

植物学报 ›› 2024, Vol. 59 ›› Issue (5): 709-725.DOI: 10.11983/CBB24043 cstr: 32102.14.CBB24043
路笃贤1,2,3,4, 张严妍1,2,3,4, 刘艳5, 李岩竣1,2,3,4, 左新秀1,2,3,4, 林金星1,2,3,4, 崔亚宁1,2,3,4,*( )
)
收稿日期:2024-03-18
接受日期:2024-06-21
出版日期:2024-09-10
发布日期:2024-08-19
通讯作者:
崔亚宁
基金资助:
Duxian Lu1,2,3,4, Yanyan Zhang1,2,3,4, Yan Liu5, Yanjun Li1,2,3,4, Xinxiu Zuo1,2,3,4, Jinxing Lin1,2,3,4, Yaning Cui1,2,3,4,*( )
)
Received:2024-03-18
Accepted:2024-06-21
Online:2024-09-10
Published:2024-08-19
Contact:
Yaning Cui
摘要: 非编码RNA (ncRNA)是一类不具备蛋白质编码能力但有多种生物学功能的RNA分子, 广泛存在于各种生物体内。随着高通量测序技术的不断完善, 大量的非编码RNA被鉴定出来, 其功能和作用机制也逐渐被阐释。大量研究表明, 非编码RNA在植物生长发育和逆境胁迫响应中发挥重要作用。尽管对某一类非编码RNA调控植物生长发育及逆境响应的总结有很多, 但缺少对非编码RNA系统而全面的总结。因此, 该文首先简要介绍非编码RNA的分类及特征, 随后重点介绍非编码RNA在植物生长发育, 如种子休眠和萌发、根和叶的生长发育、花和果实的发育以及果实成熟方面的作用, 最后对非编码RNA在逆境胁迫响应中的功能及作用机制进行总结, 旨在全面论述非编码RNA在植物生长发育和胁迫响应中的分子调控机理, 以期为改良品种、提高农林业生产的产量和品质提供参考。
路笃贤, 张严妍, 刘艳, 李岩竣, 左新秀, 林金星, 崔亚宁. 非编码RNA在植物生长发育及逆境响应中的研究进展. 植物学报, 2024, 59(5): 709-725.
Duxian Lu, Yanyan Zhang, Yan Liu, Yanjun Li, Xinxiu Zuo, Jinxing Lin, Yaning Cui. Recent Advances of Non-coding RNA in Plant Growth, Development and Stress Response. Chinese Bulletin of Botany, 2024, 59(5): 709-725.
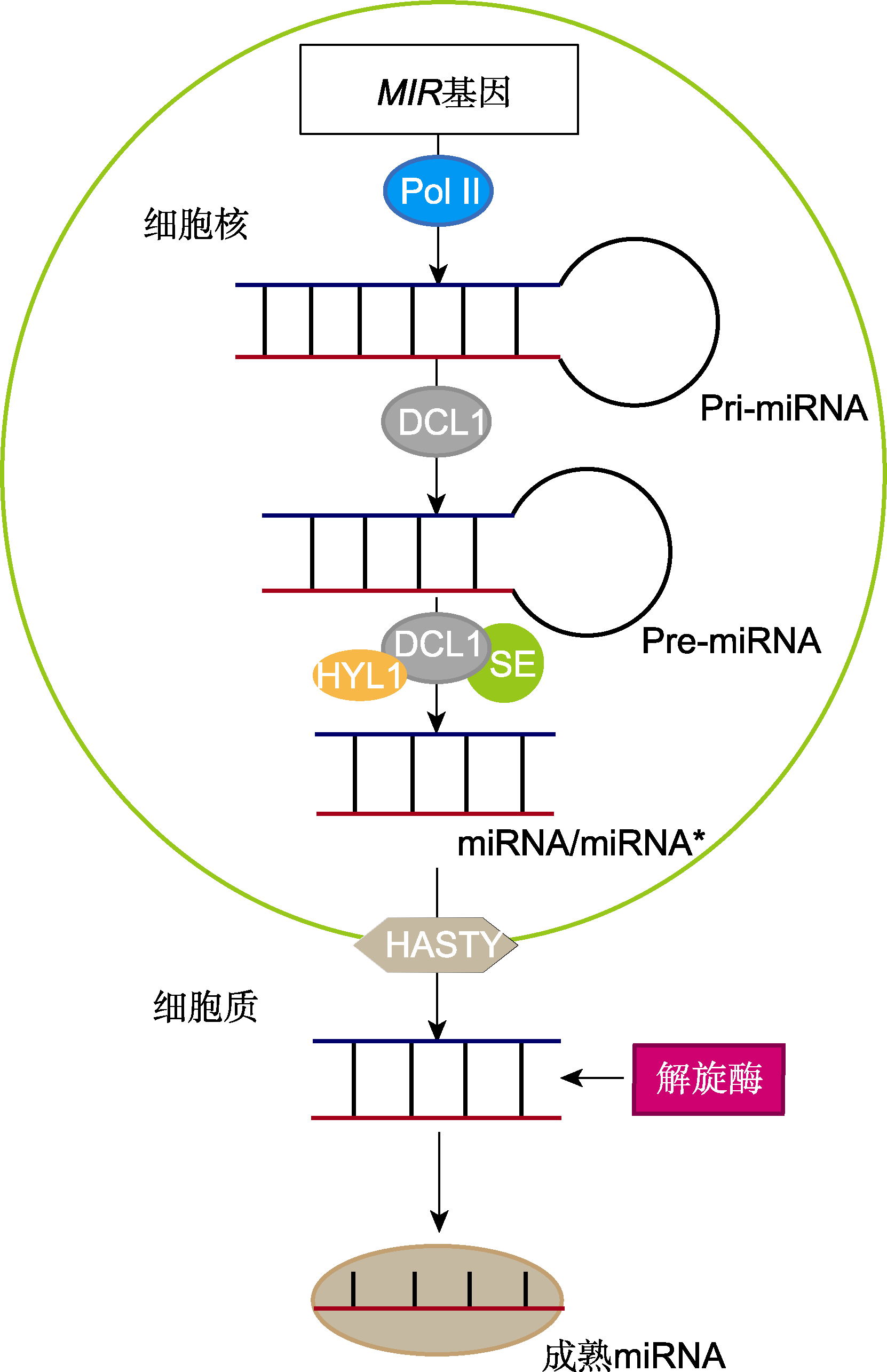
图2 微小RNA (miRNA)的形成机制 Pol II: RNA聚合酶II: Pri-miRNA: 初级miRNA: Pre-miRNA: 前体miRNA: DCL1: Dicer-like1核酸内切酶: HYL1: RNA结合蛋白Hyponastic Leaves1: SE: RNA结合蛋白SERRAT。DCL1、HYL1和SE共同组成复合体, 用来切割Pre-miRNA。
