

植物学报 ›› 2018, Vol. 53 ›› Issue (1): 104-109.DOI: 10.11983/CBB16216 cstr: 32102.14.CBB16216
收稿日期:2016-11-11
接受日期:2017-04-17
出版日期:2018-01-01
发布日期:2018-08-10
通讯作者:
王丹阳
基金资助:
Yuan Cao, Yun Yang, Huaquan Xu, Yang Liu, Danyang Wan*( )
)
Received:2016-11-11
Accepted:2017-04-17
Online:2018-01-01
Published:2018-08-10
Contact:
Danyang Wan
摘要: T-DNA突变体是研究基因功能的重要资源。高效热不对称交错PCR (hiTAIL-PCR)是克隆突变体中T-DNA插入位点侧翼序列的常用方法。然而我们发现, 利用hiTAIL-PCR克隆到的一些侧翼序列并不对应于宿主的染色体DNA序列, 而是质粒的骨架DNA片段。通过设置1组RB-S4/AC1或者LB-A4/AC1对照反应, 用PCR方法鉴定了hiTAIL-PCR扩增产物中位于T-DNA侧翼的质粒骨架片段。在后续分析中, 通过排除这些片段, 提高了利用hiTAIL-PCR获得宿主染色体DNA片段的效率。同时, 通过调整反应程序, 使得整个PCR的反应时间也大为缩短。在拟南芥(Arabidopsis thaliana) T-DNA突变体drf1侧翼序列的克隆实例中, 对照反应的引入将hiTAIL-PCR中需鉴定的22条扩增产物降至4条, 效率提高了81.8%。
曹媛, 杨云, 徐化全, 刘洋, 王丹阳. 利用PCR方法鉴定hiTAIL-PCR扩增产物中的质粒骨架片段. 植物学报, 2018, 53(1): 104-109.
Yuan Cao, Yun Yang, Huaquan Xu, Yang Liu, Danyang Wan. PCR Used to Find Plasmid Backbone Fragments in the Products of hiTAIL-PCR. Chinese Bulletin of Botany, 2018, 53(1): 104-109.
| Primers | Sequence (5'-3') |
|---|---|
| Specific primers | |
| RB-S1 | GTTATCCGCTCACAATTCCACA |
| RB-S2 | TCGGGAAACCTGTCGTGCCA |
| RB-S3 | AGAGGCGGTTTGCGTATTGGG |
| RB-S4 | AAGTCGCTGTATGTGTTTGTTTGAGA |
| LB-A1 | GGCGGACCGCTATCAGGACAT |
| LB-A2 | TTGGCTACCCGTGATATTGCTG |
| LB-A3 | GACCGCTTCCTCGTGCTTTA |
| LB-A4 | GTTACACCACAATATATCCTGCCAAGAT |
| AC1 | ACGATGGACTCCAGAG |
| DRF1-S | ACAACAGAAACAACCAAAAATAATG |
| QRT-S | TGTGCAGGAGACATCATTCC |
| QRT-A | TTTCGCATTGCCAAAGAT |
| Random primers | |
| LAD1-1 | ACGATGGACTCCAGAVNVNNNGGAA |
| LAD1-2 | ACGATGGACTCCAGABNBNNGGTT |
| LAD1-3 | ACGATGGACTCCAGAVVNVNNNCCAA |
| LAD1-4 | ACGATGGACTCCAGABDNBNNNCGGT |
| LAD1-QA | ACGATGGACTCCAGAGWWWWHWWACCT |
| LAD1-HS | ACGATGGACTCCAGAGWWWWWWDYAGG |
| LAD1-A | ACGATGGACTCCAGAGVNVNNNGGCC |
| LAD1-B | ACGATGGACTCCAGAGBNBNNGGGG |
| LAD1-C | ACGATGGACTCCAGAGVVNVNNNCCGG |
| LAD1-D | ACGATGGACTCCAGAGBDNBNNNCCCC |
| LAD1-E | ACGATGGACTCCAGAGVNVNNNCAGA |
| LAD1-F | ACGATGGACTCCAGAGVNVNNNAGAT |
表1 根据T-DNA序列设计的特异性引物以及随机引物
