

植物学报 ›› 2025, Vol. 60 ›› Issue (6): 863-874.DOI: 10.11983/CBB24156 cstr: 32102.14.CBB24156
所属专题: 虚拟专辑 | 干旱响应与适应 | 整合生物学期刊集群跨刊组建
收稿日期:2024-10-17
接受日期:2025-03-18
出版日期:2025-11-10
发布日期:2025-03-18
通讯作者:
刘香利
基金资助:
Yue Sun, Shujuan Guo, Huixian Zhao, Meng Ma, Xiangli Liu*( )
)
Received:2024-10-17
Accepted:2025-03-18
Online:2025-11-10
Published:2025-03-18
Contact:
Xiangli Liu
摘要: 14-3-3蛋白广泛参与植物生长发育、代谢和非生物逆境信号转导过程。该研究克隆了小麦(Triticum aestivum) 14-3-3蛋白TaGRF3-D基因, TaGRF3-D基因编码261个氨基酸残基, 在单子叶植物中高度保守, 其与乌拉尔图小麦(T. urartu)的TuGF14d和大麦(Hordeum vulgare)的HvGF14a氨基酸序列完全相同; TaGRF3-D启动子区含有脱落酸等激素响应元件和多个非生物胁迫响应元件。亚细胞定位结果显示, TaGRF3-D蛋白主要定位于细胞膜与细胞核。对过表达TaGRF3-D基因的拟南芥(Arabidopsis thaliana)转基因株系ABA敏感性及干旱胁迫耐受性分析发现, TaGRF3-D过表达拟南芥在PEG和ABA处理下根长显著大于野生型, 干旱胁迫后存活率显著高于野生型。进一步利用酵母双杂交(yeast two-hybrid, Y2H)实验对TaGRF3-D蛋白与小麦AREBs/ABFs (ABA-responsive element binding proteins/ABA-responsive element binding factors)蛋白进行互作分析, 结果表明TaGRF3-D蛋白与TaABF3-B、TaABF4-A、TaABF15-D、TaABF16-B、TaABF17-D和 TaABF18-B存在相互作用; 而与TaABF1-D、TaABF2-A和TabABF19-A不互作。综上表明, TaABF3-D可能通过与TaABFs蛋白互作响应ABA信号, 从而提高转基因植株对干旱胁迫的耐受性。研究结果为小麦TaGRF3-D基因逆境胁迫响应功能研究奠定了基础。
孙月, 郭树娟, 赵惠贤, 马猛, 刘香利. 小麦14-3-3蛋白TaGRF3-D基因克隆及功能分析. 植物学报, 2025, 60(6): 863-874.
Yue Sun, Shujuan Guo, Huixian Zhao, Meng Ma, Xiangli Liu. Cloning and Functional Analysis of the 14-3-3 Protein-encoding Gene TaGRF3-D in Wheat (Triticum aestivum). Chinese Bulletin of Botany, 2025, 60(6): 863-874.
| Primers | Primer sequence (5′-3′) | Primer usage |
|---|---|---|
| TaGRF3-D-F | ATGTCTACCGCTGAGGCAAC | TaGRF3-D gene cloning |
| TaGRF3-D-R | TCAGTGCCCCTCTCCCTCAG | |