Figure 2 The formation mechanism of microRNA (miRNA) Pol II: RNA polymerase II; Pri-miRNA: Primary miRNA; Pre-miRNA: Precursor miRNA; DCL1: Dicer-like1 endonuclease; HYL1: RNA binding protein Hyponastic Leaves1; SE: RNA binding protein SERRAT. DCL1, HYL1 and SE together form a complex for cutting Pre-miRNA.
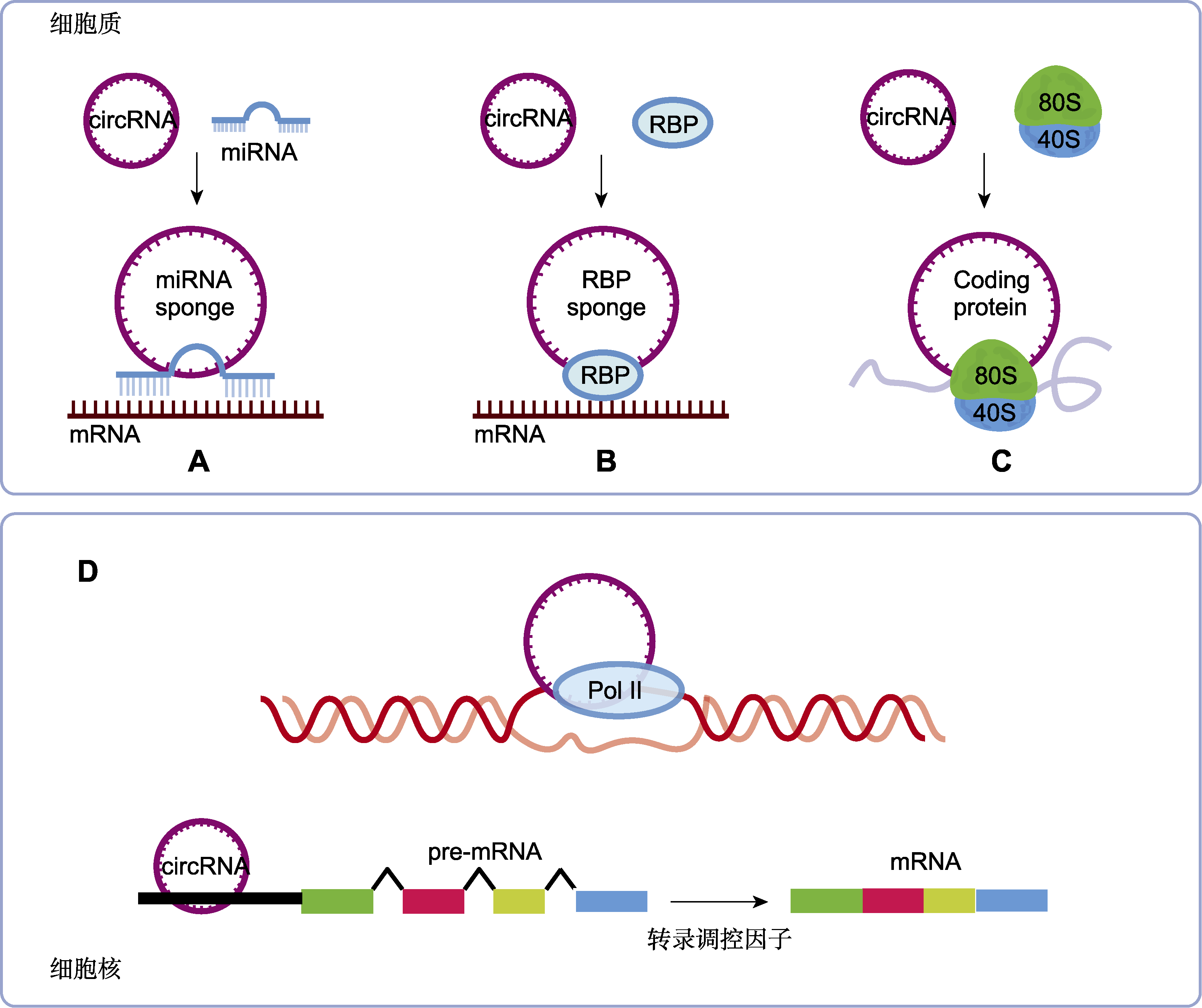
图3 环状RNA (circRNA)在生物体内的作用机制 细胞质内circRNA主要通过3种方式调控生物过程: (A) 与miRNA相互作用(circRNA通过与miRNA相互作用形成竞争性内源RNA (ceRNA), 负向调节miRNA的活性及其对下游靶基因的调控作用); (B) 与RBP (RNA结合蛋白)相互作用(RBP参与circRNA的剪接、加工和折叠等过程); (C) 编码多肽(一些circRNA具有编码多肽的潜能, 通过多肽调控相关基因的表达); (D) 细胞核内, circRNA主要通过调控转录过程调控相关基因的表达。
Figure 3 Mechanism of action of circular RNA (circRNA) in vivo In cytoplasm, circRNA regulates biological processes in three main ways: (A) Interaction with miRNA (circRNA can form competing endogenous RNA (ceRNA) by interacting with miRNA, negatively regulating the activity of miRNA and its regulatory effect on downstream target genes); (B) Interaction with RBP (RNA binding protein) (RBP is involved in the splicing, processing, folding and other processes of circRNA); (C) Encoding peptides (some circRNAs have the potential to encode peptides and regulate the expression of related genes through peptides); (D) In the nucleus, circRNA regulates the expression of related genes mainly by regulating the transcription process.
| 物种 | lncRNA | 生物学功能 | 参考文献 |
|---|---|---|---|
| 拟南芥(Arabidopsis thaliana) | APOLO | 侧根发育和避阴反应 | Mammarella et al., |
| FLAIL | 开花 | Liu et al., | |
| PUAR | 下胚轴伸长 | Zhu et al., | |
| HID1 | 种子萌发 | Wang et al., | |
| FL7 | 病原菌抗性 | Ai et al., | |
| SABC1 | 水杨酸合成和免疫 | Liu et al., | |
| lncRNA109897 | 病原菌抗性 | Zhou et al., | |
| T5120 | 根发育和硝酸盐同化 | Liu et al., | |
| 水稻(Oryza sativa) | Ef-cd | 开花 | Fang et al., |
| RIFLA | 开花 | Shin et al., | |
| MISSEN | 胚乳发育 | Zhou et al., | |
| ALEX1 | 病原菌抗性 | Yu et al., | |
| 小麦(Triticum aestivum) | VAS | 开花 | Xu et al., |
| 玉米(Zea mays) | GARR2 | 茎高和叶鞘长度等 | Li et al., |
| 苹果(Malus × domestica) | MLNC3.2MLNC4.6 | 花青素合成 | Yang et al., |
| MdLNC499 | 花青素合成 | Ma et al., | |
| MdLNC610 | 花青素合成 | Yu et al., | |
| 番茄(Solanum lycopersicum) | XLOC_1662和XLOC_033910 | 果实开裂 | Xue et al., |
| MSTRG.16920和MSTRG.7613 | 叶片衰老 | Li et al., | |
| lncRNA33732 | 晚疫病抗性 | Cui et al., | |
| 柑橘(Citrus reticulata) | XLOC_016898和XLOC_017200 | 果实成熟 | Ke et al., |
| 棉花(Gossypium hirsutum) | lnc-Ga13g0352 | 棉纤维发育 | Zheng et al., |
| MSTRG.2723.1、MSTRG.3390.1、MSTRG.48719.1和MSTRG.31176.1 | 棉纤维发育 | Zou et al., | |
| lncRNA973 | 盐胁迫反应 | Zhang et al., | |
| XH123 | 冷胁迫反应 | Cao et al., | |
| lncRNA2和lncRNA7 | 病原菌抗性 | Zhang et al., | |
| 杨树(Populus) | lncWOX5 | 不定根发育 | Qi et al., |
| MSL-lncRNAs | 性别稳定性 | Mao et al., | |
| LNC_004484、LNC_008014和 LNC010781等 | 木质部发育 | Sun et al., | |
| 毛果杨(P. trichocarpa) | Ptlinc-NAC72 | 耐盐性 | Ye et al., |
| 毛白杨(P. tomentosa) | MSTRG.22608.1和MSTRG.5634.1 | 镉胁迫反应 | Quan et al., |
| 白桦(Betula platyphylla) | LncRNA2705.1和LncRNA11415.1等 | 镉胁迫反应 | Wen et al., |
| BplncSIR1 | 盐胁迫反应 | Jia et al., | |
| 巨桉(Eucalyptus grandis) | TCONS_00004999 | 茎生长等 | Lin et al., |
表1 近5年发现的植物lncRNA及其主要功能
Table 1 lncRNA discovered in recent 5 years and their main functions in plant
| 物种 | lncRNA | 生物学功能 | 参考文献 |
|---|---|---|---|
| 拟南芥(Arabidopsis thaliana) | APOLO | 侧根发育和避阴反应 | Mammarella et al., |
| FLAIL | 开花 | Liu et al., | |
| PUAR | 下胚轴伸长 | Zhu et al., | |
| HID1 | 种子萌发 | Wang et al., | |
| FL7 | 病原菌抗性 | Ai et al., | |
| SABC1 | 水杨酸合成和免疫 | Liu et al., | |
| lncRNA109897 | 病原菌抗性 | Zhou et al., | |
| T5120 | 根发育和硝酸盐同化 | Liu et al., | |
| 水稻(Oryza sativa) | Ef-cd | 开花 | Fang et al., |
| RIFLA | 开花 | Shin et al., | |
| MISSEN | 胚乳发育 | Zhou et al., | |
| ALEX1 | 病原菌抗性 | Yu et al., | |
| 小麦(Triticum aestivum) | VAS | 开花 | Xu et al., |
| 玉米(Zea mays) | GARR2 | 茎高和叶鞘长度等 | Li et al., |
| 苹果(Malus × domestica) | MLNC3.2MLNC4.6 | 花青素合成 | Yang et al., |
| MdLNC499 | 花青素合成 | Ma et al., | |
| MdLNC610 | 花青素合成 | Yu et al., | |
| 番茄(Solanum lycopersicum) | XLOC_1662和XLOC_033910 | 果实开裂 | Xue et al., |
| MSTRG.16920和MSTRG.7613 | 叶片衰老 | Li et al., | |
| lncRNA33732 | 晚疫病抗性 | Cui et al., | |
| 柑橘(Citrus reticulata) | XLOC_016898和XLOC_017200 | 果实成熟 | Ke et al., |
| 棉花(Gossypium hirsutum) | lnc-Ga13g0352 | 棉纤维发育 | Zheng et al., |
| MSTRG.2723.1、MSTRG.3390.1、MSTRG.48719.1和MSTRG.31176.1 | 棉纤维发育 | Zou et al., | |
| lncRNA973 | 盐胁迫反应 | Zhang et al., | |
| XH123 | 冷胁迫反应 | Cao et al., | |
| lncRNA2和lncRNA7 | 病原菌抗性 | Zhang et al., | |
| 杨树(Populus) | lncWOX5 | 不定根发育 | Qi et al., |
| MSL-lncRNAs | 性别稳定性 | Mao et al., | |
| LNC_004484、LNC_008014和 LNC010781等 | 木质部发育 | Sun et al., | |
| 毛果杨(P. trichocarpa) | Ptlinc-NAC72 | 耐盐性 | Ye et al., |
| 毛白杨(P. tomentosa) | MSTRG.22608.1和MSTRG.5634.1 | 镉胁迫反应 | Quan et al., |
| 白桦(Betula platyphylla) | LncRNA2705.1和LncRNA11415.1等 | 镉胁迫反应 | Wen et al., |
| BplncSIR1 | 盐胁迫反应 | Jia et al., | |
| 巨桉(Eucalyptus grandis) | TCONS_00004999 | 茎生长等 | Lin et al., |
| 物种 | circRNA | 生物学功能 | 参考文献 |
|---|---|---|---|
| 拟南芥(Arabidopsis thaliana) | circGORK | 抗旱性 | Zhang et al., |
| 水稻(Oryza sativa) | circR5g05160 | 抗病原菌 | Fan et al., |
| Os02circ25329、Os06circ02797、Os03circ00204和Os05circ02465 | 种子萌发和耐盐性 | Zhou et al., | |
| 葡萄(Vitis vinifera) | Vv-circATS1 | 抗寒性 | Gao et al., |
| 毛果杨(Populus trichocarpa) | circ-CESA4、circ-IRX7和circ-GUX1 | 纤维素和半纤维素合成 | Liu et al., |
| 毛白杨(P. tomentosa) | circ_0003418 | 耐热性 | Song et al., |
表2 近5年发现的植物circRNA及其主要功能
Table 2 circRNA discovered in recent 5 years and their main functions in plant
| 物种 | circRNA | 生物学功能 | 参考文献 |
|---|---|---|---|
| 拟南芥(Arabidopsis thaliana) | circGORK | 抗旱性 | Zhang et al., |
| 水稻(Oryza sativa) | circR5g05160 | 抗病原菌 | Fan et al., |
| Os02circ25329、Os06circ02797、Os03circ00204和Os05circ02465 | 种子萌发和耐盐性 | Zhou et al., | |
| 葡萄(Vitis vinifera) | Vv-circATS1 | 抗寒性 | Gao et al., |
| 毛果杨(Populus trichocarpa) | circ-CESA4、circ-IRX7和circ-GUX1 | 纤维素和半纤维素合成 | Liu et al., |
| 毛白杨(P. tomentosa) | circ_0003418 | 耐热性 | Song et al., |
| [1] | Achard P, Herr A, Baulcombe DC, Harberd NP (2004). Modulation of floral development by a gibberellin-regulated microRNA. Development 131, 3357-3365. |
| [2] | Ai G, Li TL, Zhu H, Dong XH, Fu XW, Xia CY, Pan WY, Jing MF, Shen DY, Xia A, Tyler BM, Dou DL (2023). BPL3 binds the long non-coding RNA nalncFL7 to suppress FORKED-LIKE7 and modulate HAI1-mediated MPK3/6 dephosphorylation in plant immunity. Plant Cell 35, 598- 616. |
| [3] | Bao ML, Bian HW, Zha YL, Li FY, Sun YZ, Bai B, Chen ZH, Wang JH, Zhu MY, Han N (2014). miR396a-mediated basic helix-loop-helix transcription factor bHLH74 repression acts as a regulator for root growth in Arabidopsis seedlings. Plant Cell Physiol 55, 1343-1353. |
| [4] | Bernardi Y, Ponso MA, Belén F, Vegetti AC, Dotto MC (2022). MicroRNA miR394 regulates flowering time in Arabidopsis thaliana. Plant Cell Rep 41, 1375-1388. |
| [5] | Cao DC, Xu HM, Zhao YY, Deng X, Liu YX, Soppe WJJ, Lin JX (2016). Transcriptome and degradome sequencing reveals dormancy mechanisms of Cunninghamia lanceolata seeds. Plant Physiol 172, 2347-2362. |
| [6] | Cao ZY, Zhao T, Wang LY, Han J, Chen JW, Hao YP, Guan XY (2021). The lincRNA XH123 is involved in cotton cold-stress regulation. Plant Mol Biol 106, 521-531. |
| [7] | Chen J, Liu Q, Yuan LY, Shen WZ, Shi QX, Qi GJ, Chen T, Zhang ZF (2023a). Osa-miR162a enhances the resistance to the brown planthopper via α-linolenic acid metabolism in rice (Oryza sativa). J Agric Food Chem 71, 11847-11859. |
| [8] | Chen L, Luan YS, Zhai JM (2015). Sp-miR396a-5p acts as a stress-responsive genes regulator by conferring tolerance to abiotic stresses and susceptibility to Phytophthora nicotianae infection in transgenic tobacco. Plant Cell Rep 34, 2013-2025. |
| [9] | Chen L, Zhang P, Fan Y, Lu Q, Li Q, Yan JB, Muehlbauer GJ, Schnable PS, Dai MQ, Li L (2018). Circular RNAs mediated by transposons are associated with transcriptomic and phenotypic variation in maize. New Phytol 217, 1292-1306. |
| [10] | Chen YQ, Lu J, Wang WN, Fan CG, Yuan GZ, Sun JJ, Liu JY, Wang CQ (2023b). Rose long noncoding RNA lncWD83 promotes flowering by modulating ubiquitination of the floral repressor RcMYC2L. Plant Physiol 193, 2573- 2591. |
| [11] | Cheng JP, Zhang Y, Li ZW, Wang TY, Zhang XT, Zheng BL (2018). A lariat-derived circular RNA is required for plant development in Arabidopsis. Sci China Life Sci 61, 204-213. |
| [12] | Chung PJ, Park BS, Wang H, Liu J, Jang IC, Chua NH (2016). Light-inducible MiR163 targets PXMT1 transcripts to promote seed germination and primary root elongation in Arabidopsis. Plant Physiol 170, 1772-1782. |
| [13] | Conn VM, Hugouvieux V, Nayak A, Conos SA, Capovilla G, Cildir G, Jourdain A, Tergaonkar V, Schmid M, Zubieta C, Conn SJ (2017). A circRNA from SEPALLATA3 regulates splicing of its cognate mRNA through R-loop formation. Nat Plants 3, 17053. |
| [14] | Cui J, Jiang N, Meng J, Yang GL, Liu WW, Zhou XX, Ma N, Hou XX, Luan YS (2019). LncRNA33732-respiratory burst oxidase module associated with WRKY1 in tomato- Phytophthora infestans interactions. Plant J 97, 933-946. |
| [15] | Cuperus JT, Fahlgren N, Carrington JC (2011). Evolution and functional diversification of MIRNA genes. Plant Cell 23, 431-442. |
| [16] | Du QG, Wang K, Zou C, Xu C, Li WX (2018). The PILNCR1-miR399 regulatory module is important for low phosphate tolerance in maize. Plant Physiol 177, 1743-1753. |
| [17] | Fan J, Quan WL, Li GB, Hu XH, Wang Q, Wang H, Li XP, Luo XT, Feng Q, Hu ZJ, Feng H, Pu M, Zhao JQ, Huang YY, Li Y, Zhang Y, Wang WM (2020). circRNAs are involved in the rice-Magnaporthe oryzae interaction. Plant Physiol 182, 272-286. |
| [18] | Fang J, Zhang FT, Wang HR, Wang W, Zhao F, Li ZJ, Sun CH, Chen FM, Xu F, Chang SQ, Wu L, Bu QY, Wang PR, Xie JK, Chen F, Huang XH, Zhang YJ, Zhu XG, Han B, Deng XJ, Chu CC (2019). Ef-cd locus shortens rice maturity duration without yield penalty. Proc Natl Acad Sci USA 116, 18717-18722. |
| [19] | Gao Z, Li J, Luo M, Li H, Chen QJ, Wang L, Song SR, Zhao LP, Xu WP, Zhang CX, Wang SP, Ma C (2019). Characterization and cloning of grape circular RNAs identified the cold resistance-related Vv-circATS1. Plant Physiol 180, 966-985. |
| [20] | Gao ZH, Nie JT, Wang HS (2021). MicroRNA biogenesis in plant. Plant Growth Regul 93, 1-12. |
| [21] | Ghorbani A, Izadpanah K, Peters JR, Dietzgen RG, Mitter N (2018). Detection and profiling of circular RNAs in uninfected and maize Iranian mosaic virus-infected maize. Plant Sci 274, 402-409. |
| [22] | Guo GH, Liu XY, Sun FL, Cao J, Huo N, Wuda B, Xin MM, Hu ZR, Du JK, Xia R, Rossi V, Peng HR, Ni ZF, Sun QX, Yao YY (2018). Wheat miR9678 affects seed germination by generating phased siRNAs and modulating abscisic acid/gibberellin signaling. Plant Cell 30, 796-814. |
| [23] | He ZH, Lan YM, Zhou XK, Yu BJ, Zhu T, Yang F, Fu LY, Chao HY, Wang JH, Feng RX, Zuo SM, Lan WZ, Chen CL, Chen M, Zhao X, Hu KM, Chen DJ (2024). Single-cell transcriptome analysis dissects lncRNA-associated gene networks in Arabidopsis. Plant Commun 5, 100717. |
| [24] | Herbst J, Nagy SH, Vercauteren I, De Veylder L, Kunze R (2023). The long non-coding RNA LINDA restrains cellular collapse following DNA damage in Arabidopsis thaliana. Plant J 116, 1370-1384. |
| [25] | Hu G, Lei Y, Liu JF, Hao MY, Zhang ZN, Tang Y, Chen AM, Wu JH (2020). The ghr-miR164 and GhNAC100 modulate cotton plant resistance against Verticillium dahlia. Plant Sci 293, 110438. |
| [26] | Huang XP, Zhang HY, Guo R, Wang Q, Liu XZ, Kuang WG, Song HY, Liao JL, Huang YJ, Wang ZH (2021). Systematic identification and characterization of circular RNAs involved in flag leaf senescence of rice. Planta 253, 26. |
| [27] | Hyun Y, Richter R, Coupland G (2017). Competence to flower: age-controlled sensitivity to environmental cues. Plant Physiol 173, 36-46. |
| [28] | Jia YQ, Zhao HM, Niu YN, Wang YC (2023). Identification of birch lncRNAs and mRNAs responding to salt stress and characterization of functions of lncRNA. Hortic Res 10, uhac277. |
| [29] | Jia YQ, Zhao HM, Niu YN, Wang YC (2024). Long noncoding RNA from Betula platyphylla, BplncSIR1, confers salt tolerance by regulating BpNAC2 to mediate reactive oxygen species scavenging and stomatal movement. Plant Biotechnol J 22, 48-65. |
| [30] | Jiang YL, Li ZW, Liu XZ, Zhu TT, Xie K, Hou QC, Yan TW, Niu CF, Zhang SW, Yang MB, Xie RR, Wang J, Li JP, An XL, Wan XY (2021). ZmFAR1 and ZmABCG26 regulated by microRNA are essential for lipid metabolism in maize anther. Int J Mol Sci 22, 7916. |
| [31] | Jiao P, Ma RQ, Wang CL, Chen NN, Liu SY, Qu J, Guan SY, Ma YY (2022). Integration of mRNA and microRNA analysis reveals the molecular mechanisms underlying drought stress tolerance in maize (Zea mays L.). Front Plant Sci 13, 932667. |
| [32] | Jing WK, Gong FF, Liu GQ, Deng YL, Liu JQ, Yang WJ, Sun XM, Li YH, Gao JP, Zhou XF, Ma N (2023). Petal size is controlled by the MYB73/TPL/HDA19-miR159- CKX6 module regulating cytokinin catabolism in Rosa hybrida. Nat Commun 14, 7106. |
| [33] | Ke LL, Zhou ZW, Xu XW, Wang X, Liu YL, Xu YT, Huang Y, Wang ST, Deng XX, Chen LL, Xu Q (2019). Evolutionary dynamics of lincRNA transcription in nine citrus species. Plant J 98, 912-927. |
| [34] | Komatsu S, Kitai H, Suzuki HI (2023). Network regulation of microRNA biogenesis and target interaction. Cells 12, 306. |
| [35] | Koyama T, Mitsuda N, Seki M, Shinozaki K, Ohme-Takagi M (2010). TCP transcription factors regulate the activities of ASYMMETRIC LEAVES1 and miR164, as well as the auxin response, during differentiation of leaves in Arabidopsis. Plant Cell 22, 3574-3588. |
| [36] | Koyama T, Sato F, Ohme-Takagi M (2017). Roles of miR319 and TCP transcription factors in leaf development. Plant Physiol 175, 874-885. |
| [37] | Lee RC, Feinbaum RL, Ambros V (1993). The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75, 843-854. |
| [38] | Li BQ, Feng C, Zhang WH, Sun SM, Yue DD, Zhang XL, Yang XY (2023). Comprehensive non-coding RNA analysis reveals specific lncRNA/circRNA-miRNA-mRNA regulatory networks in the cotton response to drought stress. Int J Biol Macromol 253, 126558. |
| [39] | Li MZ, Si XY, Liu Y, Liu YC, Cheng X, Dai ZR, Yu XL, Ali M, Lu G (2022a). Transcriptomic analysis of ncRNA and mRNA interactions during leaf senescence in tomato. Int J Biol Macromol 222, 2556-2570. |
| [40] | Li N, Liu TT, Guo F, Yang JW, Shi YG, Wang SG, Sun DZ (2022b). Identification of long non-coding RNA-microRNA-mRNA regulatory modules and their potential roles in drought stress response in wheat (Triticum aestivum L.). Front Plant Sci 13, 1011064. |
| [41] | Li P, Yang H, Wang L, Liu HJ, Huo HQ, Zhang CJ, Liu AZ, Zhu AD, Hu JY, Lin YJ, Liu L (2019). Physiological and transcriptome analyses reveal short-term responses and formation of memory under drought stress in rice. Front Genet 10, 55. |
| [42] | Li W, Chen YD, Wang YL, Zhao J, Wang YJ (2022c). Gypsy retrotransposon-derived maize lncRNA GARR2 modulates gibberellin response. Plant J 110, 1433-1446. |
| [43] | Lin Z, Long JM, Yin Q, Wang B, Li HL, Luo JZ, Wang HC, Wu AM (2019). Identification of novel lncRNAs in Eucalyptus grandis. Ind Crop Prod 129, 309-317. |
| [44] | Liu F, Xu YR, Chang KX, Li SN, Liu ZG, Qi SD, Jia JB, Zhang M, Crawford NM, Wang Y (2019). The long noncoding RNA T5120 regulates nitrate response and assimilation in Arabidopsis. New Phytol 224, 117-131. |
| [45] | Liu H, Yuan K, Hu YY, Wang S, He QG, Feng CT, Liu JP, Wang ZH (2023a). Construction and analysis of the tapping panel dryness-related lncRNA/circRNA-miRNA-mRNA ceRNA network in latex of Hevea brasiliensis. Plant Physiol Biochem 205, 108156. |
| [46] | Liu HJ, Shen EL, Wu H, Ma WH, Chen H, Lin YJ (2022a). Trans-kingdom expression of an insect endogenous microRNA in rice enhances resistance to striped stem borer Chilo suppressalis. Pest Manag Sci 78, 770-777. |
| [47] | Liu NK, Xu YZ, Li Q, Cao YX, Yang DC, Liu SS, Wang XK, Mi YJ, Liu Y, Ding CX, Liu Y, Li Y, Yuan YW, Gao G, Chen JF, Qian WQ, Zhang XM (2022b). A lncRNA fine-tunes salicylic acid biosynthesis to balance plant immunity and growth. Cell Host Microbe 30, 1124-1138. |
| [48] | Liu X, Li DY, Zhang DL, Yin DD, Zhao Y, Ji CJ, Zhao XF, Li XB, He Q, Chen RS, Hu SN, Zhu LH (2018). A novel antisense long noncoding RNA, TWISTED LEAF, maintains leaf blade flattening by regulating its associated sense R2R3-MYB gene in rice. New Phytol 218, 774-788. |
| [49] | Liu XQ, Gao YB, Liao JK, Miao M, Chen K, Xi FH, Wei WT, Wang HH, Wang YS, Xu X, Reddy ASN, Gu LF (2021). Genome-wide profiling of circular RNAs, alternative splicing, and R-loops in stem-differentiating xylem of Populus trichocarpa. J Integr Plant Biol 63, 1294-1308. |
| [50] | Liu Y, Zhu QF, Li WY, Chen P, Xue J, Yu Y, Feng YZ (2023b). The pivotal role of noncoding RNAs in flowering time regulation. Genes 14, 2114. |
| [51] | Ma HY, Yang T, Li Y, Zhang J, Wu T, Song TT, Yao YC, Tian J (2021). The long noncoding RNA MdLNC499 bridges MdWRKY1 and MdERF109 function to regulate early- stage light-induced anthocyanin accumulation in apple fruit. Plant Cell 33, 3309-3330. |
| [52] | Mammarella MF, Lucero L, Hussain N, Muñoz-Lopez A, Huang Y, Ferrero L, Fernandez-Milmanda GL, Manavella P, Benhamed M, Crespi M, Ballare CL, Marcos JG, Cubas P, Ariel F (2023). Long noncoding RNA-mediated epigenetic regulation of auxin-related genes controls shade avoidance syndrome in Arabidopsis. EMBO J 42, e113941. |
| [53] | Mao JY, Wei SY, Chen YN, Yang YH, Yin TM (2023). The proposed role of MSL-lncRNAs in causing sex lability of female poplars. Hortic Res 10, uhad042. |
| [54] | Martin RC, Liu PP, Goloviznina NA, Nonogaki H (2010). microRNA, seeds, and Darwin? Diverse function of miRNA in seed biology and plant responses to stress. J Exp Bot 61, 2229-2234. |
| [55] | Miao CB, Wang Z, Zhang L, Yao JJ, Hua K, Liu X, Shi HZ, Zhu JK (2019). The grain yield modulator miR156 regulates seed dormancy through the gibberellin pathway in rice. Nat Commun 10, 3822. |
| [56] | Nelson BR, Makarewich CA, Anderson DM, Winders BR, Troupes CD, Wu FF, Reese AL, McAnally JR, Chen XW, Kavalali ET, Cannon SC, Houser SR, Bassel-Duby R, Olson EN (2016). A peptide encoded by a transcript annotated as long noncoding RNA enhances SERCA activity in muscle. Science 351, 271-275. |
| [57] | Qi HR, Wu L, Shen TF, Liu SA, Cai H, Ran N, Wang JL, Xu M (2023). Overexpression of the long non-coding RNA lncWOX5 negatively regulates the development of adventitious roots in Populus. Ind Crop Prod 192, 116054. |
| [58] | Qin T, Zhao HY, Cui P, Albesher N, Xiong LM (2017). A nucleus-localized long non-coding RNA enhances drought and salt stress tolerance. Plant Physiol 175, 1321- 1336. |
| [59] | Quan MY, Liu X, Xiao L, Chen PF, Song FY, Lu WJ, Song YP, Zhang DQ (2021). Transcriptome analysis and association mapping reveal the genetic regulatory network response to cadmium stress in Populus tomentosa. J Exp Bot 72, 576-591. |
| [60] | Ren YJ, Li MS, Wang WZ, Lan W, Schenke D, Cai DG, Miao Y (2021). MicroRNA840 (MIR840) accelerates leaf senescence by targeting the overlapping 3'UTRs of PPR and WHIRLY3 in Arabidopsis thaliana. Plant J 109, 126- 143. |
| [61] | Rodriguez RE, Mecchia MA, Debernardi JM, Schommer C, Weigel D, Palatnik JF (2010). Control of cell proliferation in Arabidopsis thaliana by microRNA miR396. Development 137, 103-112. |
| [62] | Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK (1976). Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci USA 73, 3852- 3856. |
| [63] | Sekhar S, Das S, Panda D, Mohanty S, Mishra B, Kumar A, Navadagi DB, Sah RP, Pradhan SK, Samantaray S, Baig MJ, Behera LB, Mohapatra T (2022). Identification of microRNAs that provide a low light stress tolerance- mediated signaling pathway during vegetative growth in rice. Plants 11, 2558. |
| [64] | Shahid S, Kim G, Johnson NR, Wafula E, Wang F, Coruh C, Bernal-Galeano V, Phifer T, Depamphilis CW, Westwood JH, Axtell MJ (2018). MicroRNAs from the parasitic plant Cuscuta campestris target host messenger RNAs. Nature 553, 82-85. |
| [65] | Shamloo-Dashtpagerdi R, Sisakht JN, Tahmasebi A (2022). MicroRNA miR1118 contributes to wheat (Triticum aestivum L.) salinity tolerance by regulating the Plasma Membrane Intrinsic Proteins1;5 (PIP1;5) gene. J Plant Physiol 278, 153827. |
| [66] | Sharma D, Tiwari M, Pandey A, Bhatia C, Sharma A, Trivedi PK (2016). MicroRNA858 is a potential regulator of phenylpropanoid pathway and plant development. Plant Physiol 171, 944-959. |
| [67] | Shin WJ, Nam AH, Kim JY, Kwak JS, Song JT, Seo HS (2022). Intronic long noncoding RNA, RICE FLOWERING ASSOCIATED (RIFLA), regulates OsMADS56-mediated flowering in rice. Plant Sci 320, 111278. |
| [68] | Silva GFFE, Silva EM, Da Silva Azevedo M, Guivin MAC, Ramiro DA, Figueiredo CR, Carrer H, Peres LEP, Nogueira FTS (2014). microRNA156-targeted SPL/SBP box transcription factors regulate tomato ovary and fruit development. Plant J 78, 604-618. |
| [69] | Song RJ, Ma SQ, Xu JJ, Ren X, Guo PL, Liu HJ, Li P, Yin F, Liu M, Wang Q, Yu L, Liu JL, Duan BW, Rahman NA, Wołczyński S, Li GM, Li XD (2023). A novel polypeptide encoded by the circular RNA ZKSCAN1 suppresses HCC via degradation of mTOR. Mol Cancer 22, 16. |
| [70] | Song YP, Bu CH, Chen PF, Liu P, Zhang DQ (2021). Miniature inverted repeat transposable elements cis-regulate circular RNA expression and promote ethylene biosynthesis, reducing heat tolerance in Populus tomentosa. J Exp Bot 72, 1978-1994. |
| [71] | Sun N, Bu YF, Wu XY, Ma XC, Yang HB, Du L, Li XJ, Xiao JW, Lin JX, Jing YP (2023). Comprehensive analysis of lncRNA-mRNA regulatory network in Populus associated with xylem development. J Plant Physiol 287, 154055. |
| [72] | Tang YJ, Qu ZP, Lei JJ, He RQ, Adelson DL, Zhu YL, Yang ZB, Wang D (2021). The long noncoding RNA FRILAIR regulates strawberry fruit ripening by functioning as a noncanonical target mimic. PLoS Genet 17, e1009461. |
| [73] | Tognacca RS, Botto JF (2021). Post-transcriptional regulation of seed dormancy and germination: current understanding and future directions. Plant Commun 2, 100169. |
| [74] | Traubenik S, Charon C, Blein T (2024). From environmental responses to adaptation: the roles of plant lncRNAs. Plant Physiol 195, 232-244. |
| [75] | Wang JW, Mao YB, Qi YJ (2016). Recent progress and prospects for plant non-coding RNAs. China Basic Sci 18(2), 22-29. (in Chinese) |
| 王佳伟, 毛颖波, 戚益军 (2016). 植物非编码RNA的研究进展与展望. 中国基础科学 18(2), 22-29. | |
| [76] | Wang WW, Liu Z, An XY, Jin YZ, Hou JM, Liu T (2022). Integrated high-throughput sequencing, microarray hybridization and degradome analysis uncovers microRNA- mediated resistance responses of maize to pathogen Curvularia lunata. Int J Mol Sci 23, 14038. |
| [77] | Wang XS, Chang XC, Jing Y, Zhao JL, Fang QW, Sun MY, Zhang YZ, Li WB, Li YG (2020). Identification and functional prediction of soybean circRNAs involved in low-temperature responses. J Plant Physiol 250, 153188. |
| [78] | Wang YJ, Wang YL, Chen YD (2020). Transposon-derived long noncoding RNA in plants. Chin Bull Bot 55, 768-776. (in Chinese) |
| 王益军, 王亚丽, 陈煜东 (2020). 转座子来源的植物长链非编码RNA. 植物学报 55, 768-776. | |
| [79] | Wang YQ, Fan YY, Fan D, Zhou XL, Jiao YT, Deng XW, Zhu DM (2023). The noncoding RNA HIDDEN TREASURE 1 promotes phytochrome B-dependent seed germination by repressing abscisic acid biosynthesis. Plant Cell 35, 700-716. |
| [80] | Wang Z, Zhu TQ, Ma WJ, Wang N, Qu GZ, Zhang SG, Wang JH (2018). Genome-wide analysis of long non-coding RNAs in Catalpa bungei and their potential function in floral transition using high-throughput sequencing. BMC Genet 19, 86. |
| [81] | Wen XJ, Ding Y, Tan ZL, Wang JX, Zhang DY, