

植物学报 ›› 2022, Vol. 57 ›› Issue (4): 457-467.DOI: 10.11983/CBB22040 cstr: 32102.14.CBB22040
石水琴1, 秦华光1, 张静静1, 韩钰1, 余淏1, 彭怡宁2, 杨邵1, 汪嘉怡1, 何光宇1, 岂泽华1, 吴文杰2, 朱星雨1, 饶玉春2,*( ), 穆丹1,*(
), 穆丹1,*( )
)
收稿日期:2022-03-07
修回日期:2022-05-30
出版日期:2022-07-01
发布日期:2022-07-14
通讯作者:
饶玉春,穆丹
作者简介:mudansmile@126.com基金资助:
Shi Shuiqin1, Qin Huaguang1, Zhang Jingjing1, Han Yu1, Yu Hao1, Peng Yining2, Yang Shao1, Wang Jiayi1, He Guangyu1, Qi Zehua1, Wu Wenjie2, Zhu Xingyu1, Rao Yuchun2,*( ), Mu Dan1,*(
), Mu Dan1,*( )
)
Received:2022-03-07
Revised:2022-05-30
Online:2022-07-01
Published:2022-07-14
Contact:
Rao Yuchun,Mu Dan
About author:First author contact:† These authors contributed equally to this paper
摘要: 植物根际微生物群落对植物健康生长有重要影响, 每种植物根际都有其特定的微生物群落。大别山五针松(Pinus dabeshanensis)被国际自然保护联盟列为濒危物种, 具有重要研究价值。该研究采用16S rRNA高通量测序技术与生物信息学方法, 对濒危植物大别山五针松根际细菌群落特征与功能进行分析。结果表明, 大别山五针松根际微生物的主要种类为变形菌门、放线菌门、酸杆菌门和疣微菌门。网络分析表明, 大别山五针松根际细菌类群存在显著相关性, 其中Bryobacter属、Bradyrhizobium属和未定义的TK10属是互作网络中的重要节点。PICRUSt1功能预测表明其微生物组功能主要为氨基酸运输和代谢、细胞壁/膜/膜生物发生以及能量产生和转换。FAPROTAX功能预测表明, 大别山五针松根际富含的优势菌群具有丰富的化学异养、纤维素水解、需氧化学异养和固氮功能, 其对植物生长发育具有重要作用。研究结果可为培育健康的大别山五针松根际微生物菌群及微生物资源的开发利用提供重要依据。
石水琴, 秦华光, 张静静, 韩钰, 余淏, 彭怡宁, 杨邵, 汪嘉怡, 何光宇, 岂泽华, 吴文杰, 朱星雨, 饶玉春, 穆丹. 濒危植物大别山五针松根际细菌群落特征与功能分析. 植物学报, 2022, 57(4): 457-467.
Shi Shuiqin, Qin Huaguang, Zhang Jingjing, Han Yu, Yu Hao, Peng Yining, Yang Shao, Wang Jiayi, He Guangyu, Qi Zehua, Wu Wenjie, Zhu Xingyu, Rao Yuchun, Mu Dan. Characteristics and Function Analysis of Rhizosphere Bacterial Community of Endangered Plant Pinus dabeshanensis. Chinese Bulletin of Botany, 2022, 57(4): 457-467.
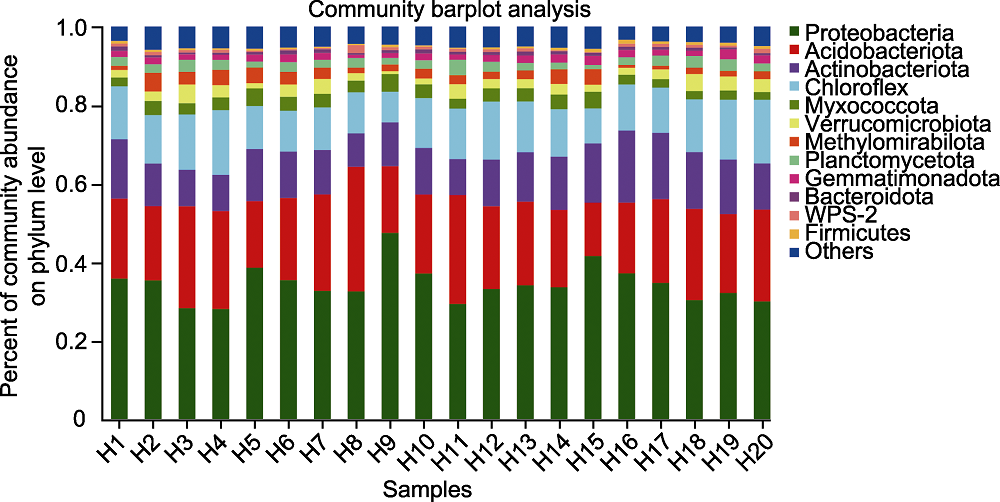
图1 大别山五针松根际微生物在门级水平上的物种组成 横坐标为样本编号, 纵坐标为该样本不同物种在门级水平上所占的比例。不同颜色的柱子代表样本中不同物种, 柱子的长短代表该物种所占比例的大小。
Figure 1 Species composition of rhizosphere microorganisms of Pinus dabeshanensis at phylum level The abscissa is the sample number, and the ordinate is the proportion of different species in the sample at phylum level. Columns in different colors represent different species in the sample, and the height of columns represents the proportion of the species.
