

植物学报 ›› 2018, Vol. 53 ›› Issue (1): 94-103.DOI: 10.11983/CBB16247 cstr: 32102.14.CBB16247
收稿日期:2016-12-13
接受日期:2017-03-29
出版日期:2018-01-01
发布日期:2018-08-10
通讯作者:
郭军战
基金资助:
Qiaoli Li, Na Yan, Qiong Song, Junzhan Guo*( )
)
Received:2016-12-13
Accepted:2017-03-29
Online:2018-01-01
Published:2018-08-10
Contact:
Junzhan Guo
摘要: 鲁桑(Morus multicaulis)是亚洲地区栽培的重要经济作物。以鲁桑品种日本胡橙为实验材料, 利用高通量测序技术对鲁桑叶绿体基因组进行测序, 获得NCBI登录号(KU355297), 并研究鲁桑的叶绿体基因组结构。结合前人对蒙桑(M. mongolica)、印度桑(M. indica)和川桑(M. notabilis)的研究结果, 对鲁桑的系统进化关系进行了探讨。研究结果表明: 鲁桑叶绿体基因组是一个典型的四部分结构, 全长159 154 bp, 共注释130个基因, 包含85个蛋白质编码基因(18个基因在反向重复区重复)、37个转运RNA (tRNA)基因和8个核糖体RNA (rRNA)基因。生物信息学分析表明, 在鲁桑中共搜索到82个SSR位点, 单核苷酸、二核苷酸、三核苷酸、四核苷酸和五核苷酸重复基序个数分别为63、7、2、9和1个, 并没有发现六核苷酸; 其中单核苷酸重复在鲁桑的叶绿体基因组SSR中占76.8%。采用MEGA 6.0软件, 通过最大似然法和近邻结合法对包括4个桑属物种在内的15个物种的叶绿体基因组序列进行聚类分析, 2种方法得到的聚类结果均为鲁桑和蒙桑聚在一起。研究结果对叶绿体基因组工程研究及桑属种间的分子标记开发和优良品种培育具有一定的参考价值。
李巧丽, 延娜, 宋琼, 郭军战. 鲁桑叶绿体基因组序列及特征分析. 植物学报, 2018, 53(1): 94-103.
Qiaoli Li, Na Yan, Qiong Song, Junzhan Guo. Complete Chloroplast Genome Sequence and Characteristics Analysis of Morus multicaulis. Chinese Bulletin of Botany, 2018, 53(1): 94-103.
| Genome feature | Morus indica | M. mongolica | M. notabilis | M. multicaulis |
|---|---|---|---|---|
| Genome size (bp) | 158484 | 158459 | 158680 | 159154 |
| LSC length (bp)/percent (%)/GC content (%) | 87386/55.14/34.1 | 87367/55.14/34.0 | 87470/55.12/34.1 | 87763/55.15/33.9 |
| SSC length (bp)/percent (%)/GC content (%) | 19742/12.46/29.4 | 19736/12.45/29.3 | 19776/12.46/29.3 | 20035/12.59/29.3 |
| IR length (bp)/percent (%)/GC content (%) | 25678/16.20/42.9 | 25678/16.20/42.9 | 25717/16.21/42.9 | 25678/16.13/42.9 |
| GC content (%) | 36.4 | 36.3 | 36.4 | 36.2 |
| Number of genes | 133 | 133 | 129 | 130 |
| Number of protein-coding genes | 88 | 88 | 84 | 85 |
表1 4种桑属植物叶绿体基因组基本特征比较
Table 1 Comparison of chloroplast genomes among four species of Morus
| Genome feature | Morus indica | M. mongolica | M. notabilis | M. multicaulis |
|---|---|---|---|---|
| Genome size (bp) | 158484 | 158459 | 158680 | 159154 |
| LSC length (bp)/percent (%)/GC content (%) | 87386/55.14/34.1 | 87367/55.14/34.0 | 87470/55.12/34.1 | 87763/55.15/33.9 |
| SSC length (bp)/percent (%)/GC content (%) | 19742/12.46/29.4 | 19736/12.45/29.3 | 19776/12.46/29.3 | 20035/12.59/29.3 |
| IR length (bp)/percent (%)/GC content (%) | 25678/16.20/42.9 | 25678/16.20/42.9 | 25717/16.21/42.9 | 25678/16.13/42.9 |