| 1304-R | GTATCTTGAAAAGCATTGAACACC | Genomic PCR detection |
| Actin-F | TCGCTGACCGTATGAGCAAAG | Actin gene semi quantitative analysis |
| Actin-R | TGTGAACGATTCCTGGACCTG | |
| TaGRF3-D-RT-F | TCCTGAACTCTCCAGACCGT | TaGRF3-D gene semi quantitative analysis |
| TaGRF3-D-RT-R | CCTCTGCGTTATCGGAGGTC | |
| TaGRF3-D-BD-F | atggccatggaggccgaattcATGTCTACCGCTGAGGCAACC | BD-TaGRF3-D vector construction |
| TaGRF3-D-BD-R | ttatgcggccgctgcaggtcgacTCAGTGCCCCTCTCCCTCA |
表1 TaGRF3-D基因克隆和载体构建引物信息
Table 1 Primer information for TaGRF3-D gene cloning and vector construction
| Primers | Primer sequence (5′-3′) | Primer usage |
|---|---|---|
| TaGRF3-D-F | ATGTCTACCGCTGAGGCAAC | TaGRF3-D gene cloning |
| TaGRF3-D-R | TCAGTGCCCCTCTCCCTCAG | |
| 1304-R | GTATCTTGAAAAGCATTGAACACC | Genomic PCR detection |
| Actin-F | TCGCTGACCGTATGAGCAAAG | Actin gene semi quantitative analysis |
| Actin-R | TGTGAACGATTCCTGGACCTG | |
| TaGRF3-D-RT-F | TCCTGAACTCTCCAGACCGT | TaGRF3-D gene semi quantitative analysis |
| TaGRF3-D-RT-R | CCTCTGCGTTATCGGAGGTC | |
| TaGRF3-D-BD-F | atggccatggaggccgaattcATGTCTACCGCTGAGGCAACC | BD-TaGRF3-D vector construction |
| TaGRF3-D-BD-R | ttatgcggccgctgcaggtcgacTCAGTGCCCCTCTCCCTCA |
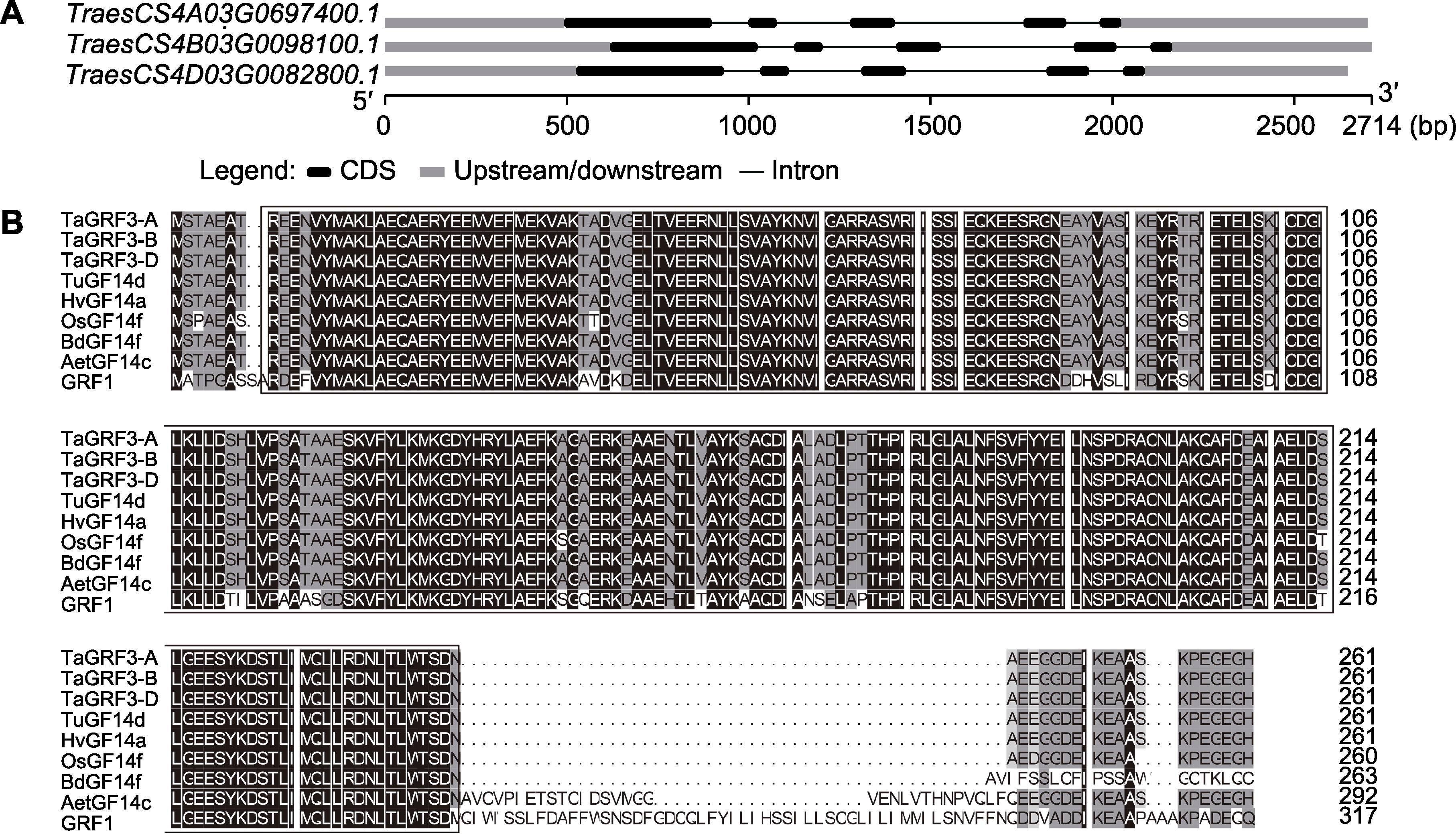
图1 TaGRF3-A/B/D基因结构分析(A)及TaGRF3-A/B/D蛋白与其它物种同源蛋白的氨基酸序列比对(B) B图中黑色方框表示14-3-3蛋白家族保守结构域。
Figure 1 Structural analysis of the TaGRF3-A/B/D gene (A) and alignment of the amino acid sequences of TaGRF3-A/B/D with those of homologous proteins of other species (B) The black box in Figure B indicates the conserved domain of the 14-3-3 protein family.
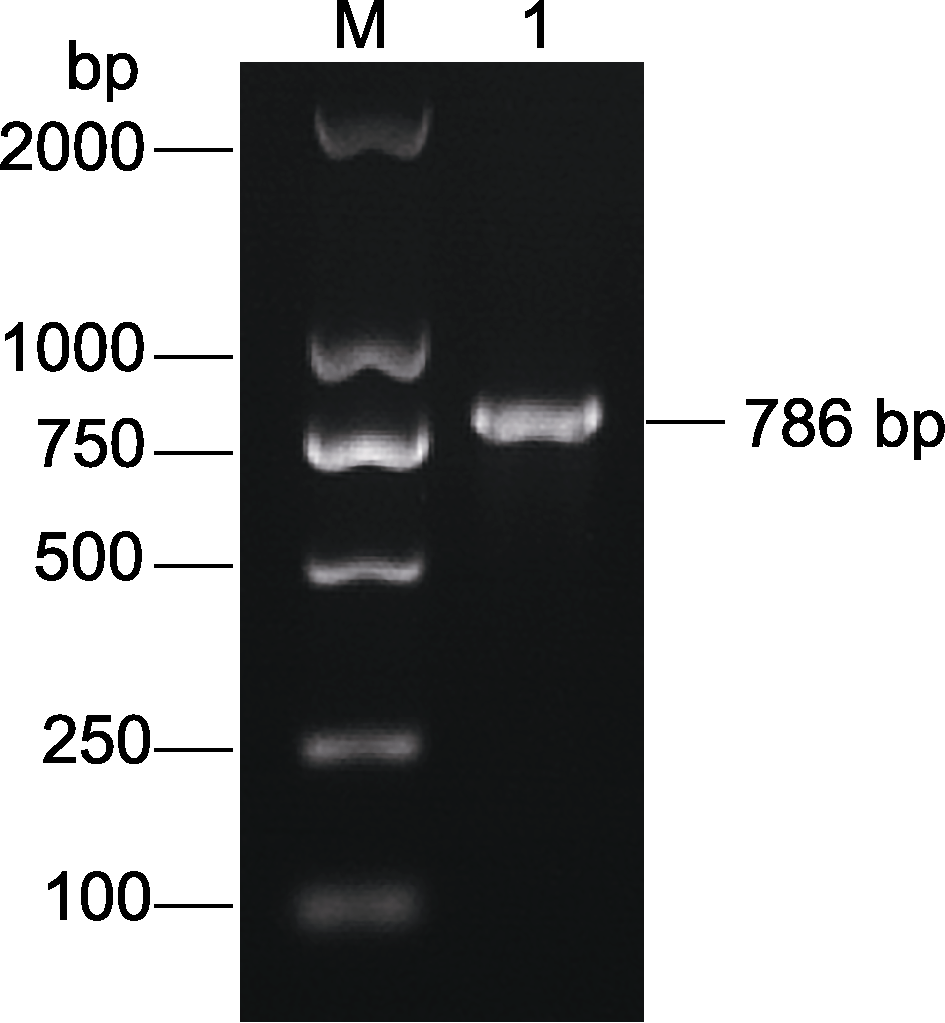
图3 TaGRF3-D cDNA PCR扩增 M: DL 2000 bp DNA marker; 1: cDNA扩增产物
Figure 3 Amplification of TaGRF3-D cDNA via PCR M: DL 2000 bp DNA marker; 1: Amplification product of cDNA
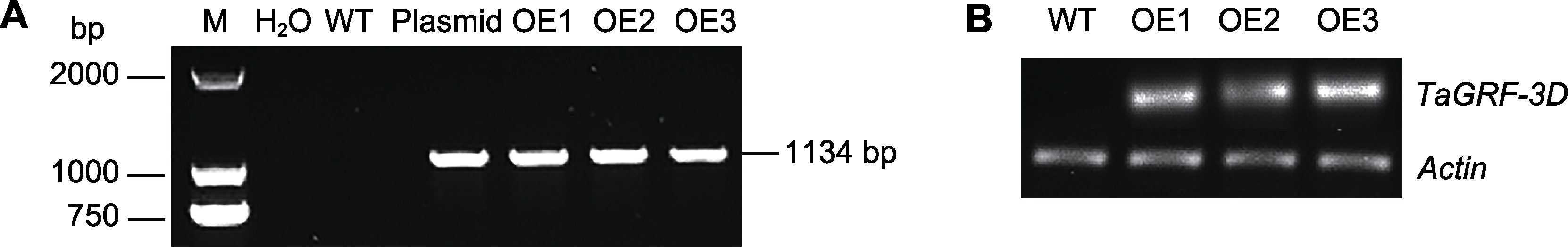
图5 TaGRF3-D转基因株系和野生型PCR鉴定(A)以及TaGRF3-D基因表达分析(B) M: DL 2000 DNA marker; H2O: 空白对照; WT: 野生型; Plasmid: pCAMBIA1304-TaGRF3-D; OE1-OE3: 转基因株系
Figure 5 PCR identification (A) and TaGRF3-D gene expression analysis (B) of the TaGRF3-D transgenic lines and the wild- type M: DL 2000 DNA marker; H2O: Blank control; WT: Wild type; Plasmid: pCAMBIA1304-TaGRF3-D; OE1-OE3: Transgenic lines