Table 1 Specific primers designed according to T-DNA sequence and random primers
| Primers | Sequence (5'-3') |
|---|---|
| Specific primers | |
| RB-S1 | GTTATCCGCTCACAATTCCACA |
| RB-S2 | TCGGGAAACCTGTCGTGCCA |
| RB-S3 | AGAGGCGGTTTGCGTATTGGG |
| RB-S4 | AAGTCGCTGTATGTGTTTGTTTGAGA |
| LB-A1 | GGCGGACCGCTATCAGGACAT |
| LB-A2 | TTGGCTACCCGTGATATTGCTG |
| LB-A3 | GACCGCTTCCTCGTGCTTTA |
| LB-A4 | GTTACACCACAATATATCCTGCCAAGAT |
| AC1 | ACGATGGACTCCAGAG |
| DRF1-S | ACAACAGAAACAACCAAAAATAATG |
| QRT-S | TGTGCAGGAGACATCATTCC |
| QRT-A | TTTCGCATTGCCAAAGAT |
| Random primers | |
| LAD1-1 | ACGATGGACTCCAGAVNVNNNGGAA |
| LAD1-2 | ACGATGGACTCCAGABNBNNGGTT |
| LAD1-3 | ACGATGGACTCCAGAVVNVNNNCCAA |
| LAD1-4 | ACGATGGACTCCAGABDNBNNNCGGT |
| LAD1-QA | ACGATGGACTCCAGAGWWWWHWWACCT |
| LAD1-HS | ACGATGGACTCCAGAGWWWWWWDYAGG |
| LAD1-A | ACGATGGACTCCAGAGVNVNNNGGCC |
| LAD1-B | ACGATGGACTCCAGAGBNBNNGGGG |
| LAD1-C | ACGATGGACTCCAGAGVVNVNNNCCGG |
| LAD1-D | ACGATGGACTCCAGAGBDNBNNNCCCC |
| LAD1-E | ACGATGGACTCCAGAGVNVNNNCAGA |
| LAD1-F | ACGATGGACTCCAGAGVNVNNNAGAT |
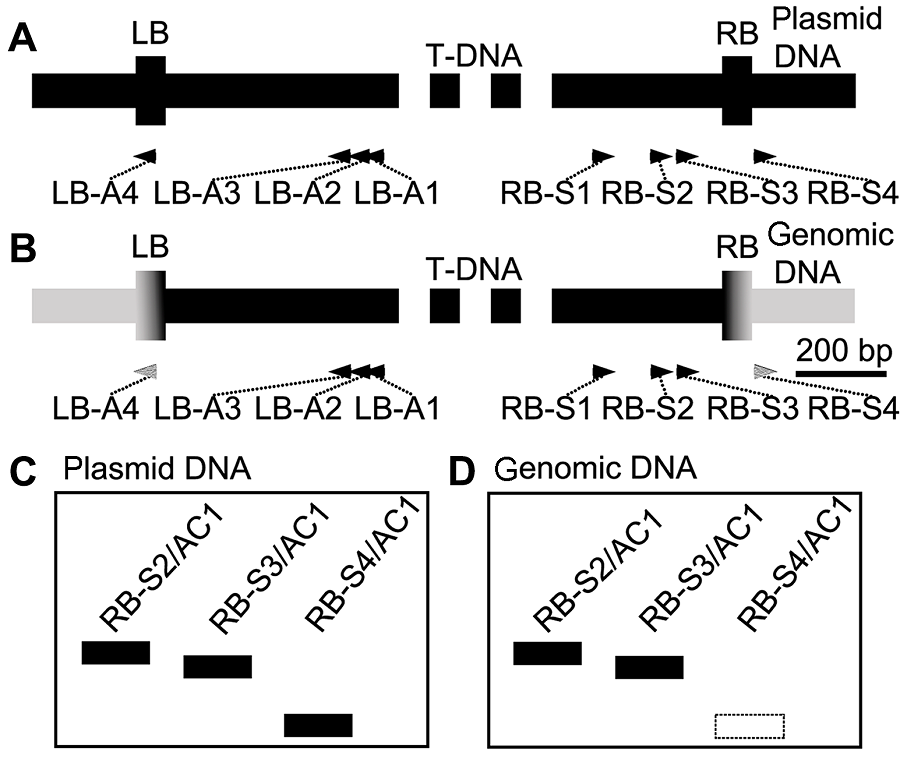
图1 设计原理(A) T-DNA质粒RB和LB处区域; (B) T-DNA质粒与染色体DNA整合后RB和LB处区域; (C) 当hiTAIL-PCR扩增质粒骨架时, RB-S2/AC1、RB-S3/AC1与RB-S4/AC1都将产生相应扩增; (D) 当hiTAIL-PCR扩增基因组DNA时, RB-S4/AC1将不产生相应扩增(虚线框示未扩增)
Figure 1 The principle of design(A) The regions of RB and LB of T-DNA plasmid; (B) The regions of RB and LB which integrate with chromosomal DNA; (C) When hiTAIL-PCR amplifies the backbone of plas- mid, the three PCR groups containing the RB-S2/AC1, RB-S3/AC1 and RB-S4/AC1 primer pairs, respectively, will all produce the positive bands; (D) When hiTAIL-PCR amplifies the genomic DNA, RB-S4/AC1 will not produce the positive bands (dashed frame)
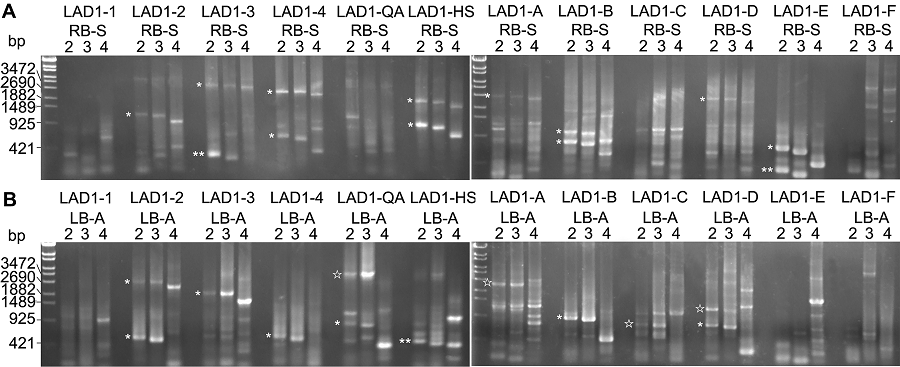
图2 拟南芥突变体drf1 T-DNA插入位点侧翼序列的克隆(A) 用RB-S系列引物扩增的第2轮PCR的结果; (B) 用LB-A系列引物扩增的第2轮PCR的结果。* 代表非特异性扩增结果; ☆代表潜在的特异性扩增结果; ** 代表来自RB之前或LB之后的T-DNA片段扩增结果。
