

植物学报 ›› 2025, Vol. 60 ›› Issue (6): 1005-1016.DOI: 10.11983/CBB25002 cstr: 32102.14.CBB25002
所属专题: 2025年集群网站期刊最受关注文章TOP10
收稿日期:2025-01-09
接受日期:2025-04-02
出版日期:2025-11-10
发布日期:2025-05-07
通讯作者:
王若涵
基金资助:
Mengsha Huang, Lingdie Kong, Miao Yu, Chang Liu, Siqin Wang, Ruohan Wang*( )
)
Received:2025-01-09
Accepted:2025-04-02
Online:2025-11-10
Published:2025-05-07
Contact:
Ruohan Wang
摘要: 三维重建技术(3D reconstruction)是利用计算机图形学和图像处理技术, 从二维图像数据中提取目标物体的几何和拓扑信息, 构建计算机可处理的三维数学模型, 从而实现物体的虚拟重建。在植物学研究中, 三维模型的构建已成为研究植物生长发育、形态结构和功能机制的有效手段, 为多尺度成像、测量和分析提供了有力支撑, 并在农业和林业领域展现出巨大的应用潜力。近年来, 随着植物三维重建技术的不断完善, 其在植物学研究中衍生出不同的应用方向, 涵盖植物形态结构建模、生长发育动态监测以及植物育种等多方面。该文综述了三维重建技术的发展历程及常见的三维重建成像技术在植物不同尺度(从器官、组织到细胞)研究中的应用, 重点阐述这些技术的基本原理及应用, 旨在为植物多模态跨尺度成像以及表型与功能研究提供理论和技术支撑, 为揭示植物生长发育规律及响应环境变化的机制提供新途径。
黄梦莎, 孔令蝶, 于淼, 刘畅, 王思钦, 王若涵. 不同尺度三维重建技术在植物研究中的发展及应用. 植物学报, 2025, 60(6): 1005-1016.
Mengsha Huang, Lingdie Kong, Miao Yu, Chang Liu, Siqin Wang, Ruohan Wang. Development and Application of 3D Reconstruction Technology at Different Scales in Plant Research. Chinese Bulletin of Botany, 2025, 60(6): 1005-1016.

图1 三维重建技术在植物学研究中的应用场景趋势分析 节点表示三维重建技术在植物学中的应用场景, 流量数据基于2011-2024年植物三维重建相关的484篇文献, 分流表示应用场景的变化。
Figure 1 Trends in the application scenarios of 3D reconstruction technology in botanical research The node represents the application scenarios of 3D reconstruction technology in the field of botany, and the flow data is derived from 484 articles on plant 3D reconstruction published from 2011 to 2024, with the diversions indicating shifts in application scenarios over time.
| 尺度 | 植物物种 | 方法 | 应用部位 | 参考文献 |
|---|---|---|---|---|
| 器官 | 水稻(Oryza sativa) | Micro-CT | 茎 | 吴迪, |
| 银杏(Ginkgo biloba)、拟南芥(Arabidopsis thaliana)和榆树(Ulmus pumila) | Micro-CT、LSFM、Auto CUTS-SEM和X-CT | 种子 | 马灵玉, | |
| 柑橘(Citrus reticulata ‘Bu Zhi Huo’)、苹果(Malus pumila)、梨(Pyrus spp.)、茄子(Solanum melongena)、萝卜(Raphanus sativus)和葡萄(Vitis vinifera) | X-CT | 果实 | Herremans et al., | |
| 拟南芥(A. thaliana)和大麦(Hordeum vulgare) | X-CT | 花 | Ijiri et al., | |
| 组织 | 雪松(Cedrus deodara)、苹果(M. pumila)和月季(Rosa hybrida) | FIB-SEM和X-CT | 输导组织 | Herppich et al., |
| 拟南芥(A. thaliana) | CLSM | 分生组织 | Gómez-Felipe and de Folter, | |
| 橡树(Quercus palustris) | X-CT | 保护组织 | Morisset et al., | |