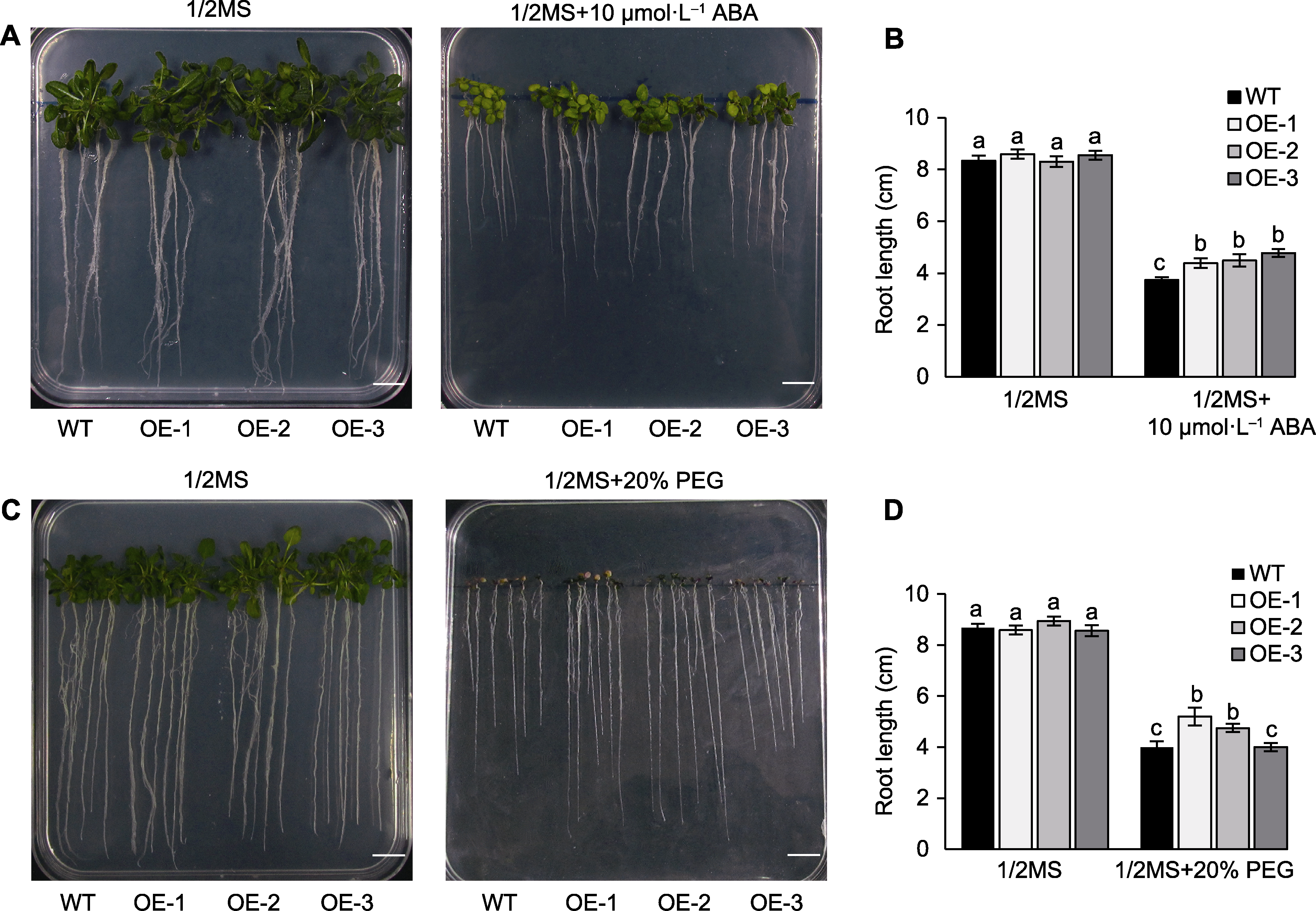
图6 脱落酸(ABA)和聚乙二醇(PEG)处理下TaGRF3-D转基因拟南芥根长分析 (A) ABA处理下的表型; (B) ABA处理下的根长统计; (C) PEG处理下的表型; (D) PEG处理下的根长统计。不同小写字母表示根据ANOVA分析结果(n=3)数据间存在显著性差异(P<0.05)。WT: 野生型; Bars=1 cm
Figure 6 Analysis of the root length of the TaGRF3-D Arabidopsis thaliana transgenic lines under abscisic acid (ABA) and polyethylene glycol (PEG) treatment (A) Growth phenotype under ABA treatment; (B) Root length statistics under ABA treatment; (C) Growth phenotype under PEG treatment; (D) Root length statistics under PEG treatment. Different lowercase letters indicate significant differences (P<0.05) among the data according to the results of ANOVA analysis (n=3). WT: Wild type; Bars=1 cm
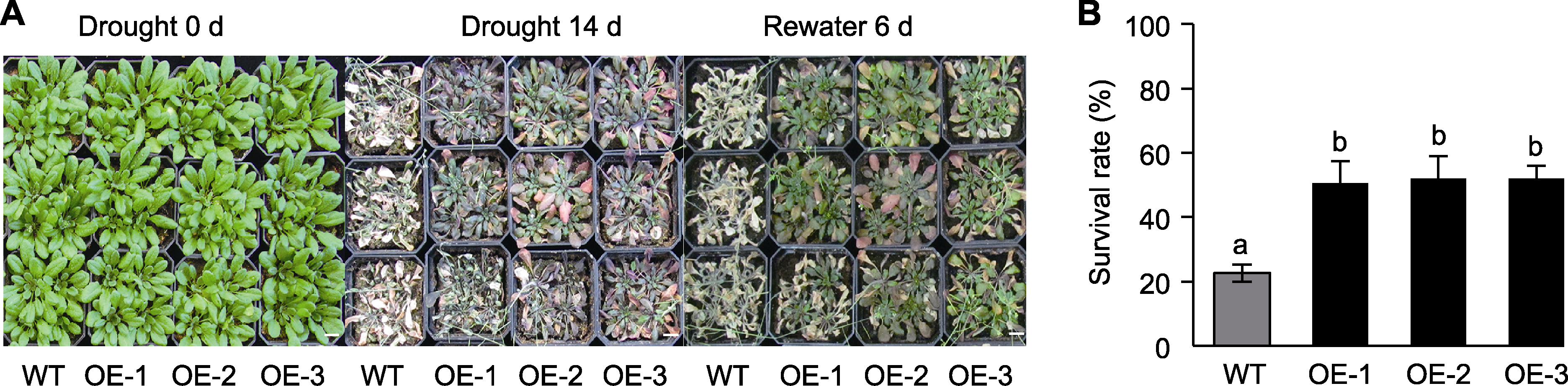
图7 干旱胁迫下TaGRF3-D转基因拟南芥和野生型(WT)的表型(A)和存活率(B) 不同小写字母表示根据ANOVA分析结果(n=3)数据间存在显著性差异(P<0.05)。Bars=1 cm
Figure 7 Phenotype (A) and survival rate (B) of the TaGRF3-D transgenic Arabidopsis thaliana and the wild type (WT) under drought stress Different lowercase letters indicate significant differences (P<0.05) among the data according to the results of ANOVA (n=3). Bars=1 cm
| [1] |
Aitken A (2006). 14-3-3 proteins: a historic overview. Semin Cancer Biol 16, 162-172.
DOI PMID |
| [2] |
Campo S, Peris-Peris C, Montesinos L, Peñas G, Messeguer J, San Segundo B (2012). Expression of the maize ZmGF14-6 gene in rice confers tolerance to drought stress while enhancing susceptibility to pathogen infection. J Exp Bot 63, 983-999.
DOI URL |
| [3] |
Choi HI, Hong JH, Ha JO, Kang JY, Kim SY (2000). ABFs, a family of ABA-responsive element binding factors. J Biol Chem 275, 1723-1730.
DOI PMID |
| [4] |
Fu HA, Subramanian RR, Masters SC (2000). 14-3-3 proteins: structure, function, and regulation. Annu Rev Pharmacol Toxicol 40, 617-647.
PMID |
| [5] |
Gietz RD, Woods RA (2002). Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol 350, 87-96.
PMID |
| [6] | Guo SJ, Sun Y, Zheng HY, Zhao HX, Ma M, Liu XL (2023). Genome-wide identification and expression analysis of ABF/AREB/ABI5 gene family in wheat (Triticum aestivum). J Agric Biotechnol 31, 667-681. (in Chinese) |
| 郭树娟, 孙月, 郑昊元, 赵惠贤, 马猛, 刘香利 (2023). 小麦ABF/AREB/ABI5基因家族全基因组鉴定与表达分析. 农业生物技术学报 31, 667-681. | |
| [7] | He Y, Zhang Y, Chen LH, Wu CL, Luo QC, Zhang F, Wei QH, Li KX, Chang JL, Yang GX, He GY (2017). A member of the 14-3-3 gene family in Brachypodium distachyon, BdGF14d, confers salt tolerance in transgenic tobacco plants. Front Plant Sci 8, 340. |
| [8] |
Ho SL, Huang LF, Lu CA, He SL, Wang CC, Yu SP, Chen J, Yu SM (2013). Sugar starvation- and GA-inducible calcium-dependent protein kinase 1 feedback regulates GA biosynthesis and activates a 14-3-3 protein to confer drought tolerance in rice seedlings. Plant Mol Biol 81, 347-361.
DOI URL |
| [9] |
Huang Y, Wang WS, Yu H, Peng JH, Hu ZR, Chen L (2022). The role of 14-3-3 proteins in plant growth and response to abiotic stress. Plant Cell Rep 41, 833-852.
DOI |
| [10] |
Jiang W, Tong T, Li W, Huang ZH, Chen G, Zeng FR, Riaz A, Amoanimaa-Dede H, Pan R, Zhang WY, Deng FL, Chen ZH (2023). Molecular evolution of plant 14-3-3 proteins and function of Hv14-3-3A in stomatal regulation and drought tolerance. Plant Cell Physiol 63, 1857-1872.
DOI URL |
| [11] |
Johnson RR, Shin M, Shen JQ (2008). The wheat PKABA1-interacting factor TaABF1 mediates both abscisic acid- suppressed and abscisic acid-induced gene expression in bombarded aleurone cells. Plant Mol Biol 68, 93-103.
DOI URL |
| [12] |
Kaundal A, Ramu VS, Oh S, Lee S, Pant B, Lee HK, Rojas CM, Senthil-Kumar M, Mysore KS (2017). GENERAL CONTROL NONREPRESSIBLE4 degrades 14-3-3 and the RIN4 complex to regulate stomatal aperture with implications on nonhost disease resistance and drought tolerance. Plant Cell 29, 2233-2248.
DOI URL |
| [13] |
Kobayashi F, Maeta E, Terashima A, Takumi S (2008). Positive role of a wheat HvABI5 ortholog in abiotic stress response of seedlings. Physiol Plant 134, 74-86.
DOI PMID |
| [14] |
Latz A, Becker D, Hekman M, Müller T, Beyhl D, Marten I, Eing C, Fischer A, Dunkel M, Bertl A, Rapp UR, Hedrich R (2007). TPK1, a Ca2+-regulated Arabidopsis vacuole two-pore K+ channel is activated by 14-3-3 proteins. Plant J 52, 449-459.