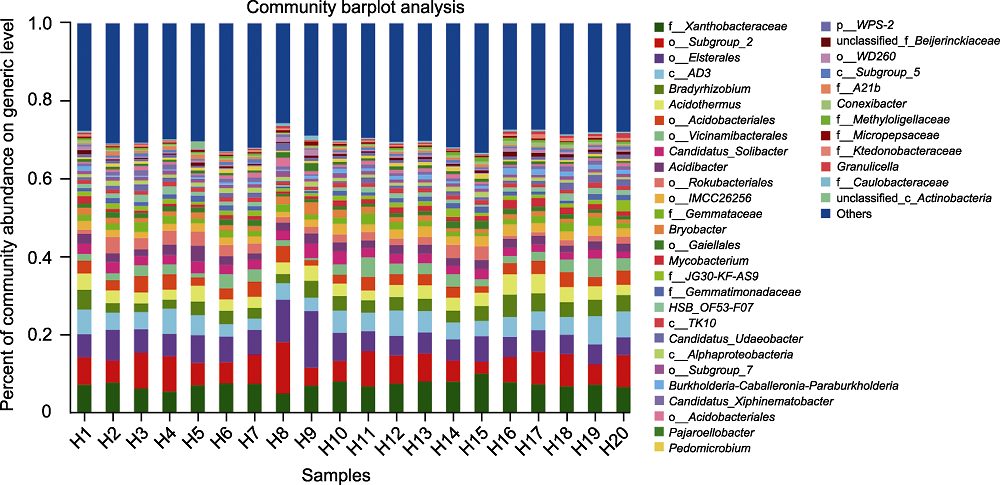
图2 大别山五针松根际微生物在属级水平上的物种组成 横坐标为样本编号, 纵坐标为该样本不同物种在属级水平上所占比例, 不同颜色柱子代表样本中不同的物种, 柱子长短代表该物种所占比例的大小。
Figure 2 Species composition of rhizosphere microorganisms of Pinus dabeshanensis at generic level The abscissa is the sample number, and the ordinate is the proportion of different species in the sample at generic level. Columns in different colors represent different species in the sample, and the height of columns represents the proportion of the species.
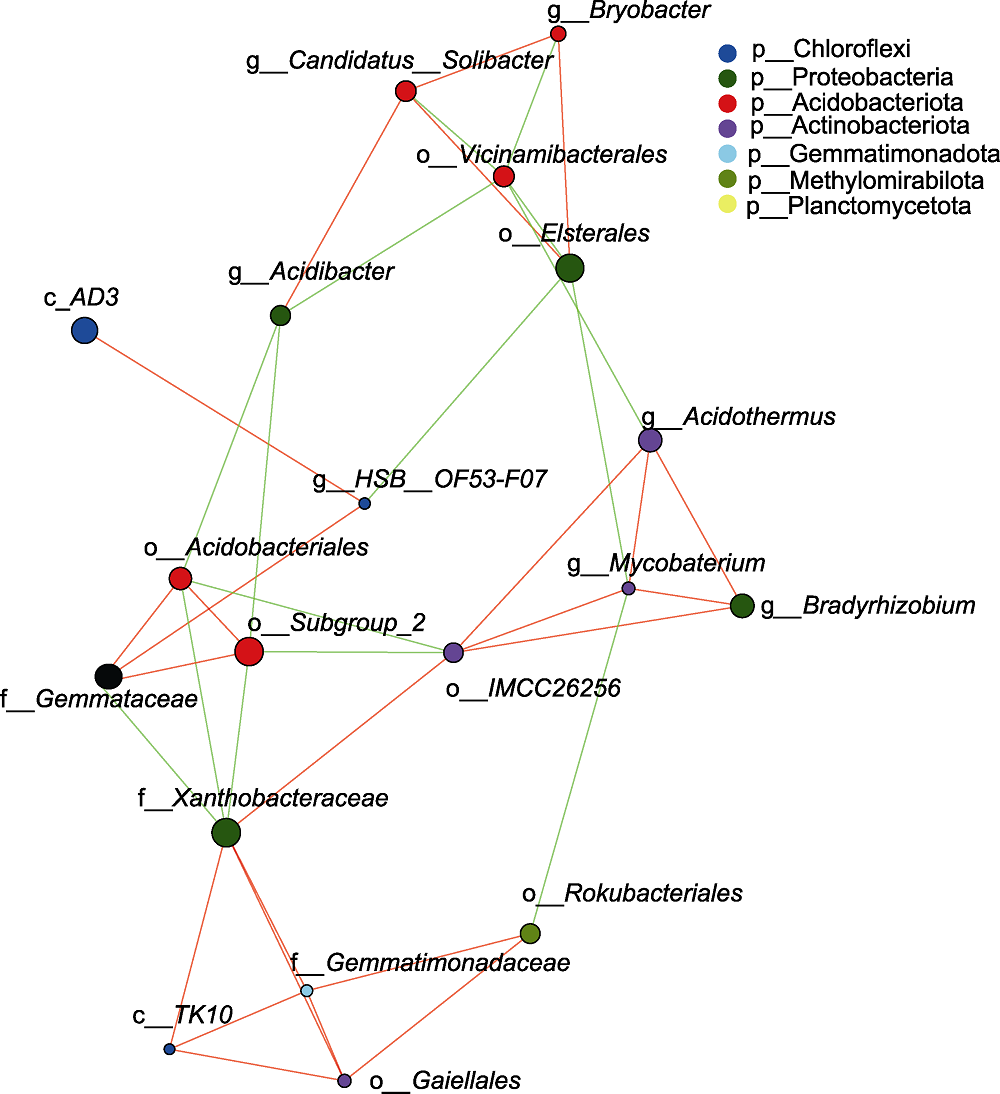
图3 大别山五针松根际微生物网络分析 节点的大小与注释到的物种丰度比例呈正相关。红色线表示正相关, 绿色线表示负相关。连接线越粗, 表示物种间相关性越高。物种间连接线越多, 表示物种间关系越密切。
Figure 3 Network analysis of rhizosphere microorganisms of Pinus dabeshanensis The size of nodes correlated with the proportion of species abundance annotated. The red line indicates