Figure 2 The cloning of flanking sequence at the T-DNA insertion site of Arabidopsis mutant drf1(A) The second round results amplified by PCR with RB-S serial primers; (B) The second round results amplified by PCR with LB-A serial primers. * display the non-specific amplification results; ☆ represent the potential specific amplification results; ** show the amplification results from the T-DNA region before RB or after LB.
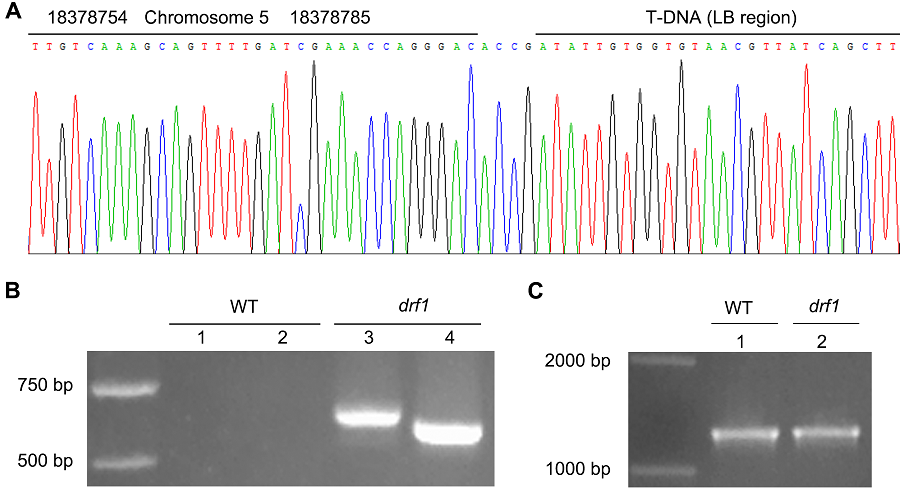
图3 拟南芥突变体drf1侧翼序列的PCR验证(A) 部分测序结果; (B) 用引物对DRF1-S/LB-A2 (1,3)及DRF1-S/LB-A3 (2,4)扩增野生型(WT)与drf1突变体基因组的结果; (C) 用引物对QRT-S/QRT-A扩增野生型(WT)与drf1突变体基因组的结果
Figure 3 The PCR confirmation for Arabidopsis mutant drf1 flanking sequence(A) The partial sequencing result; (B) The results amplifying the genomic DNA from wild type (WT) and drf1 mutant with primer pairs of DRF1-S/LB-A2 (1,3) or DRF1-S/LB-A3 (2,4); (C) The PCR results with primer pairs of QRT-S/QRT-A to amplifying WT and drf1 mutant genomic DNA
| [1] | Abe A, Kosugi S, Yoshida K, Natsume S, Takagi H, Kanzaki H, Matsumura H, Yoshida K, Mitsuoka C, Tamiru M, Innan H, Cano L, Kamoun S, Terauchi R (2012). Genome sequencing reveals agronomically important loci in rice using MutMap.Nat Biotechnol 30, 174-178. |
| [2] | Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, Gadrinab C, Heller C, Jeske A, Koesema E, Meyers CC, Parker H, Prednis L, Ansari Y, Choy N, Deen H, Geralt M, Hazari N, Hom E, Karnes M, Mulholland C, Ndubaku R, Schmidt I, Guzman P, Aguilar-Henonin L, Schmid M, Weigel D, Carter DE, Marchand T, Risseeuw E, Brogden D, Zeko A, Crosby WL, Berry CC, Ecker JR (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301, 653-657. |
| [3] | Coulondre C, Miller JH (1977). Genetic studies of the lac repressor: III. Additional correlation of mutational sites with specific amino acid residues. J Mol Biol 117, 525-567. |
| [4] | Hellens RP, Edwards EA, Leyland NR, Bean S, Mullineaux PM (2000). pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol Biol 42, 819-832. |
| [5] | Jander G (2006). Gene identification and cloning by molecular marker mapping.Methods Mol Biol 323, 115-126. |