DOI PMID |
| [15] |
Latz A, Mehlmer N, Zapf S, Mueller TD, Wurzinger B, Pfister B, Csaszar E, Hedrich R, Teige M, Becker D (2013). Salt stress triggers phosphorylation of the Arabidopsis vacuolar K+ channel TPK1 by calcium-dependent protein kinases (CDPKs). Mol Plant 6, 1274-1289.
DOI URL |
| [16] |
Li FF, Mei FM, Zhang YF, Li SM, Kang ZS, Mao HD (2020). Genome-wide analysis of the AREB/ABF gene lineage in land plants and functional analysis of TaABF3 in Arabidopsis. BMC Plant Biol 20, 558.
DOI |
| [17] |
Lu G, DeLisle AJ, de Vetten NC, Ferl RJ (1992). Brain proteins in plants: an Arabidopsis homolog to neurotransmitter pathway activators is part of a DNA binding complex. Proc Natl Acad Sci USA 89, 11490-11494.
PMID |
| [18] |
Ma YM, Wu ZY, Dong JF, Zhang SH, Zhao JL, Yang TF, Yang W, Zhou L, Wang J, Chen JS, Liu Q, Liu B (2023). The 14-3-3 protein OsGF14f interacts with OsbZIP23 and enhances its activity to confer osmotic stress tolerance in rice. Plant Cell 35, 4173-4189.
DOI URL |
| [19] |
Ren YR, Yang YY, Zhang R, You CX, Zhao Q, Hao YJ (2019). MdGRF11, an apple 14-3-3 protein, acts as a positive regulator of drought and salt tolerance. Plant Sci 288, 110219.
DOI URL |
| [20] |
Schoonheim PJ, Costa Pereira DD, De Boer AH (2009). Dual role for 14-3-3 proteins and ABF transcription factors in gibberellic acid and abscisic acid signaling in barley (Hordeum vulgare) aleurone cells. Plant Cell Environ 32, 439-447.
DOI URL |
| [21] |
Shao WN, Chen W, Zhu XG, Zhou XY, Jin YY, Zhan C, Liu GS, Liu X, Ma DF, Qiao YL (2021). Genome-wide identification and characterization of wheat 14-3-3 genes unravels the role of TaGRF6-A in salt stress tolerance by binding MYB transcription factor. Int J Mol Sci 22, 1904.
DOI URL |
| [22] | Shen YX (2018). Identification and Expression Analysis of the 14-3-3 Gene Family in Triticum aestivum (L.). Master’s thesis. Yangling: Northwest A&F University. pp. 30-36. (in Chinese) |
| 申玉霞 (2018). 小麦14-3-3蛋白基因家族鉴定和表达分析. 硕士论文. 杨凌: 西北农林科技大学. pp. 30-36. | |
| [23] | Sun XL, Sun MZ, Jia BW, Chen C, Qin ZW, Yang KJ, Shen Y, Meiping Z, Mingyang C, Zhu YM (2015). A 14-3-3 family protein from wild soybean (Glycine soja) regulates ABA sensitivity in Arabidopsis. PLoS One 10, e0146163. |
| [24] |
Yang L, You J, Wang YP, Li JZ, Quan WL, Yin MZ, Wang QF, Chan ZL (2017). Systematic analysis of the G-box factor 14-3-3 gene family and functional characterization of GF14a in Brachypodium distachyon. Plant Physiol Biochem 117, 1-11.
DOI URL |
| [25] |
Zhang XR, Henriques R, Lin SS, Niu QW, Chua NH (2006). Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat Protoc 1, 641-646.
DOI |
| [26] |
Zhang Y, He Y, Zhao HY, Wang Y, Wu CL, Zhao YZ, Xue HN, Zhu QD, Zhang JL, Ou XQ (2024). The 14-3-3 protein BdGF14a increases the transcriptional regulation activity of BdbZIP62 to confer drought and salt resistance in tobacco. Plants 13, 245.
DOI URL |
| [27] |
Zhang Y, He Y, Zhao HY, Zhang Y, Yang J, Ou XQ, Zhang JL, Zhu QD (2023). A 14-3-3 protein-encoding gene, BdGF14g, confers better drought tolerance by regulating ABA biosynthesis and signaling. Plants 12, 3975.