positive correlations and the green line indicates negative correlations. The thicker the connecting line, the higher the correlation between species. The more connecting lines between species, the closer the relationship between species.
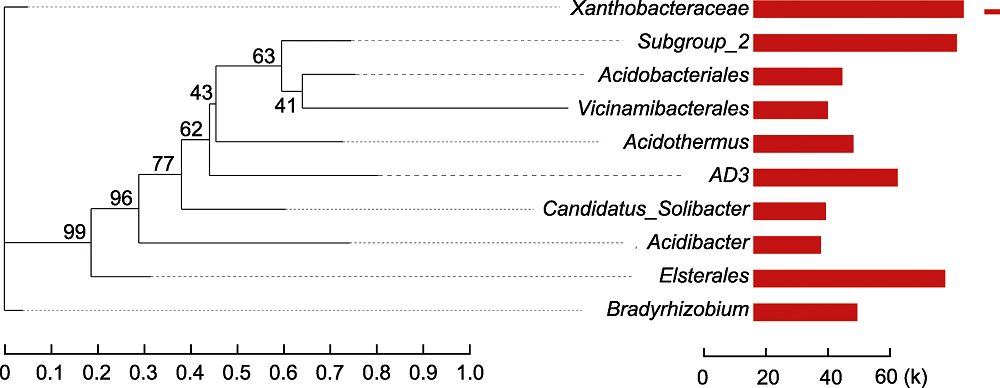
图4 大别山五针松根际微生物进化分析 系统发生进化树中每条分支代表一类物种, 根据物种所属的高级分类学水平对分支进行着色, 分支长度为2个物种间的进化距离, 即物种的差异程度。柱状图展示物种在不同分组中的reads占比。
Figure 4 Phylogenetic analysis of rhizosphere microorganisms of Pinus dabeshanensis Each branch of phylogenetic evolutionary tree represents a species. The branches are colored according to the high taxonomic level of the species. The branch length is the phylogenetic distance between the two species, that is, the degree of species difference. The bar chart shows the proportion of reads of species in different groups.
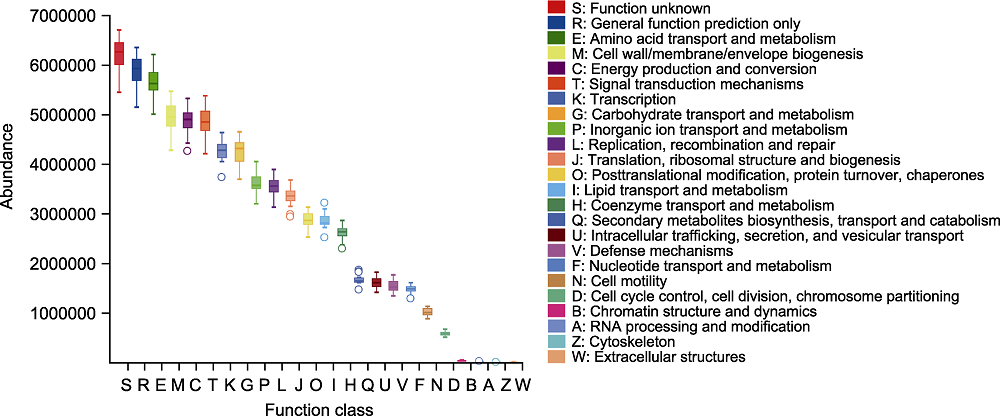
图5 大别山五针松根际微生物组蛋白相邻类的聚簇(COG)功能分类及统计分析 横坐标代表COG二级功能编号, 纵坐标代表功能丰度。
Figure 5 Classification and statistical analysis of cluster of orthologous groups of proteins (COG) function of rhizosphere microbiome of Pinus dabeshanensis The abscissa represents COG secondary function number, and the ordinate represents function abundance.
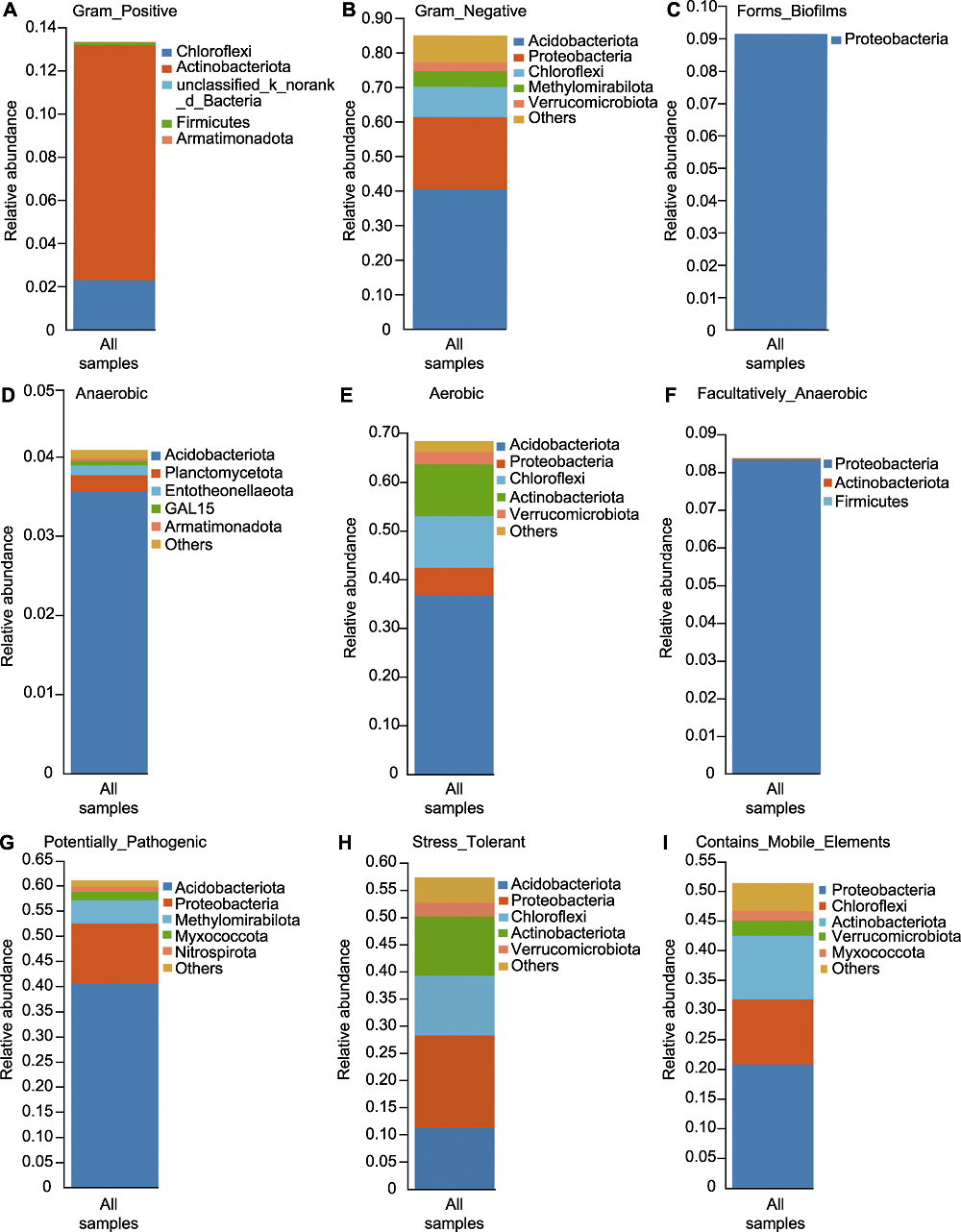
图6 大别山五针松根际微生物组物种-表型贡献度 (A) 革兰氏阳性表型; (B) 革兰氏阴性表型; (C) 生物膜形成表型; (D) 厌氧表型; (E) 需氧表型; (F) 兼性厌氧表型; (G) 致病性表型; (H) 耐压力表型; (I) 移动元件表型。物种-表型贡献度表示特定表型的主要物种组成, 即反映物种与表型的对应关系。横坐标为所有样品, 不同颜色图例代表不同的物种, 纵坐标为该样本中不同物种对此表型的贡献度。
Figure 6 Species-phenotype contribution of rhizosphere microbiome of Pinus dabeshanensis (A) Gram positive phenotype; (B) Gram negative phenotype; (C) Biofilm forming phenotype; (D) Anaerobic phenotype; (E) Aerobic phenotype; (F) Facultatively anaerobic phenotype; (G) Pathogenic phenotype; (H) Stress tolerant phenotype; (I) Mobile element containing phenotype. The species-phenotype