| 橡树(Q. palustris) | CLSM和X-CT | 营养组织 | Fink, | |
| 细胞 | 拟南芥(A. thaliana)、狗尾草(Setaria viridis)、青杄(Picea wilsonii)、红松(Pinus densiflora)和黑松(P. thunbergii) | ODT、ssTEM、 FIB-SEM和CLSM | 叶肉细胞、花粉粒细胞、下胚轴细胞和雄蕊细胞 | Jackson et al., |
表1 植物不同尺度(器官、组织和细胞)三维重建成像方法与应用
Table 1 Three-dimensional reconstruction imaging methods and applications of plants at different scales (organ, tissue, and cell)
| 尺度 | 植物物种 | 方法 | 应用部位 | 参考文献 |
|---|---|---|---|---|
| 器官 | 水稻(Oryza sativa) | Micro-CT | 茎 | 吴迪, |
| 银杏(Ginkgo biloba)、拟南芥(Arabidopsis thaliana)和榆树(Ulmus pumila) | Micro-CT、LSFM、Auto CUTS-SEM和X-CT | 种子 | 马灵玉, | |
| 柑橘(Citrus reticulata ‘Bu Zhi Huo’)、苹果(Malus pumila)、梨(Pyrus spp.)、茄子(Solanum melongena)、萝卜(Raphanus sativus)和葡萄(Vitis vinifera) | X-CT | 果实 | Herremans et al., | |
| 拟南芥(A. thaliana)和大麦(Hordeum vulgare) | X-CT | 花 | Ijiri et al., | |
| 组织 | 雪松(Cedrus deodara)、苹果(M. pumila)和月季(Rosa hybrida) | FIB-SEM和X-CT | 输导组织 | Herppich et al., |
| 拟南芥(A. thaliana) | CLSM | 分生组织 | Gómez-Felipe and de Folter, | |
| 橡树(Quercus palustris) | X-CT | 保护组织 | Morisset et al., | |
| 橡树(Q. palustris) | CLSM和X-CT | 营养组织 | Fink, | |
| 细胞 | 拟南芥(A. thaliana)、狗尾草(Setaria viridis)、青杄(Picea wilsonii)、红松(Pinus densiflora)和黑松(P. thunbergii) | ODT、ssTEM、 FIB-SEM和CLSM | 叶肉细胞、花粉粒细胞、下胚轴细胞和雄蕊细胞 | Jackson et al., |
| 方法 | 分辨率 | 成像速度 | 样品制备 | 定位水平 | 参考文献 |
|---|---|---|---|---|---|
| LSCM | 200-500 nm | 秒级/分级, 快 | 需荧光标记和透明化处理 | 细胞、亚细胞和分子水平 | 李叶等, |
| X-CT | 250 μm以下 | 分级, 较快 | 需稳定固定 | 器官、组织、细胞和亚细胞水平 | Du et al., |
| Micro-CT | 50 μm以下 | 分级/小时级, 较快, 越精细越慢且越清晰 | 采用冷冻或化学固定 | 器官、组织、细胞和亚细胞水平 | Wang et al., |
| ODT | 200-600 nm | 秒级/分级, 快 | 无需荧光标记, 透明化处理 | 器官、组织、细胞和亚细胞水平 | He et al., |
| FIB-SEM | 1-10 nm | 小时级, 慢 | 制备方法复杂, 常温树脂包埋方法、高压冷冻及冷冻等 | 细胞及亚细胞水平 | Heymann et al., |
| TEM | 0.1-2 nm | 小时级, 慢 | 化学固定、常温脱水、包埋、切片、染色后观察 | 细胞、亚细胞、分子和原子水平 | Drummy et al., |
表2 植物中6种常见的三维重建成像方法比较分析
Table 2 Comparative analysis of six common 3D reconstruction imaging methods in plants
| 方法 | 分辨率 | 成像速度 | 样品制备 | 定位水平 | 参考文献 |
|---|---|---|---|---|---|
| LSCM | 200-500 nm | 秒级/分级, 快 | 需荧光标记和透明化处理 | 细胞、亚细胞和分子水平 | 李叶等, |
| X-CT | 250 μm以下 | 分级, 较快 | 需稳定固定 | 器官、组织、细胞和亚细胞水平 | Du et al., |