| [6] | Jander G, Norris SR, Rounsley SD, Bush DF, Levin IM, Last RL (2002). Arabidopsis map-based cloning in the post-genome era.Plant Physiol 129, 440-450. |
| [7] | Kleinboelting N, Huep G, Appelhagen I, Viehoever P, Li Y, Weisshaar B (2015). The structural features of thousands of T-DNA insertion sites are consistent with a double- strand break repair-based insertion mechanism.Mol Plant 8, 1651-1664. |
| [8] | Liu YG, Chen YL (2007). High-efficiency thermal asymmetric interlaced PCR for amplification of unknown flanking sequences.Biotechniques 43, 649-650. |
| [9] | Liu YG, Mitsukawa N, Oosumi T, Whittier RF (1995). Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J 8, 457-463. |
| [10] | Martineau B, Voelker TA, Sanders RA (1994). On defining T-DNA.Plant Cell 6, 1032-1033. |
| [11] | Mayerhofer R, Koncz-Kalman Z, Nawrath C, Bakkeren G, Crameri A, Angelis K, Redei GP, Schell J, Hohn B, Koncz C (1991). T-DNA integration: a mode of illegitimate recombination in plants.EMBO J 10, 697-704. |
| [12] | Mueller PR, Wold B (1989). In vivo footprinting of a muscle specific enhancer by ligation mediated PCR.Science 246, 780-786. |
| [13] | Nan GL, Walbot V (2009). Plasmid rescue: recovery of flanking genomic sequences from transgenic transposon insertion sites.Methods Mol Biol 526, 101-109. |
| [14] | Riley J, Butler R, Ogilvie D, Finniear R, Jenner D, Powell S, Anand R, Smith JC, Markham AF (1990). A novel, rapid method for the isolation of terminal sequences from yeast artificial chromosome (YAC) clones.Nucleic Acids Res 18, 2887-2890. |
| [15] | Stachel SE, Timmerman B, Zambryski P (1987). Activation of Agrobacterium tumefaciens vir gene expression generates multiple single-stranded T-strand molecules from the pTiA6 T-region: requirement for 5' virD gene products. EMBO J 6, 857-863. |
| [16] | Triglia T, Peterson MG, Kemp DJ (1988). A procedure for in vitro amplification of DNA segments that lie outside the boundaries of known sequences. Nucleic Acids Res 16, 8186. |
| [17] | Tzfira T, Li JX, Lacroix B, Citovsky V (2004). Agrobacterium T-DNA integration: molecules and models.Trends Ge- net 20, 375-383. |
| [18] | Wang HR, Fang J, Liang CZ, He MH, Li QY, Chu CC (2011). Computation-assisted SiteFinding-PCR for isolating flanking sequence tags in rice.Biotechniques 51, 421-423. |
| [19] | Zhou ZW, Ma HY, Qu LJ, Xie F, Ma QW, Ren ZR (2012). Establishment of an improved high-efficiency thermal asym- metric interlaced PCR for identification of genomic integration sites mediated by phiC31 integrase.World J Microbiol Biotechnol 28, 1295-1299. |
| [1] | 韩大勇, 李海燕, 张维, 杨允菲. 东北碱化草甸芦苇匍匐型分株超速生长过程及生理机制[J]. 植物生态学报, 2025, 49(2): 320-330. |
| [2] | 王子阳, 刘升学, 杨志蕊, 秦峰. 玉米抗旱性的遗传解析[J]. 植物学报, 2024, 59(6): 883-902. |
| [3] | 吴锁伟, 安学丽, 万向元. 玉米雄性不育机理及其在工程核不育制种中的应用[J]. 植物学报, 2024, 59(6): 932-949. |
| [4] | 何璐梅, 马伯军, 陈析丰. 植物执行者抗病基因研究进展[J]. 植物学报, 2024, 59(4): 671-680. |
| [5] | 廖丹, 王艺彤, 雷晶晶, 王映霓, 张新娜, 王娟. 雌雄异株克隆植物髭脉槭对种间竞争的性别差异响应[J]. 植物生态学报, 2024, 48(12): 1623-1636. |
| [6] | 杨凯如, 贾绮玮, 金佳怡, 叶涵斐, 王盛, 陈芊羽, 管易安, 潘晨阳, 辛德东, 方媛, 王跃星, 饶玉春. 水稻黄绿叶调控基因YGL18的克隆与功能解析[J]. 植物学报, 2022, 57(3): 276-287. |
| [7] | 王霞, 严维, 周志勤, 常振仪, 郑敏婷, 唐晓艳, 吴建新. 水稻雄性不育突变体ms102的鉴定和基因定位[J]. 植物学报, 2022, 57(1): 42-55. |
| [8] | 杨丽婷, 谢燕燕, 左珂怡, 徐森, 谷瑞, 陈双林, 郭子武. 分株比例对异质光环境下美丽箬竹克隆系统光合生理的影响[J]. 植物生态学报, 2022, 46(1): 88-101. |
| [9] | 尚江源, 淳雁, 李学勇. 水稻穗长基因PAL3的克隆及自然变异分析[J]. 植物学报, 2021, 56(5): 520-532. |
| [10] | 陆静, 陈赢男, 尹佟明. 木本植物性别决定基因研究进展[J]. 植物学报, 2021, 56(1): 90-103. |
| [11] | 叶学华, 薛建国, 谢秀芳, 黄振英. 外部干扰对根茎型克隆植物甘草自然种群植株生长及主要药用成分含量的影响[J]. 植物生态学报, 2020, 44(9): 951-961. |
| [12] | 张婵, 安宇梦, Yun JÄSCHKE, 王林林, 周知里, 王力平, 杨永平, 段元文. 青藏高原及周边高山地区的植物繁殖生态学研究进展[J]. 植物生态学报, 2020, 44(1): 1-21. |
| [13] | 陈析丰,刘亚萍,马伯军. 图位克隆技术新型遗传学实验教学项目的设计与实践[J]. 植物学报, 2019, 54(6): 797-803. |
| [14] | 杨德卫,王莫,韩利波,唐定中,李生平. 水稻稻瘟病抗性基因的克隆、育种利用及稻瘟菌无毒基因研究进展[J]. 植物学报, 2019, 54(2): 265-276. |
| [15] | 李建军, 刘恋, 陈迪马, 许丰伟, 程军回, 白永飞. 底座入土深度和面积对典型草原土壤呼吸测定结果的影响[J]. 植物生态学报, 2019, 43(2): 152-164. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||