contribution shows the main species composition of a specific phenotype, it reflects the corresponding relationship between species and phenotype. The abscissa represents all samples, different color represents different species, and the ordinate is the contribution of different species in the sample to this phenotype.
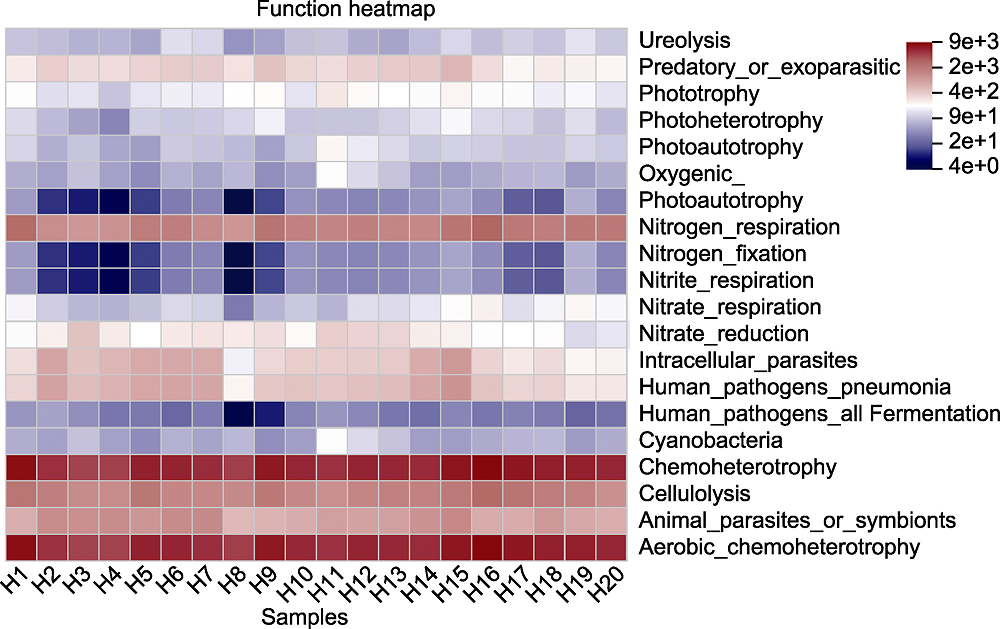
图7 大别山五针松根际微生物主要物种功能热图 横坐标为样本编号, 纵坐标为功能名称, 色块颜色梯度展示样本中不同功能的丰度变化情况。右侧为颜色梯度代表的数值。
Figure 7 Functional heatmap of main species of rhizosphere microorganisms of Pinus dabeshanensis The abscissa is the sample number, the ordinate is the function name, and the color block gradient shows the abundance changes of different functions in the sample. The right side is the value represented by the color gradient.
| [1] | 宋健, 张海剑, 刘莉, 杜立新, 柳健虎, 曹伟平 (2020). 高通量测序分析高温覆膜对韭菜根际微生物多样性的影响. 中国生物防治学报 36, 938-945. |
| [2] | 滕泽栋, 李敏, 朱静, 宋明阳 (2017). 野鸭湖湿地芦苇根际微生物多样性与磷素形态关系. 环境科学 38, 4589-4597. |
| [3] |
王孝林, 王二涛 (2019). 根际微生物促进水稻氮利用的机制. 植物学报 54, 285-287.
