业科学 50, 1655-1665.
DOI |
| [14] |
杨亚蒙, 焦健, 樊秀彩, 张颖, 姜建福, 李民, 刘崇怀 (2019). 桑叶葡萄叶绿体基因组及其特征分析. 园艺学报 46, 635-648.
DOI |
| [15] | 杨艳婷 (2018). 羊草叶绿体全基因组分析及分子标记开发. 硕士论文. 扬州: 扬州大学. pp. 55-65. |
| [16] |
赵月梅, 杨振艳, 赵永平, 李筱玲, 赵志新, 赵桂仿 (2019). 木犀科植物叶绿体基因组结构特征和系统发育关系. 植物学报 54, 441-454.
DOI |
| [17] | 周会, 荆胜利, 李刚, 张磊, 覃瑞, 刘虹 (2014). 叶绿体基因组分析在植物系统发育中的应用. 植物学研究 3, 1-9. |
| [18] |
Cesca TG, Faqueti LG, Rocha LW, Meira NA, Meyre-Silva C, De Souza MM, Quintão NLM, Silva RML, Filho VC, Bresolin TMB (2012). Antinociceptive, anti-inflammatory and wound healing features in animal models treated with a semisolid herbal medicine based on Aleurites moluccana L. Willd. Euforbiaceae standardized leaf extract: semisolid herbal. J Ethnopharmacol 143, 355-362.
DOI PMID |
| [19] |
Daniell H, Lin CS, Yu M, Chang WJ (2016). Chloroplast genomes: diversity, evolution, and applications in genetic engineering. Genome Biol 17, 134.
DOI PMID |
| [20] |
Fukuda Y, Tomita M, Washio T (1999). Comparative study of overlapping genes in the genomes of Mycoplasma gen- italium and Mycoplasma pneumoniae. Nucleic Acids Res 27, 1847-1853.
PMID |
| [21] |
Jin JJ, Yu WB, Yang JB, Song Y, dePamphilis CW, Yi TS, Li DZ (2020). GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol 21, 241.
DOI |
| [22] |
Krause K (2008). From chloroplasts to “cryptic” plastids: evolution of plastid genomes in parasitic plants. Curr Genet 54, 111-121.
DOI URL |
| [23] | Li PR, Zhang SJ, Li F, Zhang SF, Zhang H, Wang XW, Sun RF, Bonnema G, Borm TJA (2017a). A phylogenetic analysis of chloroplast genomes elucidates the relationships of the six economically important Brassica species comprising the triangle of U. Front Plant Sci 8, 111. |
| [24] |
Li Z, Long HX, Zhang L, Liu ZM, Cao HP, Shi MW, Tan XF (2017b). The complete chloroplast genome sequence of tung tree (Vernicia fordii): organization and phylogenetic relationships with other angiosperms. Sci Rep 7, 1869.
DOI |
| [25] | Liu L, Hao ZZ, Liu YY, Wei XX, Cun YZ, Wang XQ (2014). Phylogeography of Pinus armandii and its relatives: heterogeneous contributions of geography and climate changes to the genetic differentiation and diversification of Chinese white pines. PLoS One 9, e85920. |
| [26] | Martin G, Baurens FC, Cardi C, Aury JM, D’Hont A (2013). The complete chloroplast genome of banana (Musa acuminata, Zingiberales): insight into plastid monocotyledon evolution. PLoS One 8, e67350. |
| [27] | Nie XJ, Lv SZ, Zhang YX, Du XH, Wang L, Biradar SS, Tan XF, Wan FH, Song WN (2012). Complete chloroplast genome sequence of a major invasive species, crofton weed (Ageratina adenophora). PLoS One 7, e36869. |
| [28] |
Niu YF, Hu YS, Zheng C, Liu ZY, Liu J (2020). The complete chloroplast genome of Hevea camargoana. Mitochondrial DNA Part B 5, 607-608.
DOI URL |
| [29] |
Quintão NLM, Pastor MVD, de-Souza Antonialli C, da Silva GF, Rocha LW, Berté TE, de Souza MM, Meyre- Silva C, Lucinda-Silva RM, Bresolin TMB, Filho VC (2019). Aleurites moluccanus and its main active constituent, the flavonoid 2″-O-rhamnosylswertisin, in experimental model of rheumatoid arthritis. J Ethnopharmacol 235, 248-254.
DOI PMID |
| [30] |
Radunz A, He P, Schmid GH (1998). Analysis of the seed lipids of Aleurites montana. Z Naturforsch C 53, 305-310.
DOI URL |
| [31] |
Reback RG, Kapgate DK, Wurdack K, Manchester SR (2022). Fruits of euphorbiaceae from the late cretaceous Deccan intertrappean beds of India. Int J Plant Sci 183, 128-138.