DOI URL |
| [28] |
Zhang Y, Zhao HY, Zhou SY, He Y, Luo QC, Zhang F, Qiu D, Feng JL, Wei QH, Chen LH, Chen MJ, Chang JL, Yang GX, He GY (2018). Expression of TaGF14b, a 14-3- 3 adaptor protein gene from wheat, enhances drought and salt tolerance in transgenic tobacco. Planta 248, 117-137.
DOI PMID |
| [29] |
Zhao X, Li F, Li K (2021). The 14-3-3 proteins: regulators of plant metabolism and stress responses. Plant Biol 23, 531-539.
DOI URL |
| [30] | Zhou Y, Li BY, Li XB (2012). Roles of 14-3-3 proteins in regulating plant development. Chin Bull Bot 47, 55-64. (in Chinese) |
|
周颖, 李冰樱, 李学宝 (2012). 14-3-3蛋白对植物发育的调控作用. 植物学报 47, 55-64.
DOI |
| [1] | 樊蓓, 任敏, 王延峰, 党峰峰, 陈国梁, 程国亭, 杨金雨, 孙会茹. 番茄SlWRKY45转录因子在响应低温和干旱胁迫中的功能(长英文摘要)[J]. 植物学报, 2025, 60(2): 186-203. |
| [2] | 赵来鹏, 王柏柯, 杨涛, 李宁, 杨海涛, 王娟, 闫会转. SlHVA22l基因调节番茄耐旱性[J]. 植物学报, 2024, 59(4): 558-573. |
| [3] | 张盈川, 吴晓明玉, 陶保龙, 陈丽, 鲁海琴, 赵伦, 文静, 易斌, 涂金星, 傅廷栋, 沈金雄. Bna-miR43介导甘蓝型油菜响应干旱胁迫[J]. 植物学报, 2023, 58(5): 701-711. |
| [4] | 邝嘉怡, 李洪清, 沈文锦, 高彩吉. 基于TurboID的植物蛋白邻近标记实验方法[J]. 植物学报, 2021, 56(5): 584-593. |
| [5] | 车永梅, 孙艳君, 卢松冲, 侯丽霞, 范欣欣, 刘新. AtMYB77促进NO合成参与调控干旱胁迫下拟南芥侧根发育[J]. 植物学报, 2021, 56(4): 404-413. |
| [6] | 李佳馨, 李霞, 谢寅峰. 外源海藻糖增强高表达转玉米C4型PEPC水稻耐旱性的机制[J]. 植物学报, 2021, 56(3): 296-314. |
| [7] | 徐重益. 植物中验证蛋白相互作用的Pull-down和Co-IP技术[J]. 植物学报, 2020, 55(1): 62-68. |
| [8] | 张彤,郭亚璐,陈悦,马金姣,兰金苹,燕高伟,刘玉晴,徐珊,李莉云,刘国振,窦世娟. 水稻OsPR10A的表达特征及其在干旱胁迫应答过程中的功能[J]. 植物学报, 2019, 54(6): 711-722. |
| [9] | 郜怀峰,张亚飞,王国栋,孙希武,贺月,彭福田,肖元松. 钼在桃树干旱胁迫响应中的作用解析[J]. 植物学报, 2019, 54(2): 227-236. |
| [10] | 宋爱华, 张文斌, 孙姝兰, 李凌飞, 王小菁. 非洲菊原生质体制备及瞬时转化系统的建立[J]. 植物学报, 2017, 52(4): 511-519. |
| [11] | 景艳军, 林荣呈. 我国植物光信号转导研究进展概述[J]. 植物学报, 2017, 52(3): 257-270. |
| [12] | 刘小龙, 李霞, 钱宝云. 外源Ca2+对PEG处理下转C4型PEPC基因水稻光合生理的调节[J]. 植物学报, 2015, 50(2): 206-216. |
| [13] | 苗红霞, 王园, 徐碧玉, 刘菊华, 贾彩红, 张建斌, 王卓, 孙佩光, 金志强. 香蕉MaASR1基因的抗干旱作用[J]. 植物学报, 2014, 49(5): 548-559. |
| [14] | 许凤莲, 李姣姣, 杜春晖, 王娜, 张素巧. 一个具有S位点的水稻类受体激酶OsSRL参与干旱胁迫反应[J]. 植物学报, 2012, 47(5): 474-482. |
| [15] | 刘焱, 邢立静, 李俊华, 戴绍军. 水稻含有B-box锌指结构域的OsBBX25蛋白参与植物对非生物胁迫的响应[J]. 植物学报, 2012, 47(4): 366-378. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||