DOI |
| [4] | 王亚茹, 田宝玉, 张碧尧, 范競文, 戈峰, 王国红 (2021). 饵料蛋白水平对德国小蠊肠道细菌群落的影响. 生态学报 41, 5495-5505. |
| [5] | Agler MT, Ruhe J, Kroll S, Morhenn C, Kim ST, Weigel D, Kemen EM (2016). Microbial hub taxa link host and abiotic factors to plant microbiome variation. PLoS Biol 14, e1002352. |
| [6] |
Babalola OO, Emmanuel OC, Adeleke BS, Odelade KA, Nwachukwu BC, Ayiti OE, Adegboyega TT, Igiehon NO (2021). Rhizosphere microbiome cooperations: strategies for sustainable crop production. Curr Microbiol 78, 1069-1085.
DOI URL |
| [7] |
Bano S, Wu XG, Zhang XJ (2021). Towards sustainable agriculture: rhizosphere microbiome engineering. Appl Microbiol Biotechnol 105, 7141-7160.
DOI URL |
| [8] |
Berendsen RL, Pieterse CMJ, Bakker PAHM (2012). The rhizosphere microbiome and plant health. Trends Plant Sci 17, 478-486.
DOI PMID |
| [9] |
Bulgarelli D, Garrido-Oter R, Münch PC, Weiman A, Dröge J, Pan Y, McHardy AC, Schulze-Lefert P (2015). Structure and function of the bacterial root microbiota in wild and domesticated barley. Cell Host Microbe 17, 392-403.
DOI PMID |
| [10] |
Campbell BJ, Engel AS, Porter ML, Takai K (2006). The versatile ε-proteobacteria: key players in sulphidic habitats. Nat Rev Microbiol 4, 458-468.
PMID |
| [11] |
Dai ZM, Su WQ, Chen HH, Barberán A, Zhao HC, Yu MJ, Yu L, Brookes PC, Schadt CW, Chang SX, Xu JM (2018). Long-term nitrogen fertilization decreases bacterial diversity and favors the growth of Actinobacteria and Proteobacteria in agro-ecosystems across the globe. Glob Chang Biol 24, 3452-3461.
DOI URL |
| [12] |
Delgado-Baquerizo M, Grinyer J, Reich PB, Singh BK, Allen E (2016). Relative importance of soil properties and microbial community for soil functionality: insights from a microbial swap experiment. Funct Ecol 30, 1862-1873.
DOI URL |
| [13] |
Durán P, Thiergart T, Garrido-Oter R, Agler M, Kemen E, Schulze-Lefert P, Hacquard S (2018). Microbial interkingdom interactions in roots promote Arabidopsis survival. Cell 175, 973-983.
DOI URL |
| [14] |
Edgar RC (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460-2461.
DOI URL |
| [15] | Eren AM, Vineis JH, Morrison HG, Sogin ML (2013). A filtering method to generate high quality short reads using illumina paired-end technology. PLoS One 8, e66643. |
| [16] |
Fierer N (2017). Embracing the unknown: disentangling the complexities of the soil microbiome. Nat Rev Microbiol 15, 579-590.
DOI PMID |
| [17] | Fu LK, Chin CM (1992). China Plant Red Data Book:Rare and Endangered Plants. Beijing: Science Press. pp. 46-47. |
| [18] |
Jacoby RP, Chen L, Schwier M, Koprivova A, Kopriva S (2020). Recent advances in the role of plant metabolites in shaping the root microbiome. F1000Res 9, 151.
DOI URL |
| [19] |
Keswani C, Prakash O, Bharti N, Vilchez JI, Sansinenea E, Lally RD, Borriss R, Singh SP, Gupta VK, Fraceto LF, de Lima R, Singh HB (2019). Re-addressing the biosafety issues of plant growth promoting rizobacteria. Sci Total Environ 690, 841-852.
DOI |
| [20] |
Li YB, Li Q, Chen SF (2021). Diazotroph Paenibacillus triticisoli BJ-18 drives the variation in bacterial, diazotrophic and fungal communities in the rhizosphere and root/shoot endosphere of maize. Int J Mol Sci 22, 1460.
DOI URL |
| [21] |
Ling N, Song Y, Raza W, Huang QW, Guo SW, Shen QR (2015). The response of root-associated bacterial commu- nity to the grafting of watermelon. Plant Soil 391, 253-264.
DOI URL |
| [22] | Lucas G, Synge H (1978). The IUCN Plant Red Data Book: Comprising Red Data Sheets on Two Hundred-fifty Selected Plants Threatened on a World Scale. Gland: IUCN. pp. 365-366. |
| [23] |
Lundberg DS, Lebeis SL, Paredes SH, Yourstone S, Geh- ring J, Malfatti S, Tremblay J, Engelbrektson A, Kunin V, del Rio TG, Edgar RC, Eickhorst T, Ley RE, Hugenholtz P, Tringe SG, Dangl JL (2012). Defining the core Arabidopsis thaliana root microbiome. Nature 488, 86-90.