DOI URL |
| [32] | Shaw J, Lickey EB, Schilling EE, Small RL (2007). Comparison of whole chloroplast genome sequences to choose noncoding regions for phylogenetic studies in angiosperms: the tortoise and the hare III. Am J Bot 94, 275-288. |
| [33] |
Song Y, Dong WP, Liu B, Xu C, Yao X, Gao J, Corlett RT (2015). Comparative analysis of complete chloroplast genome sequences of two tropical trees Machilus yunnanensis and Machilus balansae in the family Lauraceae. Front Plant Sci 6, 662.
DOI PMID |
| [34] |
Villarante NR, Davila RAE, Sumalapao DEP (2018). Removal of lead (ΙΙ) by Lumbang, Aleurites moluccana activated carbon carboxymethylcellulose composite crosslinked with epichlorohydrin. Orient J Chem 34, 693-703.
DOI URL |
| [35] |
Villarante NR, Ibarrientos CH (2021). Physicochemical characterization of candlenut (Aleurites moluccana)-derived biodiesel purified with deed eutectic solvents. J Oleo Sci 70, 113-123.
DOI PMID |
| [36] |
Wang YL, Jian X, Wang S (2022). Characterization of the complete chloroplast genome of Rupr. (Euphorbiaceae). Mitochondrial DNA Part B 7, 1550-1552.
DOI URL |
| [37] | Zhang QY, Chen X, Guo MB, Guo R, Xu YP, Yang M, Guo HY (2017). Screening and development of chloroplast polymorphic molecular markers on wild hemp (Cannabis sativa L.). Mol Plant Breed 15, 979-985. |
| [1] | 王传永, 庄典, 宋正达, 翟恒华, 李乃伟, 张凡. 黑果腺肋花楸叶绿体全基因组的结构和比较分析及系统进化推断[J]. 植物学报, 2025, 60(4): 1-0. |
| [2] | 孙亚君. 何谓高等或低等生物——澄清《物种起源》所蕴含的生物等级性的涵义及其成立性[J]. 生物多样性, 2025, 33(1): 24394-. |
| [3] | 艾妍雨, 胡海霞, 沈婷, 莫雨轩, 杞金华, 宋亮. 附生维管植物多样性及其与宿主特征的相关性: 以哀牢山中山湿性常绿阔叶林为例[J]. 生物多样性, 2024, 32(5): 24072-. |
| [4] | 吕燕文, 王子韵, 肖钰, 何梓晗, 吴超, 胡新生. 谱系分选理论与检测方法的研究进展[J]. 生物多样性, 2024, 32(4): 23400-. |
| [5] | 曹可欣, 王敬雯, 郑国, 武鹏峰, 李英滨, 崔淑艳. 降水格局改变及氮沉降对北方典型草原土壤线虫多样性的影响[J]. 生物多样性, 2024, 32(3): 23491-. |
| [6] | 王斌, 钟艺倩, 杨美雪, 吴淼锐, 王艳萍, 陆芳, 陶旺兰, 李健星, 赵弘明, 刘晟源, 向悟生, 李先琨. 喀斯特季节性雨林优势树种叶片非结构性碳水化合物空间变异及生态驱动因素[J]. 生物多样性, 2024, 32(12): 24325-. |
| [7] | 杨向林, 赵彩云, 李俊生, 种方方, 李文金. 植物入侵导致群落谱系结构更加聚集: 以广西国家级自然保护区草本植物为例[J]. 生物多样性, 2024, 32(11): 24175-. |
| [8] | 王振宇, 黄志群. 亚热带27种木本植物叶片性状对植食作用的影响: 验证生长-防御权衡假说[J]. 植物生态学报, 2024, 48(11): 1501-1509. |
| [9] | 何林君, 杨文静, 石宇豪, 阿说克者莫, 范钰, 王国严, 李景吉, 石松林, 易桂花, 彭培好. 火烧干扰下植物群落系统发育和功能多样性对紫茎泽兰入侵的影响[J]. 生物多样性, 2024, 32(11): 24269-. |
| [10] | 李庆多, 栗冬梅. 全球蝙蝠巴尔通体流行状况分析[J]. 生物多样性, 2023, 31(9): 23166-. |
| [11] | 褚振州, 古丽巴哈尔·依斯拉木, 屈泽众, 田新民. 同域分布的3种木蓼属植物叶绿体基因组比较[J]. 植物学报, 2023, 58(3): 417-432. |
| [12] | 宋会银, 胡征宇, 刘国祥. 绿藻门小球藻科的分类学研究进展[J]. 生物多样性, 2023, 31(2): 22083-. |
| [13] | 李治中, 彭帅, 王青锋, 李伟, 梁士楚, 陈进明. 中国海菜花属植物隐种多样性[J]. 生物多样性, 2023, 31(2): 22394-. |
| [14] | 朱瑞良, 马晓英, 曹畅, 曹子寅. 中国苔藓植物多样性研究进展[J]. 生物多样性, 2022, 30(7): 22378-. |
| [15] | 王婷, 舒江平, 顾钰峰, 李艳清, 杨拓, 徐洲锋, 向建英, 张宪春, 严岳鸿. 中国石松类和蕨类植物多样性研究进展[J]. 生物多样性, 2022, 30(7): 22381-. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||