| GC content (%) | 36.4 | 36.3 | 36.4 | 36.2 |
| Number of genes | 133 | 133 | 129 | 130 |
| Number of protein-coding genes | 88 | 88 | 84 | 85 |
| Function | Gene group | Gene name | |||
|---|---|---|---|---|---|
| Self-replication | Ribosomal RNA genes | rrn4 | rrn5 | rrn16 | rrn23 |
| Transfer RNA genes | trnA-UGC trnF-GAA trnH-GUG trnL-CAA trnN-GUU trnR-UCU trnT-GGU trnW-CCA | trnC-GCA trnfM-CAU trnI-CAU trnL-UAA trnP-UGG trnS-GCU trnT-UGU trnY-GUA | trnD-GUC trnG-GCC trnI-GAU trnL-UAG trnQ-UUG trnS-GGA trnV-GAC | trnE-UUC trnG-UCC trnK-UUU trnM-CAU trnR-ACG trnS-UGA trnV-UAC | |
| Small subunit of ribosome | rps2 rps8 rps15 | rps3 rps11 rps16* | rps4 rps12 rps18 | rps7 rps14 rps19 | |
| Lange subunit of ribosome | rpl2* rpl22 rpl36 | rpl14 rpl23 | rpl16* rpl32 | rpl20 rpl33 | |
| RNA polymerase subunits | rpoA | rpoB | rpoC1* | rpoC2 | |
| NADH dehydrogenase | ndhA* ndhE ndhI | ndhB* ndhF ndhJ | ndhC ndhG ndhK | ndhD ndhH | |
| Photosynthesis | Photosystem I | psaA psaJ | psaB | psaC | psaI |
| Photosystem II | psbA psbE psbJ psbN | psbB psbF psbK psbT | psbC psbH psbL psbZ | psbD psbI psbM | |
| Cytochrome b/f complex | petA petL | petB* petN | petD* | petG | |
| ATP synthase | atpA atpH | atpB atpI | atpE | atpF* | |
| ATP Protease | rbcl | ||||
| Large subunit of rubisco | matK | ||||
| Maturase | clpP* | ||||
| Envelope membrane protein | cemA | ||||
| Other genes | Subunit of acetyl-CoA-carboxylase | accD | |||
| C-type cytochrome synthesis | ccsA | ||||
| Unknown function | Hypothetical chloroplast reading frames | yf1 | ycf3* | ycf4 | ycf15 |
| ORFs | ycf2 ycf68* | ||||
表2 鲁桑叶绿体基因组注释基因信息
Table 2 Genes present in the chloroplast genome of Morus multicaulis
| Function | Gene group | Gene name | |||
|---|---|---|---|---|---|
| Self-replication | Ribosomal RNA genes | rrn4 | rrn5 | rrn16 | rrn23 |
| Transfer RNA genes | trnA-UGC trnF-GAA trnH-GUG trnL-CAA trnN-GUU trnR-UCU trnT-GGU trnW-CCA | trnC-GCA trnfM-CAU trnI-CAU trnL-UAA trnP-UGG trnS-GCU trnT-UGU trnY-GUA | trnD-GUC trnG-GCC trnI-GAU trnL-UAG trnQ-UUG trnS-GGA trnV-GAC | trnE-UUC trnG-UCC trnK-UUU trnM-CAU trnR-ACG trnS-UGA trnV-UAC | |
| Small subunit of ribosome | rps2 rps8 rps15 | rps3 rps11 rps16* | rps4 rps12 rps18 | rps7 rps14 rps19 | |
| Lange subunit of ribosome | rpl2* rpl22 rpl36 | rpl14 rpl23 | rpl16* rpl32 | rpl20 rpl33 | |
| RNA polymerase subunits | rpoA | rpoB | rpoC1* | rpoC2 | |