DOI URL |
| [24] |
Mendes LW, Kuramae EE, Navarrete AA, van Veen JA, Tsai SM (2014). Taxonomical and functional microbial community selection in soybean rhizosphere. ISME J 8, 1577-1587.
DOI PMID |
| [25] | Nihorimbere V, Ongena M, Smargiassi M, Thonart P (2011). Beneficial effect of the rhizosphere microbial community for plant growth and health. Biotechnol Agron Soc Environ 15, 327-337. |
| [26] |
Noman M, Ahmed T, Ijaz U, Shahid M, Azizullah, Li DY, Manzoor I, Song FM (2021). Plant-microbiome crosstalk: dawning from composition and assembly of microbial community to improvement of disease resilience in plants. Int J Mol Sci 22, 6852.
DOI URL |
| [27] |
Qu Q, Zhang ZY, Peijnenburg WJGM, Liu WY, Lu T, Hu BL, Chen JM, Chen J, Lin ZF, Qian HF (2020). Rhizosphere microbiome assembly and its impact on plant growth. J Agric Food Chem 68, 5024-5038.
DOI URL |
| [28] |
Reinhold-Hurek B, Bϋnger W, Burbano CS, Sabale M, Hurek T (2015). Roots shaping their microbiome: global hotspots for microbial activity. Annu Rev Phytopathol 53, 403-424.
DOI PMID |
| [29] |
Sarkar J, Chakraborty B, Chakraborty U (2018). Plant growth promoting rhizobacteria protect wheat plants against temperature stress through antioxidant signaling and reducing chloroplast and membrane injury. J Plant Growth Regul 37, 1396-1412.
DOI URL |
| [30] |
Tang HM, Xiao XP, Li C, Pan XC, Cheng KK, Li WY, Wang K (2020). Microbial carbon source utilization in rice rhizosphere and nonrhizosphere soils with short-term manure N input rate in paddy field. Sci Rep 10, 6487.
DOI URL |
| [31] |
Tian L, Chang JJ, Shi SH, Ji L, Zhang JF, Sun Y, Li XJ, Li XJ, Xie HW, Cai YH, Chen DZ, Wang JL, van Veen JA, Kuramae EE, Tran LSP, Tian CJ (2022). Comparison of methane metabolism in the rhizomicrobiomes of wild and related cultivated rice accessions reveals a strong impact of crop domestication. Sci Total Environ 803, 150131.
DOI URL |
| [32] |
Tiepo AN, Hertel MF, Rocha SS, Calzavara AK, De Oliveira ALM, Pimenta JA, Oliveira HC, Bianchini E, Stolf-Moreira R (2018). Enhanced drought tolerance in seedlings of Neotropical tree species inoculated with plant growth-promoting bacteria. Plant Physiol Biochem 130, 277-288.
DOI URL |
| [33] |
Trivedi P, Leach JE, Tringe SG, Sa TM, Singh BK (2021). Plant-microbiome interactions: from community assembly to plant health. Nat Rev Microbiol 18, 607-621.
DOI URL |
| [34] |
Ward NL, Challacombe JF, Janssen PH, Henrissat B, Coutinho PM, Wu M, Xie G, Haft DH, Sait M, Badger J, Barabote RD, Bradley B, Brettin TS, Brinkac LM, Bruce D, Creasy T, Daugherty SC, Davidsen TM, DeBoy RT, Detter JC, Dodson RJ, Durkin AS, Ganapathy A, Gwinn-Giglio M, Han CS, Khouri H, Kiss H, Kothari SP, Madupu R, Nelson KE, Nelson WC, Paulsen I, Penn K, Ren QH, Rosovitz MJ, Selengut JD, Shrivastava S, Sullivan SA, Tapia R, Thompson LS, Watkins KL, Yang Q, Yu CH, Zafar N, Zhou LW, Kuske CR (2009). Three genomes from the phylum Acidobacteria provide insight into the lifestyles of these microorganisms in soils. Appl Environ Microbiol 75, 2046-2056.
DOI URL |
| [35] |
Warrad M, Hassan YM, Mohamed MSM, Hagagy N, Al- Maghrabi OA, Selim S, Saleh AM, AbdElgawad H (2020). A bioactive fraction from Streptomyces sp. enhances maize tolerance against drought stress. J Microbiol Biotechnol 30, 1156-1168.
DOI URL |
| [36] |
Xu J, Zhang YZ, Zhang PF, Trivedi P, Riera N, Wang YY, Liu X, Fan GY, Tang JL, Coletta-Filho HD, Cubero J, Deng XL, Ancona V, Lu ZJ, Zhong BL, Roper MC, Capote N, Catara V, Pietersen G, Vernière C, Al-Sadi AM, Li L, Yang F, Xu X, Wang J, Yang HM, Jin T, Wang N (2018). The structure and function of the global citrus rhizosphere microbiome. Nat Commun 9, 4894.