| Micro-CT | 50 μm以下 | 分级/小时级, 较快, 越精细越慢且越清晰 | 采用冷冻或化学固定 | 器官、组织、细胞和亚细胞水平 | Wang et al., |
| ODT | 200-600 nm | 秒级/分级, 快 | 无需荧光标记, 透明化处理 | 器官、组织、细胞和亚细胞水平 | He et al., |
| FIB-SEM | 1-10 nm | 小时级, 慢 | 制备方法复杂, 常温树脂包埋方法、高压冷冻及冷冻等 | 细胞及亚细胞水平 | Heymann et al., |
| TEM | 0.1-2 nm | 小时级, 慢 | 化学固定、常温脱水、包埋、切片、染色后观察 | 细胞、亚细胞、分子和原子水平 | Drummy et al., |
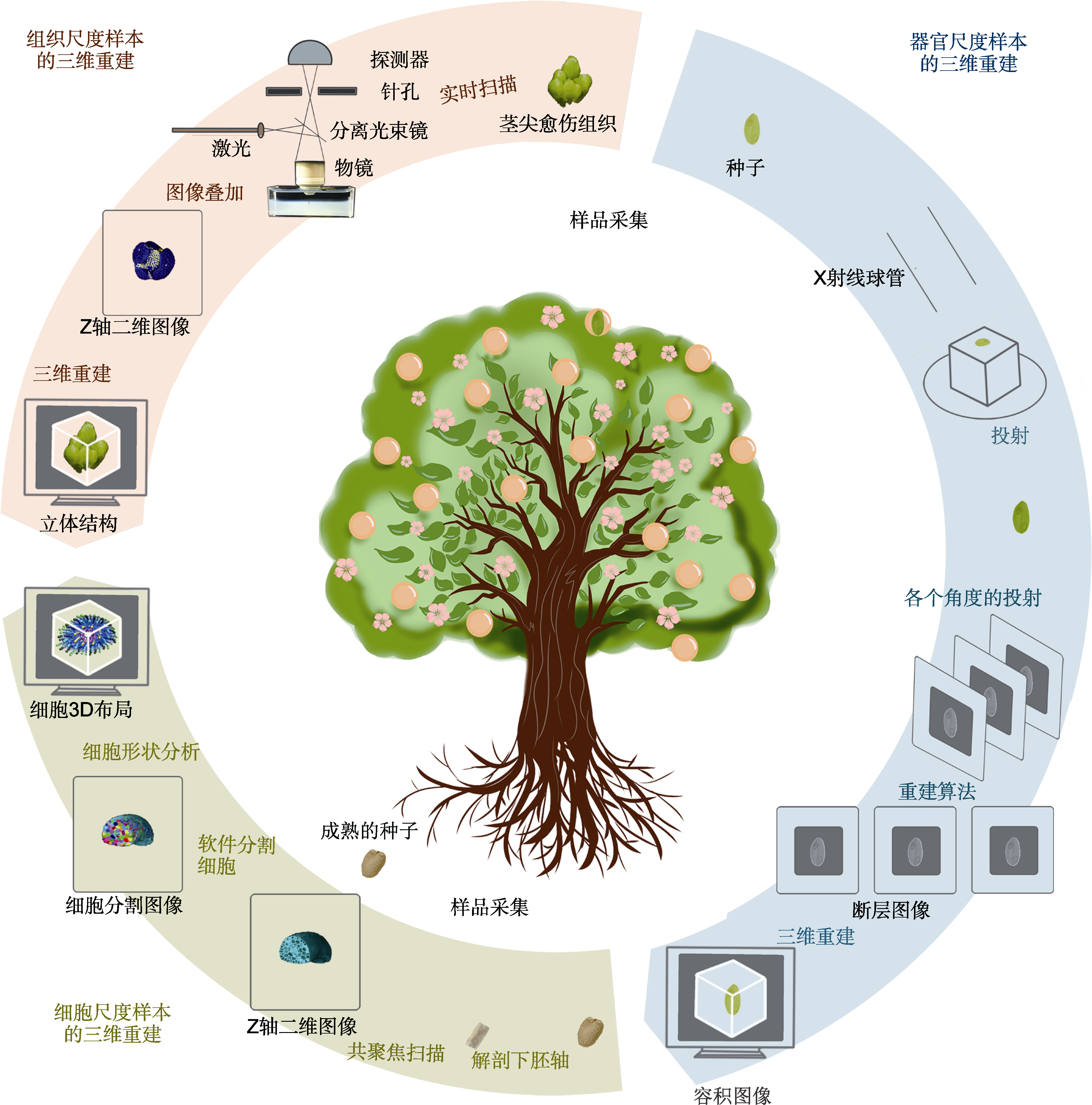
图2 不同尺度样本的三维重建流程(Jackson et al., 2017; Tracy et al., 2017; Gómez-Felipe and de Folter, 2019) 器官尺度以基于显微CT成像技术(Micro-CT)的三维重建为例, 组织和细胞尺度以基于共聚焦显微成像技术的三维重建为例。
Figure 2 Flow chart of 3D reconstruction of samples at different scales (Jackson et al., 2017; Tracy et al., 2017; Gómez-Felipe and de Folter, 2019) The 3D reconstruction based on micro-computed tomography (Micro-CT) imaging technology is used as an example at the organ scale, and the 3D reconstruction based on confocal microscopy is used as an example at the tissue and cell scales.
| [1] |
Arshad MA, Jubery T, Afful J, Jignasu A, Balu A, Ganapathysubramanian B, Sarkar S, Krishnamurthy A (2024). Evaluating neural radiance fields for 3D plant geometry reconstruction in field conditions. Plant Phenomics 6, 0235.
DOI URL |
| [2] |
Besl PJ, McKay ND (1992). A method for registration of 3-D shapes. IEEE Trans Pattern Anal Mach Intell 14, 239-256.
DOI URL |
| [3] |
Bushby AJ, P'Ng KMY, Young RD, Pinali C, Knupp C, Quantock AJ (2011). Imaging three-dimensional tissue architectures by focused ion beam scanning electron microscopy. Nat Protoc 6, 845-858.
DOI PMID |
| [4] | Cai JR, Liang XX, Xu Q, Xia ZY, Sun L, Ma LX (2024). Detecting volumetric edible rate of thick-skinned citrus using X-ray three-dimensional reconstruction. Trans Chin Soc Agric Eng 40, 293-300. (in Chinese) |
| 蔡健荣, 梁小祥, 许骞, 夏中岩, 孙力, 马立鑫 (2024). 采用X射线三维重构技术检测厚皮柑橘的体积可食率. 农业工程学报 40, 293-300. | |
| [5] | Chen XJ (2015). Research on computer vision technology based on OpenCV. Comput Knowl Technol 11(30), 137-138, 141. (in Chinese) |
| 陈雪娇 (2015). 基于OpenCV的计算机视觉技术研究. 电脑知识与技术 11(30), 137-138, 141. | |
| [6] |
Collevatti RG, Castañeda M, Silva-Caminha SAF, Jaramillo C (2024). Application of confocal laser microscopy for identification of modern and fossil pollen grains, an example in palm Mauritiinae. Rev Palaeobot Palynol 327, 105140.
DOI URL |
| [7] |
Cooley JW, Tukey JW (1965). An algorithm for the machine calculation of complex Fourier series. Math Comput 19, 297-301.
DOI URL |
| [8] |
Crumpton-Taylor M, Grandison S, Png KMY, Bushby AJ, Smith AM (2012). Control of starch granule numbers in Arabidopsis chloroplasts. Plant Physiol 158, 905-916.
DOI PMID |
| [9] |
Cui Y, Cao WH, He YL, Zhao Q, Wakazaki M, Zhuang XH, Gao JY, Zeng YL, Gao CJ, Ding Y, Wong HY, Wong WS, Lam HK, Wang PF, Ueda T, Rojas-Pierce M, Toyooka K, Kang BH, Jiang LW (2019). A whole-cell electron tomography model of vacuole biogenesis in Arabidopsis root cells. Nat Plants 5, 95-105.
DOI |
| [10] | Davies HE, Wathen CG, Gleeson FV (2011). The risks of radiation exposure related to diagnostic imaging and how to minimise them. BMJ 342, d947. |
| [11] |
Drummy LF, Yang JY, Martin DC (2004). Low-voltage electron microscopy of polymer and organic molecular thin films. Ultramicroscopy 99, 247-256.
PMID |
| [12] |
Du Z, Hu YG, Ali Buttar N, Mahmood A (2019). X-ray computed tomography for quality inspection of agricultural products: a review. Food Sci Nutr 7, 3146-3160.
DOI PMID |
| [13] |
Fink S (2006). The use of laser scanning confocal microscopy in the study of wound healing phenomena in plants. Isr J Plant Sci 54, 265-271.
DOI URL |
| [14] |
Gao T, Zhu FY, Paul P, Sandhu J, Doku HA, Sun JX, Pan Y, Staswick P, Walia H, Yu HF (2021). Novel 3D imaging systems for high-throughput phenotyping of plants. Remote Sens 13, 2113.
DOI URL |
| [15] | Gómez-Felipe A, de Folter S (2019). A simple protocol for imaging floral tissues of Arabidopsis with confocal microscopy. In: de Folter, ed. Plant MicroRNAs. New York: Humana Press. pp. 187-195. |
| [16] |
Guo JS, Wang G, Xie L, Wang XQ, Feng LC, Guo WB, Tao XR, Humbel BM, Zhang ZK, Hong J (2023). Three-dimensional analysis of membrane structures associated with tomato spotted wilt virus infection. Plant Cell Environ 46, 650-664.
DOI URL |
| [17] |
He YP, Zhou NS, Ziemczonok M, Wang YJ, Lei L, Duan LT, Zhou RJ (2023). Standardizing image assessment in optical diffraction tomography. Opt Lett 48, 395-398.
DOI PMID |
| [18] |
Hériché M, Arnould C, Wipf D, Courty PE (2022). Imaging plant tissues: advances and promising clearing practices. Trends Plant Sci 27, 601-615.
DOI URL |
| [19] |
Herppich WB, Matsushima U, Graf W, Zabler S, Dawson M, Choinka G, Manke I (2015). Synchrotron X-ray CT of rose peduncles-evaluation of tissue damage by radiation. Mater Test 57, 59-63.
DOI URL |
| [20] |
Herremans E, Melado-Herreros A, Defraeye T, Verlinden B, Hertog M, Verboven P, Val J, Fernández-Valle ME, Bongaers E, Estrade P, Wevers M, Barreiro P, Nicolaï BM (2014). Comparison of X-ray CT and MRI of watercore disorder of different apple cultivars. Postharvest Biol Technol 87, 42-50.
DOI URL |
| [21] |
Herremans E, Verboven P, Verlinden BE, Cantre D, Abera M, Wevers M, Nicolaï BM (2015). Automatic analysis of the 3-D microstructure of fruit parenchyma tissue using X-ray micro-CT explains differences in aeration. BMC Plant Biol 15, 264.
DOI PMID |
| [22] |
Heymann JAW, Hayles M, Gestmann I, Giannuzzi LA, Lich B, Subramaniam S (2006). Site-specific 3D imaging of cells and tissues with a dual beam microscope. J Struct Biol 155, 63-73.
PMID |
| [23] |
House A, Balkwill K (2013). FIB-SEM: an additional technique for investigating internal structure of pollen walls. Microsc Microanal 19, 1535-1541.
DOI PMID |
| [24] |
Hu ZJ, Liu JZ, Shen SY, Wu WQ, Yuan JB, Shen WW, Ma LY, Wang GC, Yang SY, Xu XP, Cui YN, Li ZC, Shen LJ, Li LL, Bian JH, Zhang X, Han H, Lin JX (2024). Large-volume fully automated cell reconstruction generates a cell atlas of plant tissues. Plant Cell 36, 4840-4861.