| NADH dehydrogenase | ndhA* ndhE ndhI | ndhB* ndhF ndhJ | ndhC ndhG ndhK | ndhD ndhH | |
| Photosynthesis | Photosystem I | psaA psaJ | psaB | psaC | psaI |
| Photosystem II | psbA psbE psbJ psbN | psbB psbF psbK psbT | psbC psbH psbL psbZ | psbD psbI psbM | |
| Cytochrome b/f complex | petA petL | petB* petN | petD* | petG | |
| ATP synthase | atpA atpH | atpB atpI | atpE | atpF* | |
| ATP Protease | rbcl | ||||
| Large subunit of rubisco | matK | ||||
| Maturase | clpP* | ||||
| Envelope membrane protein | cemA | ||||
| Other genes | Subunit of acetyl-CoA-carboxylase | accD | |||
| C-type cytochrome synthesis | ccsA | ||||
| Unknown function | Hypothetical chloroplast reading frames | yf1 | ycf3* | ycf4 | ycf15 |
| ORFs | ycf2 ycf68* | ||||
| Codon | Amino acid | Number | Codon | Amino acid | Number |
|---|---|---|---|---|---|
| GGG | Gly(G) | 494 | TGG | Trp(W) | 684 |
| GGA | Gly(G) | 759 | TGA | Stop | 1032 |
| GGT | Gly(G) | 599 | TGT | Cys(C) | 725 |
| GGC | Gly(G) | 350 | TGC | Cys(C) | 435 |
| GAG | Glu(E) | 550 | TAG | Stop | 786 |
| GAA | Glu(E) | 1368 | TAA | Stop | 1306 |
| GAT | Asp(D) | 1064 | TAT | Try(Y) | 1624 |
| GAC | Asp(D) | 425 | TAC | Try(Y) | 690 |
| GTG | Val(V) | 418 | TTG | Leu(L) | 1073 |
| GTA | Val(V) | 728 | TTA | Leu(L) | 1250 |
| GTT | Val(V) | 792 | TTT | Phe(F) | 2343 |
| GTC | Val(V) | 430 | TTC | Phe(F) | 1471 |
| GCG | Ala(A) | 249 | TCG | Ser(S) | 578 |
| GCA | Ala(A) | 430 | TCA | Ser(S) | 979 |
| GCT | Ala(A) | 511 | TCT | Ser(S) | 1273 |
| GCC | Ala(A) | 321 | TCC | Ser(S) | 864 |
| AGG | Arg(R) | 596 | CGG | Arg(R) | 350 |
| AGA | Arg(R) | 1044 | CGA | Arg(R) | 596 |
| AGT | Ser(S) | 718 | CGT | Arg(R) | 363 |
| AGC | Ser(S) | 478 | CGC | Arg(R) | 236 |
| AAG | Lys(K) | 1039 | CAG | Gln(Q) | 440 |
| AAA | Lys(K) | 2280 | CAA | Gln(Q) | 1013 |
| AAT | Asn(N) | 1883 | CAT | His(H) | 945 |
| AAC | Asn(N) | 728 | CAC | His(H) | 362 |
| ATG | Met(M) | 855 | CTG | Leu(L) | 489 |
| ATA | Ile(I) | 1729 | CTA | Leu(L) | 799 |
| ATT | Ile(I) | 1965 | CTT | Leu(L) | 1065 |
| ATC | Ile(I) | 1083 | CTC | Leu(L) | 581 |
| ACG | Thr(T) | 399 | CCG | Pro(P) | 400 |
| ACA | Thr(T) | 689 | CCA | Pro(P) | 738 |
| ACT | Thr(T) | 690 | CCT | Pro(P) | 730 |
| ACC | Thr(T) | 587 | CCC | Pro(P) | 580 |
表3 鲁桑密码子信息
Table 3 Codon usage in Morus multicaulis
| Codon | Amino acid | Number | Codon | Amino acid | Number |
|---|---|---|---|---|---|
| GGG | Gly(G) | 494 | TGG | Trp(W) | 684 |
| GGA | Gly(G) | 759 | TGA | Stop | 1032 |
| GGT | Gly(G) | 599 | TGT | Cys(C) | 725 |
| GGC | Gly(G) | 350 | TGC | Cys(C) | 435 |
| GAG | Glu(E) | 550 | TAG | Stop | 786 |
| GAA | Glu(E) | 1368 | TAA | Stop | 1306 |
| GAT | Asp(D) | 1064 | TAT | Try(Y) | 1624 |
| GAC | Asp(D) | 425 | TAC | Try(Y) | 690 |
| GTG | Val(V) | 418 | TTG | Leu(L) | 1073 |
| GTA | Val(V) | 728 | TTA | Leu(L) | 1250 |
| GTT | Val(V) | 792 | TTT | Phe(F) | 2343 |
| GTC | Val(V) | 430 | TTC | Phe(F) | 1471 |
| GCG | Ala(A) | 249 | TCG | Ser(S) | 578 |
| GCA | Ala(A) | 430 | TCA | Ser(S) | 979 |
| GCT | Ala(A) | 511 | TCT | Ser(S) | 1273 |
| GCC | Ala(A) | 321 | TCC | Ser(S) | 864 |
| AGG | Arg(R) | 596 | CGG | Arg(R) | 350 |
| AGA | Arg(R) | 1044 | CGA | Arg(R) | 596 |
| AGT | Ser(S) | 718 | CGT | Arg(R) | 363 |
| AGC | Ser(S) | 478 | CGC | Arg(R) | 236 |
| AAG | Lys(K) | 1039 | CAG | Gln(Q) | 440 |
| AAA | Lys(K) | 2280 | CAA | Gln(Q) | 1013 |
| AAT | Asn(N) | 1883 | CAT | His(H) | 945 |
| AAC | Asn(N) | 728 | CAC | His(H) | 362 |
| ATG | Met(M) | 855 | CTG | Leu(L) | 489 |
| ATA | Ile(I) | 1729 | CTA | Leu(L) | 799 |
| ATT | Ile(I) | 1965 | CTT | Leu(L) | 1065 |
| ATC | Ile(I) | 1083 | CTC | Leu(L) | 581 |
| ACG | Thr(T) | 399 | CCG | Pro(P) | 400 |
| ACA | Thr(T) | 689 | CCA | Pro(P) | 738 |
| ACT | Thr(T) | 690 | CCT | Pro(P) | 730 |
| ACC | Thr(T) | 587 | CCC | Pro(P) | 580 |
| Length (bp) | Number | Morus multicaulis | M. mongolica |
|---|---|---|---|
| A10 | 10 | 2142, 3980, 5079, 5977, 29067, 49740, 68616, 68631, 114154 (ndhF), 116262 | 3760, 4859, 28847, 38118, 113758 (ndhF), 115866 |
| A11 | 3 | 9589, 62837, 87467 | 1921, 5757, 9371, 62504, 81011 |
| A12 | 3 | 4830, 53982, 85376 | 13368, 38142, 53676, 84178, 87070 |
| A13 | 1 | 13596 | 4609, 73766 |
| A14 | 1 | 128163 | 127468 |
| A15 | 1 | 74160 | |
| A16 | 1 | 8990 | |
| A17 | 8772 | ||
| T10 | 20 | 66, 5258, 8582, 9802, 14098, 14919, 24357, 30672, 30938, 54024, 57098 (atpB), 62610, 66927, 68743, 70892, 73958, 83130, 116784, 130487 (ycf1), 132244 (ycf1) | 5038, 7036, 9584, 24137, 30452, 30718, 53718, 56773 (atpB), 62277, 70506, 73564, 82753, 116369, 121665, 129792 (ycf1), 131549 (ycf1) |
| T11 | 6 | 513, 34264, 69552, 78684, 122351, 131346 (ycf1) | 293, 8363, 57218, 59233, 66594, 68126, 69166, 74280, 78285, 130651 (ycf1) |
| T12 | 5 | 27617 (rpoB), 57549, 59565, 72471, 85809 | 12476, 13966, 27397 (rpoB), 34035, 85411 |
| T13 | 5 | 12703, 13286, 68491, 81352, 128585 | 8996, 13058, 51524, 72085, 127890 |
| T14 | 5 | 9213, 51829, 63865, 74676, 86927 | 63532, 80953 |
| T16 | 49162, 86528 | ||
| T17 | 1 | 49475 | |
| T19 | 1 | 116631 | 116235 |
| AT5 | 1 | 11566 (ndhF) | 115270 (ndhF) |
| AT6 | 2 | 118643, 118871 | 10589, 49643 |
| TA6 | 2 | 5522, 21234 (rpoC2) | 5302, 21009 (rpoC2), 118243 |
| TC5 | 1 | 645927 (cemA) | 64259 (cemA) |
| TTC4 | 1 | 70909 | 70523 |
| AAT4 | 1 | 128565 | 127870 |
| ATTT3 | 1 | 62140 | |
| ATTT4 | 1 | 14187 | 13957, 61807 |
| AAAT3 | 2 | 24056 (rpoC1), 46731 (ycf3) | 23831(rpoC1), 46414 (ycf3) |
| TATT3 | 1 | 24388 (rpoC1) | 24168 (rpoC1) |
| ATTA3 | 2 | 33980, 116443 | 33751, 116047 |
| TCTT3 | 1 | 111575 | 111179 |
| AAAG3 | 1 | 135331 | 134636 |
| AAGGA3 | 1 | 14021 (atpF) | 13792 (atpF) |
| ATTTC3 | 24071 |
表4 鲁桑和蒙桑中简单重复序列(SSR)位点对比
Table 4 Comparison of simple sequence repeats (SSR) loci in Morus multicaulis and M. mongolica
| Length (bp) | Number | Morus multicaulis | M. mongolica |
|---|---|---|---|
| A10 | 10 | 2142, 3980, 5079, 5977, 29067, 49740, 68616, 68631, 114154 (ndhF), 116262 | 3760, 4859, 28847, 38118, 113758 (ndhF), 115866 |
| A11 | 3 | 9589, 62837, 87467 | 1921, 5757, 9371, 62504, 81011 |
| A12 | 3 | 4830, 53982, 85376 | 13368, 38142, 53676, 84178, 87070 |
| A13 | 1 | 13596 | 4609, 73766 |
| A14 | 1 | 128163 | 127468 |
| A15 | 1 | 74160 | |
| A16 | 1 | 8990 | |
| A17 | 8772 | ||
| T10 | 20 | 66, 5258, 8582, 9802, 14098, 14919, 24357, 30672, 30938, 54024, 57098 (atpB), 62610, 66927, 68743, 70892, 73958, 83130, 116784, 130487 (ycf1), 132244 (ycf1) | 5038, 7036, 9584, 24137, 30452, 30718, 53718, 56773 (atpB), 62277, 70506, 73564, 82753, 116369, 121665, 129792 (ycf1), 131549 (ycf1) |
| T11 | 6 | 513, 34264, 69552, 78684, 122351, 131346 (ycf1) | 293, 8363, 57218, 59233, 66594, 68126, 69166, 74280, 78285, 130651 (ycf1) |
| T12 | 5 | 27617 (rpoB), 57549, 59565, 72471, 85809 | 12476, 13966, 27397 (rpoB), 34035, 85411 |
| T13 | 5 | 12703, 13286, 68491, 81352, 128585 | 8996, 13058, 51524, 72085, 127890 |
| T14 | 5 | 9213, 51829, 63865, 74676, 86927 | 63532, 80953 |
| T16 | 49162, 86528 | ||
| T17 | 1 | 49475 | |
| T19 | 1 | 116631 | 116235 |
| AT5 | 1 | 11566 (ndhF) | 115270 (ndhF) |
| AT6 | 2 | 118643, 118871 | 10589, 49643 |
| TA6 | 2 | 5522, 21234 (rpoC2) | 5302, 21009 (rpoC2), 118243 |
| TC5 | 1 | 645927 (cemA) | 64259 (cemA) |