DOI PMID |
| [37] |
Zhu ZK, Ge TD, Hu YJ, Zhou P, Wang TT, Shibistova O, Guggenberger G, Su YR, Wu JS (2017). Fate of rice shoot and root residues, rhizodeposits, and microbial assimilated carbon in paddy soil-part 2: turnover and microbial utilization. Plant Soil 416, 243-257.
DOI URL |
| [38] |
Zhu ZK, Ge TD, Liu SL, Hu YJ, Ye RZ, Xiao ML, Tong CL, Kuzyakov Y, Wu JS (2018). Rice rhizodeposits affect organic matter priming in paddy soil: the role of N fertilization and plant growth for enzyme activities, CO2 and CH4 emissions. Soil Biol Biochem 116, 369-377.
DOI URL |
| [1] | 易木荣, 卢萍, 彭勇, 汤勇, 许久恒, 尹浩萍, 张路杨, 翁晓东, 底明晓, 雷隽, 卢宸祺, 曹如君, 戴年华, 占德洋, 童媚, 楼智明, 丁永刚, 柴静, 车静. 北潦河金家水支流江西大鲵野外种群现状及栖息地评估[J]. 生物多样性, 2025, 33(4): 24145-. |
| [2] | 丁扬, 冯英群, 张金羽, 王博. 动物对濒危特有种大别山五针松种子的捕食和散布[J]. 生物多样性, 2024, 32(3): 23401-. |
| [3] | 罗正明, 刘晋仙, 张变华, 周妍英, 郝爱华, 杨凯, 柴宝峰. 不同退化阶段亚高山草甸土壤原生生物群落多样性特征及驱动因素[J]. 生物多样性, 2023, 31(8): 23136-. |
| [4] | 张仲富, 王四海, 杨卫, 陈剑. 蒜头果根际细菌群落结构与功能特征对其健康状态的响应[J]. 植物生态学报, 2023, 47(7): 1020-1031. |
| [5] | 毛莹儿, 周秀梅, 王楠, 李秀秀, 尤育克, 白尚斌. 毛竹扩张对杉木林土壤细菌群落的影响[J]. 生物多样性, 2023, 31(6): 22659-. |
| [6] | 白雪, 李玉靖, 景秀清, 赵晓东, 畅莎莎, 荆韬羽, 刘晋汝, 赵鹏宇. 谷子及其根际土壤微生物群落对铬胁迫的响应机制[J]. 植物生态学报, 2023, 47(3): 418-433. |
| [7] | 赵雯, 王丹丹, 热依拉·木民, 黄开钏, 刘顺, 崔宝凯. 阿尔山地区兴安落叶松林土壤微生物群落结构[J]. 生物多样性, 2023, 31(2): 22258-. |
| [8] | 夏凡, 杨婧, 李建, 史洋, 盖立新, 黄文华, 张经纬, 杨南, 高福利, 韩莹莹, 鲍伟东. 北京地区四个豹猫亚种群肠道菌群的组成[J]. 生物多样性, 2022, 30(9): 22103-. |
| [9] | 姬云瑞, 韦雪蕾, 张国锋, 向明贵, 王永超, 龚仁琥, 胡杨, 李迪强, 刘芳. 湖北五峰后河国家级自然保护区鸟类多样性[J]. 生物多样性, 2022, 30(7): 21475-. |
| [10] | 刘艳艳, 刘畅, 魏晓新. 我国及周边地区松属白松亚组系统学研究进展和保护现状[J]. 生物多样性, 2022, 30(2): 21344-. |
| [11] | 孙翌昕, 李英滨, 李玉辉, 李冰, 杜晓芳, 李琪. 高通量测序技术在线虫多样性研究中的应用[J]. 生物多样性, 2022, 30(12): 22266-. |
| [12] | 高程, 郭良栋. 微生物物种多样性、群落构建与功能性状研究进展[J]. 生物多样性, 2022, 30(10): 22429-. |
| [13] | 夏呈强, 李毅, 党延茹, 察倩倩, 贺晓艳, 秦启龙. 中印度洋与南海西部表层海水细菌多样性[J]. 生物多样性, 2022, 30(1): 21407-. |
| [14] | 陆奇丰, 黄至欢, 骆文华. 极小种群濒危植物广西火桐、丹霞梧桐的叶绿体基因组特征[J]. 生物多样性, 2021, 29(5): 586-595. |
| [15] | 杨永. 中国裸子植物红色名录评估(2021版)[J]. 生物多样性, 2021, 29(12): 1599-1606. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||