DOI URL |
| [25] | Ijiri T, Yoshizawa S, Yokota H, Igarashi T (2014). Flower modeling via X-ray computed tomography. ACM Trans Graphics 33, 48. |
| [26] | Jackson MDB, Xu H, Duran-Nebreda S, Stamm P, Bassel GW (2017). Topological analysis of multicellular complexity in the plant hypocotyl. eLife 6, e26023. |
| [27] |
Janes G, Von Wangenheim D, Cowling S, Kerr I, Band L, French AP, Bishopp A(2018). Cellular patterning of Arabidopsis roots under low phosphate conditions. Front Plant Sci 9, 735.
DOI URL |
| [28] |
Janssen S, Verboven P, Nugraha B, Wang Z, Boone M, Josipovic I, Nicolaï BM (2020). 3D pore structure analysis of intact ‘Braeburn’ apples using X-ray micro-CT. Postharvest Biol Technol 159, 111014.
DOI URL |
| [29] | Jia X, Sun F, Ji G (2022). Advances in cryo-focused ion beam-scanning electron microscopy imaging technology. Chin Bull Bot 57, 24-29. (in Chinese) |
|
贾星, 孙飞, 季刚 (2022). 冷冻聚焦离子束-扫描电镜成像技术研究进展. 植物学报 57, 24-29.
DOI |
|
| [30] |
Jiang ZG (2016). How many species are there on Earth? Chin Sci Bull 61, 2337-2343. (in Chinese)
DOI URL |
| 蒋志刚 (2016). 地球上有多少物种? 科学通报 61, 2337-2343. | |
| [31] | Jin D, Zhou RJ, Yaqoob Z, So PTC (2017). Tomographic phase microscopy: principles and applications in bioimaging. J Opt Soc Am B 34, B64-B77. |
| [32] |
Karahara I, Yamauchi D, Uesugi K, Mineyuki Y (2023). Three-dimensional visualization of plant tissues and organs by X-ray micro-computed tomography. Microscopy (Oxf) 72, 310-325.
DOI URL |
| [33] |
Katsevich A (2002). Analysis of an exact inversion algorithm for spiral cone-beam CT. Phys Med Biol 47, 2583-2597.
PMID |
| [34] |
Kim G, Lee S, Shin S, Park Y (2018). Three-dimensional label-free imaging and analysis of Pinus pollen grains using optical diffraction tomography. Sci Rep 8, 1782.
DOI |
| [35] |
Kim K, Chung JM, Lee S, Jung HS (2015). The effects of electron beam exposure time on transmission electron microscopy imaging of negatively stained biological samples. Appl Microsc 45, 150-154.
DOI URL |
| [36] | Li CH, Tian YF, Yan SG (2020). Laser scanning confocal microscopy technology and application. Exp Sci Technol 18(4), 33-38. (in Chinese) |
| 李成辉, 田云飞, 闫曙光 (2020). 激光扫描共聚焦显微成像技术与应用. 实验科学与技术 18(4), 33-38. | |
| [37] | Li DX (2004). New progress in transmission electron microscopy I. Development and application of transmission electron microscopy and related components. J Chin Electron Microsc Soc 3, 269-277. (in Chinese) |
| 李斗星 (2004). 透射电子显微学的新进展I. 透射电子显微镜及相关部件的发展及应用. 电子显微学报 3, 269-277. | |
| [38] | Li L, Chen ZQ, Zhang L, Xing YX (2006). Xiaochuan Pan’s new BPF-type algorithms for computed tomography image reconstruction. CT Theory Appl 15(3), 68-73. (in Chinese) |
| 李亮, 陈志强, 张丽, 邢宇翔 (2006). 潘晓川教授的反投影滤波(BPF)新型重建算法介绍. CT理论与应用研究 15 (3), 68-73. | |
| [39] | Li Q, Li RR, Qiang Y, Cheng YB, Wang T (2023). Research and progress of artificial intelligence in medical CT image reconstruction. J Taiyuan Univ Technol 54, 1-16. (in Chinese) |
| 李青, 李润睿, 强彦, 成煜斌, 王涛 (2023). 人工智能在医学CT图像重建中的研究进展. 太原理工大学学报 54, 1-16. | |
| [40] |
Li XX, Ji G, Chen X, Ding W, Sun L, Xu W, Han H, Sun F (2017). Large scale three-dimensional reconstruction of an entire Caenorhabditis elegans larva using AutoCUTS- SEM. J Struct Biol 200, 87-96.
DOI URL |
| [41] | Li Y, Huang HP, Lin PQ, Cui YM, Li QF, Zheng YQ (2015). The fundamentals and techniques in laser scanning confocal microscopy. J Chin Electron Microsc Soc 34, 169-176. (in Chinese) |
| 李叶, 黄华平, 林培群, 崔艳梅, 李勤奋, 郑勇奇 (2015). 激光扫描共聚焦显微镜的基本原理及其使用技巧. 电子显微学报 34, 169-176. | |
| [42] | Lu J, Meng GL, Yu LZ (2023). Imaging techniques and application of super-resolution laser scanning confocal microscope. Exp Sci Technol 21, 25-29. (in Chinese) |
| 路姣, 孟国龙, 余凌竹 (2023). 超高分辨率激光扫描共聚焦显微镜的成像技术与应用. 实验科学与技术 21, 25-29. | |
| [43] |
Luo LY, Jiang XT, Yang Y, Samy ERA, Lefsrud M, Hoyos-Villegas V, Sun SP (2023). Eff-3DPSeg: 3D organ-level plant shoot segmentation using annotation-effi-cient deep learning. Plant Phenomics 5, 0080.