| TTC4 | 1 | 70909 | 70523 |
| AAT4 | 1 | 128565 | 127870 |
| ATTT3 | 1 | 62140 | |
| ATTT4 | 1 | 14187 | 13957, 61807 |
| AAAT3 | 2 | 24056 (rpoC1), 46731 (ycf3) | 23831(rpoC1), 46414 (ycf3) |
| TATT3 | 1 | 24388 (rpoC1) | 24168 (rpoC1) |
| ATTA3 | 2 | 33980, 116443 | 33751, 116047 |
| TCTT3 | 1 | 111575 | 111179 |
| AAAG3 | 1 | 135331 | 134636 |
| AAGGA3 | 1 | 14021 (atpF) | 13792 (atpF) |
| ATTTC3 | 24071 |
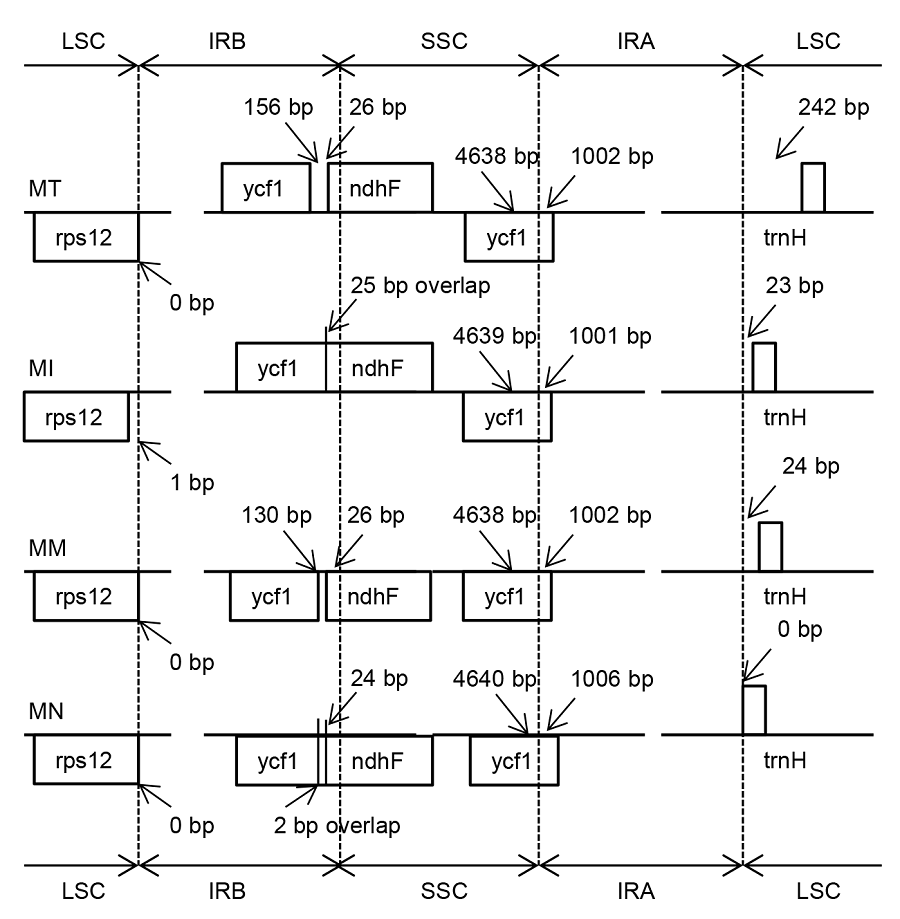
图2 4个桑属物种叶绿体基因组反向重复区(IR)、大单拷贝区(LSC)和小单拷贝区(SSC)边界比对MT: 鲁桑, MI: 印度桑; MM: 蒙桑; MN: 川桑
Figure 2 Comparison of the junction between inverted repeat region (IR), large single copy-region (LSC) and small single copy- region (SSC) of chloroplast genome among four Morus species MT: Morus multicaulis; MI: M. indica; MM: M. mongolica; MN: M. notabilis
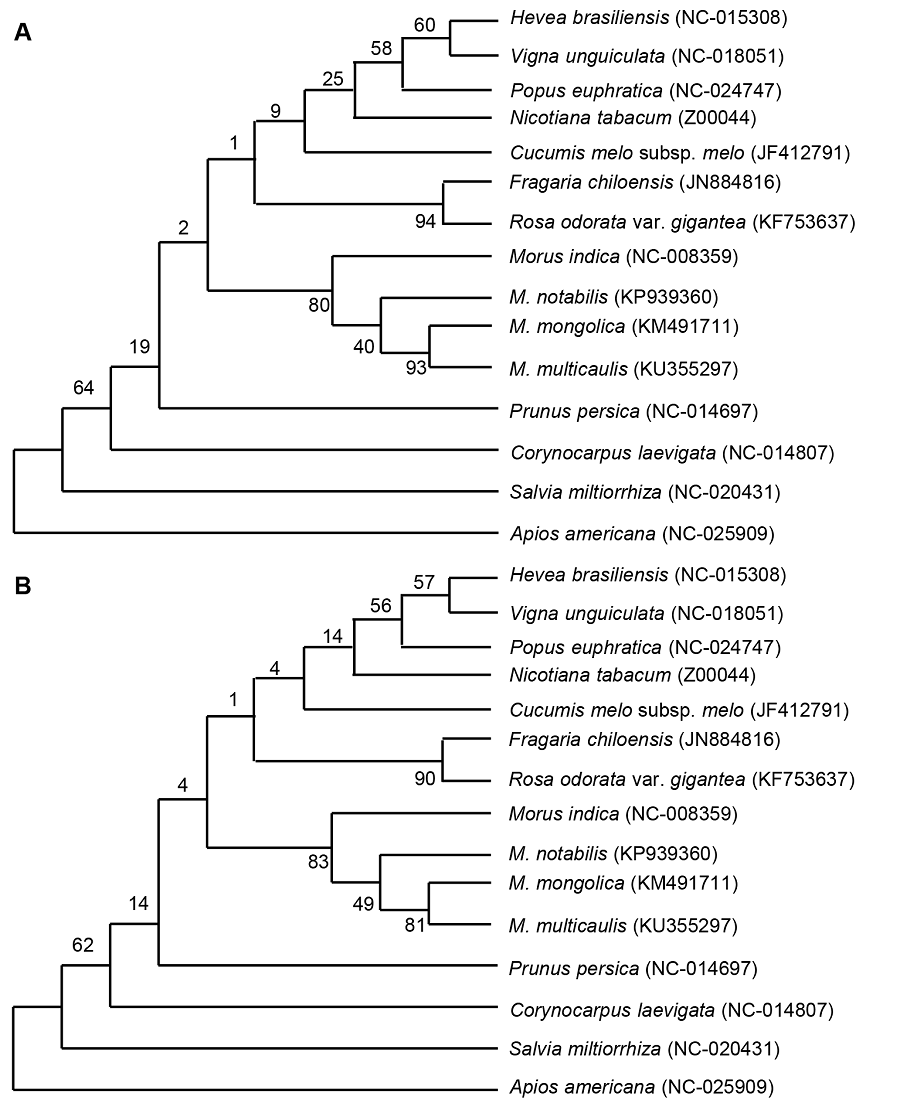
图3 基于叶绿体全基因组的桑属4个物种及其近缘种的最大似然法(ML) (A)和近邻结合法(NJ) (B)聚类结果
Figure 3 Cluster analysis of four species of Morus using complete chloroplast genome sequence by the maximum likelihood (ML) method (A) and neighbor-joining (NL) method (B)
| [1] | 冯丽春, 杨光伟, 余茂德, 张孝勇, 向怀祥 (1997). 利用RAPD对桑属植物种间亲缘关系的研究. 中国农业科学 30, 52-56. |
| [2] | 黄瑶, 李朝銮, 马诚, 吴乃虎 (1994). 叶绿体DNA及其在植物系统学研究中的应用. 植物学通报 11(2), 11-25. |
| [3] | 徐军望, 冯德江, 宋贵生, 魏晓丽, 陈蕾, 伍晓丽, 李旭刚, 朱桢 (2003). 水稻EPSP合酶第一内含子增强外源基因的表达. 中国科学(C辑) 33, 224-230. |
| [4] | 闫化学, 于杰 (2010). DNA条形码技术在植物中的研究现状. 植物学报 45, 102-108. |
| [5] | 周德贵, 赵琼一, 付崇允, 李宏, 蔡学飞, 罗达, 周少川 (2008). 新一代测序技术及其对水稻分子设计育种的影响. 分子植物育种 6, 619-630. |
| [6] | Allender CJ, Allainguillaume J, Lynn J, King GJ (2007). Simple sequence repeats reveal uneven distribution of genetic diversity in chloroplast genomes of Brassica ole- racea L. and(n=9) wild relatives. Theor Appl Genet 114, 609-618. |
| [7] | Chen C, Zhou W, Huang Y, Wang ZZ (2015). The complete chloroplast genome sequence of the mulberry Morus notabilis(Moreae). Mitochondrial DNA Part A 27, 2856-2857. |
| [8] | Flannery ML, Mitchell FJG, Coyne S, Kavanagh TA, Burke JI, Salamin N, Dowding P, Hodkinson TR (2006). Plastid genome characterisation in Brassica and Brassicaceae using a new set of nine SSRs. Theor Appl Genet 113, 1221-1231. |
| [9] | George B, Bhatt BS, Awasthi M, George B, Singh AK (2015). Comparative analysis of microsatellites in chloroplast genomes of lower and higher plants.Curr Genet 61, 665-677. |
| [10] | Hebert PD, Ratnasingham S, de Waard JR (2003). Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species. Proc R Soc Lond B 270, S96-S99. |
| [11] | Huang YY, Matzke AJM, Matzke M (2013). Complete sequence and comparative analysis of the chloroplast genome of coconut palm ( Cocos nucifera). PLoS One 8, e74736. |
| [12] | Jansen RK, Raubeson LA, Boore JL, dePamphilis CW, Chumley TW, Haberle RC, Wyman SK, Alverson AJ, Peery R, Herman SJ, Fourcade HM, Kuehl JV, McNeal JR, Leebens-Mack J, Cui LY (2005). Methods for obtaining and analyzing whole chloroplast genome sequen- ces.Methods Enzymol 395, 348-384. |
| [13] | Jiao Y, Jia HM, Li XW, Jia HJ, Chen Z, Wang GY, Chai CY, van de Weg E, Gao ZS (2012). Development of simple sequence repeat (SSR) markers from a genome survey of Chinese Bayberry ( Myrica rubra). BMC Genomics 13, 201. |
| [14] | Katti MV, Ranjekar PK, Gupta VS (2001). Differential distribution of simple sequence repeats in eukaryotic genome sequences.Mol Biol Evol 18, 1161-1167. |
| [15] | Kaundun SS, Matsumoto S (2002). Heterologous nuclear and chloroplast microsatellite amplification and variation in tea, Camellia sinensis. Genome 45, 1041-1048. |
| [16] | Kong WQ, Yang JH (2015). The complete chloroplast genome sequence of Morus mongolica and a comparative analysis within the Fabidae clade. Curr Genet 62, 165-172. |
| [17] | Leigh FJ, Mackay I, Oliveira HR, Gosman NE, Horsnell RA, Jones H, White J, Powell W, Brown TA (2013). Using diversity of the chloroplast genome to examine evolutionary history of wheat species.Genet Resour Crop Evol 60, 1831-1842. |
| [18] | Leseberg CH, Duvall MR (2009). The complete chloroplast genome of Coix lacryma-jobi and a comparative molecular evolutionary analysis of plastomes in cereals. J Mol Evol 69, 311-318. |
| [19] | Nazareno AG, Carlsen M, Lohmann LG (2015). Complete chloroplast genome of Tanaecium tetragonolobum: the first Bignoniaceae plastome. PLoS One 10, e0129930. |
| [20] | Plunkett GM, Downie SR (2000). Expansion and contraction of the chloroplast inverted repeat in Apiaceae subfamily Apioideae.Syst Bot 25, 648-667. |
| [21] | Rajendrakumar P, Biswal AK, Balachandran SM, Srinivasarao K, Sundaram RM (2007). Simple sequence repeats in organellar genomes of rice: frequency and distribution in genic and intergenic regions.Bioinformatics 23, 1-4. |
| [22] | Ravi V, Khurana JP, Tyagi AK, Khurana P (2006). The chloroplast genome of mulberry: complete nucleotide sequence, gene organization and comparative analysis.Tree Genet Genomes 3, 49-59. |
| [23] | Ruhlman TA, Jansen RK (2014). The plastid genomes of flowering plants.Methods Mol Biol 1132, 3-38. |
| [24] | Shaw J, Lickey EB, Schilling EE, Small RL (2007). Comparison of whole chloroplast genome sequences to choose noncoding regions for phylogenetic studies in angiosperms: the tortoise and the hare lll.Am J Bot 94, 275-288. |
| [25] | Temnykh S, DeClerck G, Lukashova A, Lipovich L, Cartinhour S, McCouch S (2001). Computational and experimental analysis of microsatellites in rice ( Oryza sativa L.): frequency, length variation, transposon associations, and genetic marker potential. Genome Res 11, 1441-1452. |
| [26] | Zhang HY, Li C, Miao HM, Xiong SJ (2013). Insights from the complete chloroplast genome into the evolution of Se- samum indicum L. PLoS One 8, e80508. |
| [1] | 王传永, 庄典, 宋正达, 翟恒华, 李乃伟, 张凡. 黑果腺肋花楸叶绿体全基因组的结构和比较分析及系统进化推断[J]. 植物学报, 2025, 60(4): 1-0. |
| [2] | 尚华丹, 张楚晴, 王梅, 裴文娅, 李国宏, 王鸿斌. 中国杨树害虫物种多样性及其地理分布[J]. 生物多样性, 2025, 33(2): 24370-. |
| [3] | 张锋, Richard Dormatey, 刘寅笃, 李成举, 王云姣, 张春利, 张莹, 范又方, 姚攀锋, 毕真真, 刘玉汇, 白江平, 孙超. 耐亚磷酸盐马铃薯的筛选与评价[J]. 植物学报, 2024, 59(4): 544-557. |
| [4] | 吴相獐, 雷富民, 单壹壹, 于晶. 上海城市公园苔藓植物多样性分布格局及其环境影响因子[J]. 生物多样性, 2024, 32(2): 23364-. |
| [5] | 罗正明, 刘晋仙, 张变华, 周妍英, 郝爱华, 杨凯, 柴宝峰. 不同退化阶段亚高山草甸土壤原生生物群落多样性特征及驱动因素[J]. 生物多样性, 2023, 31(8): 23136-. |
| [6] | 张仲富, 王四海, 杨卫, 陈剑. 蒜头果根际细菌群落结构与功能特征对其健康状态的响应[J]. 植物生态学报, 2023, 47(7): 1020-1031. |
| [7] | 毛莹儿, 周秀梅, 王楠, 李秀秀, 尤育克, 白尚斌. 毛竹扩张对杉木林土壤细菌群落的影响[J]. 生物多样性, 2023, 31(6): 22659-. |
| [8] | 白雪, 李玉靖, 景秀清, 赵晓东, 畅莎莎, 荆韬羽, 刘晋汝, 赵鹏宇. 谷子及其根际土壤微生物群落对铬胁迫的响应机制[J]. 植物生态学报, 2023, 47(3): 418-433. |
| [9] | 褚振州, 古丽巴哈尔·依斯拉木, 屈泽众, 田新民. 同域分布的3种木蓼属植物叶绿体基因组比较[J]. 植物学报, 2023, 58(3): 417-432. |
| [10] | 包金波, 丁志杰, 苗浩宇, 李雪丽, 任书贤, 焦若岩, 李浩, 邓茜茜, 李英姿, 田新民. 石栗叶绿体基因组研究[J]. 植物学报, 2023, 58(2): 248-260. |
| [11] | 赵雯, 王丹丹, 热依拉·木民, 黄开钏, 刘顺, 崔宝凯. 阿尔山地区兴安落叶松林土壤微生物群落结构[J]. 生物多样性, 2023, 31(2): 22258-. |
| [12] | 夏凡, 杨婧, 李建, 史洋, 盖立新, 黄文华, 张经纬, 杨南, 高福利, 韩莹莹, 鲍伟东. 北京地区四个豹猫亚种群肠道菌群的组成[J]. 生物多样性, 2022, 30(9): 22103-. |
| [13] | 石水琴, 秦华光, 张静静, 韩钰, 余淏, 彭怡宁, 杨邵, 汪嘉怡, 何光宇, 岂泽华, 吴文杰, 朱星雨, 饶玉春, 穆丹. 濒危植物大别山五针松根际细菌群落特征与功能分析[J]. 植物学报, 2022, 57(4): 457-467. |
| [14] | 孙翌昕, 李英滨, 李玉辉, 李冰, 杜晓芳, 李琪. 高通量测序技术在线虫多样性研究中的应用[J]. 生物多样性, 2022, 30(12): 22266-. |
| [15] | 周楷玲, 赵玉金, 白永飞. 基于Sentinel-2A数据的东北森林植物多样性监测方法研究[J]. 植物生态学报, 2022, 46(10): 1251-1267. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||