DOI URL |
| [44] | Ma LY (2021). Multiscale 3D Reconstruction of Ginkgo Biloba Embryo and Arabidopsis Thaliana Seed. PhD dissertation. Beijing: Beijing Forestry University. pp. 1-101. (in Chinese) |
| 马灵玉 (2021). 银杏种胚和拟南芥种子多尺度三维重构研究. 博士论文. 北京: 北京林业大学. pp. 1-101. | |
| [45] |
Ma LY, Hu ZJ, Shen WW, Zhang YY, Wang GC, Chang B, Lu JK, Cui YN, Xu HM, Feng Y, Jin B, Zhang X, Wang L, Lin JX (2024). Three-dimensional reconstruction and multiomics analysis reveal a unique pattern of embryogenesis in Ginkgo biloba. Plant Physiol 196, 95-111.
DOI URL |
| [46] | Ma LY, Qi XH, Hu ZJ, Shen WW, Wang GC, Zhang BL, Zhang X, Lin JX (2022). Applications of optical clearing technique in multi-scale imaging. Chin Bull Bot 57, 98-110. (in Chinese) |
|
马灵玉, 祁晓红, 胡子建, 沈微微, 王广超, 张柏林, 张曦, 林金星 (2022). 光学透明技术在植物多尺度成像中的应用. 植物学报 57, 98-110.
DOI |
|
| [47] |
Maizel A, Von Wangenheim D, Federici F, Haseloff J, Stelzer EHK (2011). High-resolution live imaging of plant growth in near physiological bright conditions using light sheet fluorescence microscopy. Plant J 68, 377-385.
DOI URL |
| [48] |
Masters BR, Gonzalez RC, Woods R (2009). Book review: digital image processing, third edn. J Biomed Opt 14, 029901.
DOI URL |
| [49] |
Masyutin AG, Tarasova EK, Onishchenko GE, Erokhina MV (2023). Identifying carbon nanoparticles in biological samples by means of transmission electron microscopy. Bull Russ Acad Sci Phys 87, 1443-1448.
DOI |
| [50] |
Montenegro-Johnson TD, Stamm P, Strauss S, Topham AT, Tsagris M, Wood ATA, Smith RS, Bassel GW (2015). Digital single-cell analysis of plant organ development using 3D Cell Atlas. Plant Cell 27, 1018-1033.
DOI URL |
| [51] |
Morisset JB, Mothe F, Colin F (2012). Observation of Quercus petraea epicormics with X-ray CT reveals strong pith-to-bark correlations: silvicultural and ecological implications. Forest Ecol Manag 278, 127-137.
DOI URL |
| [52] |
Nugraha B, Verboven P, Janssen S, Wang Z, Nicolaï BM (2019). Non-destructive porosity mapping of fruit and vegetables using X-ray CT. Postharvest Biol Technol 150, 80-88.
DOI URL |
| [53] |
Ovečka M, Sojka J, Tichá M, Komis G, Basheer J, Marchetti C, Šamajová O, Kuběnová L, Šamaj J (2022). Imaging plant cells and organs with light-sheet and super-resolution microscopy. Plant Physiol 188, 683-702.
DOI PMID |
| [54] |
Piovesan A, Vancauwenberghe V, Van De Looverbosch T, Verboven P, Nicolaï B (2021). X-ray computed tomography for 3D plant imaging. Trends Plant Sci 26, 1171-1185.
DOI PMID |
| [55] | Qi XH (2022). Multiscale 3D Reconstruction, Metabolism and Transcriptome Analysis of Ulmus Pumila Seeds During Late-maturation Stage. PhD dissertation. Beijing: Beijing Forestry University. pp. 1-123. (in Chinese) |
| 祁晓红 (2022). 榆树种子发育后期多尺度三维重构及代谢和转录组分析研究. 博士论文. 北京: 北京林业大学. pp. 1-123. | |
| [56] |
Qi XH, Chen LL, Hu ZJ, Shen WW, Xu HM, Ma LY, Wang GC, Jing YP, Wang XD, Zhang BL, Lin JX (2022). Cytology, transcriptomics, and mass spectrometry imaging reveal changes in late-maturation elm (Ulmus pumila) seeds. J Plant Physiol 271, 153639.
DOI URL |
| [57] | Roberts LG (1963). Machine Perception of Three-dimensional Solids. PhD dissertation. Cambridge: Massachusetts Institute of Technology. pp. 1-82. |
| [58] |
Serra L, Tan S, Robinson S, Langdale JA (2022). Flip- Flap: a simple dual-view imaging method for 3D reconstruction of thick plant samples. Plants 11, 506.
DOI URL |
| [59] | Shen RH (2021). New Methods for Three-dimensional Reconstruction in the Transmission Electron Microscope. PhD dissertation. Changsha: Hunan University. pp. 1-124. (in Chinese) |
| 沈若涵 (2021). 透射电子显微镜中的三维重构新方法. 博士论文. 长沙: 湖南大学. pp. 1-124. | |
| [60] | Shi BR, Huang XP, Fu XL, Wang BJ (2022). Advances in the plant multicellular network analysis. Chin J Biotechnol 38, 2798-2810. (in Chinese) |
| 施般若, 黄小萍, 付秀荣, 王邦俊 (2022). 植物多细胞网络分析研究进展. 生物工程学报 38, 2798-2810. | |
| [61] |
Silveira SR, Le Gloanec C, Gómez-Felipe A, Routier-Kierzkowska AL, Kierzkowski D (2022). Live-imaging provides an atlas of cellular growth dynamics in the stamen. Plant Physiol 188, 769-781.
DOI URL |
| [62] |
Stevens KA (2012). The vision of David Marr. Perception 41, 1061-1072.
PMID |
| [63] |
Tracy SR, Gómez JF, Sturrock CJ, Wilson ZA, Ferguson AC (2017). Non-destructive determination of floral staging in cereals using X-ray micro computed tomography (μCT). Plant Methods 13, 9.
DOI URL |
| [64] |
Trueba S, Théroux-Rancourt G, Earles JM, Buckley TN, Love D, Johnson DM, Brodersen C (2022). The three- dimensional construction of leaves is coordinated with water use efficiency in conifers. New Phytol 233, 851-861.
DOI URL |
| [65] |
Truernit E, Bauby H, Dubreucq B, Grandjean O, Runions J, Barthélémy J, Palauqui JC (2008). High-resolution whole-mount imaging of three-dimensional tissue organization and gene expression enables the study of phloem development and structure in Arabidopsis. Plant Cell 20, 1494-1503.
DOI URL |
| [66] |
Van De Looverbosch T, Bhuiyan MHR, Verboven P, Dierick M, Van Loo D, De Beenbouwer J, Sijbers J, Nicolaï B (2020). Nondestructive internal quality inspection of pear fruit by X-ray CT using machine learning. Food Control 113, 107170.
DOI URL |
| [67] | Wang J, Wang J, Guo J, Feng Y, Li XX, Zhang JG, Jiang XM, Yin YF, Li S (2022). The three-dimensional structure of bordered pit in xylem of Cedrus deodara base on focused ion beam scanning electron microscopy. J Chin Electron Microsc Soc 41, 66-71. (in Chinese) |
| 王静, 王杰, 郭娟, 冯韵, 李喜霞, 张建国, 姜笑梅, 殷亚方, 李姗 (2022). 基于聚焦离子束-扫描电子显微技术的雪松木质部具缘纹孔三维重构. 电子显微学报 41, 66-71. | |
| [68] |
Wang Q, Huang YG, Ren ZJ, Zhang XX, Ren J, Su JQ, Zhang C, Tian J, Yu YJ, Gao GF, Li LG, Kong ZS (2020). Transfer cells mediate nitrate uptake to control root nodule symbiosis. Nat Plants 6, 800-808.
DOI PMID |
| [69] |
Wang Z, Herremans E, Janssen S, Cantre D, Verboven P, Nicolaï B (2018). Visualizing 3D food microstructure using tomographic methods: advantages and disadvantages. Annu Rev Food Sci Technol 9, 323-343.
DOI URL |
| [70] |
Wang Z, Verboven P, Nicolai B (2017). Contrast-enhanced 3D micro-CT of plant tissues using different impregnation techniques. Plant Methods 13, 105.
DOI PMID |
| [71] |
Wei DG, Jacobs S, Modla S, Zhang S, Young CL, Cirino R, Caplan J, Czymmek K (2012). High-resolution three- dimensional reconstruction of a whole yeast cell using focused-ion beam scanning electron microscopy. BioTechniques 53, 41-48.
DOI URL |
| [72] |
Wen WL, Wang JL, Zhao YX, Wang CY, Liu K, Chen B, Wang YQ, Duan MX, Guo XY (2024). 3D morphological feature quantification and analysis of corn leaves. Plant Phenomics 6, 0225.
DOI URL |
| [73] | Wu D (2019). Nondestructive Extraction of Rice Tiller Traits Based on Micro-CT. PhD dissertation. Wuhan: Huazhong Agricultural University. pp. 1-128. (in Chinese) |
| 吴迪 (2019). 基于Micro-CT的水稻茎部性状无损提取关键技术研究. 博士论文. 武汉: 华中农业大学. pp. 1-128. | |
| [74] |
Xiao Z, Stait-Gardner T, Willis SA, Price WS, Moroni FJ, Pagay V, Tyerman SD, Schmidtke LM, Rogiers SY (2021). 3D visualisation of voids in grapevine flowers and berries using X-ray micro computed tomography. Aust J Grape Wine Res 27, 141-148.
DOI URL |
| [75] |
Yamakawa S, Kato Y, Taniguchi M, Oi T (2023). Intracellular positioning of mesophyll chloroplasts following to aggregative movement in Setaria viridis analysed three- dimensionally with a confocal laser scanning microscope. Flora 306, 152364.
DOI URL |
| [76] |
Zechmann B, Möstl S, Zellnig G (2022). Volumetric 3D reconstruction of plant leaf cells using SEM, ion milling, TEM, and serial sectioning. Planta 255, 118.
DOI PMID |
| [77] | Zhang K, Zhang Y, Hu ZJ, Ji G, Sun F (2010). Development and frontier of electron microscopy 3D reconstruction. Acta Biophys Sin 26, 533-559. (in Chinese) |
| 张凯, 张艳, 胡仲军, 季刚, 孙飞 (2010). 电子显微三维重构技术发展与前沿. 生物物理学报 26, 533-559. | |
| [78] |
Zhang X, Man Y, Zhuang XH, Shen JB, Zhang Y, Cui YN, Yu M, Xing JJ, Wang GC, Lian N, Hu ZJ, Ma LY, Shen WW, Yang SY, Xu HM, Bian JH, Jing YP, Li XJ, Li RL, Mao TL, Jiao YL, Sodmergen, Ren HY, Lin JX (2021). Plant multiscale networks: charting plant connectivity by multi-level analysis and imaging techniques. Sci China Life Sci 64, 1392-1422.
DOI PMID |
| [79] |
Zhou N, Sun JS, Zhang RN, Ye R, Li JJ, Bai ZD, Zhou S, Chen Q, Zuo C (2023). Quasi-isotropic high-resolution Fourier ptychographic diffraction tomography with opposite illuminations. ACS Photonics 10, 2461-2466.
DOI URL |
| [80] | Zhu YY (2023). 3D reconstruction of ancient building structure scene based on computer image recognition. Int J Inf Technol Syst Approach 16, 1-14. |
| [1] | 林淼. 玉米雄穗分枝数的激素“密码”[J]. 植物学报, 2026, 61(1): 1-0. |
| [2] | 张靓靓, 汪贤挺, 陈勇, 朱逸凡, 卢新元, Zaitseva Svetlana Mikhailovna, 杨海芸. 国家一级保护野生植物银缕梅组培快繁体系的建立[J]. 植物学报, 2026, 61(1): 1-0. |
| [3] | 郭馨怡, 杨珍, 王佳, 林雁冰, 上官周平, 樊妙春. 我国旱区乡土豆科植物根际促生菌资源的挖掘和应用潜力[J]. 植物学报, 2026, 61(1): 1-0. |
| [4] | 牛钰凡, 陈玲玲, 苏晨, 王雷. 成花素FT:环境感知与发育调控的信号枢纽[J]. 植物学报, 2026, 61(1): 0-0. |
| [5] | 周熠玮, 范燕萍. 基于PTR-ToF-MS实时高效检测植物特定香气挥发性化合物含量[J]. 植物学报, 2026, 61(1): 0-0. |
| [6] | 张瑞 赵忠. 细胞壁动态调控及其对植物发育的影响[J]. 植物学报, 2026, 61(1): 0-0. |
| [7] | 赵洁, 李静, 李雨欣, 黄奕, 杨杰, 李霞. 活性氧在植物种子休眠释放和萌发中的作用研究进展[J]. 植物学报, 2025, 60(6): 978-992. |
| [8] | 罗号东, 刘勇波. 植物卷须发生及调控机制研究进展[J]. 植物学报, 2025, 60(6): 993-1004. |
| [9] | 穆丹, 龙洪婕, 陈静怡, 胡心怡, 方世涛, 周琪, 步佳豪, 白雅茹, 张原蓉, 尚钰祺, 石旌男, 赫倩文, 毕晨悦, 张欣燕, 秦华光, 饶玉春, 孙廷哲. 基于“手作+课程包”的植物科普实践模式探索[J]. 植物学报, 2025, 60(6): 1017-1027. |
| [10] | 江亚楠, 徐雨青, 魏毅铤, 陈钧, 张容菀, 赵蓓蓓, 林宇翔, 饶玉春. 水稻抗病调控机制研究进展[J]. 植物学报, 2025, 60(5): 734-748. |
| [11] | 王笑, 徐昌文, 钱虹萍, 李思博, 林金星, 崔亚宁. 植物细胞壁参与免疫反应的机制及其原位非标记成像方法[J]. 植物学报, 2025, 60(5): 773-785. |
| [12] | 刘德水, 岳宁, 刘玉乐. 植物免疫机制新突破[J]. 植物学报, 2025, 60(5): 669-678. |
| [13] | 徐羽丰, 周冕. 植物免疫的转录后调控[J]. 植物学报, 2025, 60(5): 704-721. |
| [14] | 吴玉俊, 李英菊, 罗巧玉, 马永贵. 光控植物免疫: 从光信号通路到免疫应答的调控网络[J]. 植物学报, 2025, 60(5): 786-803. |
| [15] | 肖银燕, 于华, 万里. 植物免疫研究: 机制突破和应用创新[J]. 植物学报, 2025, 60(5): 693-703. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||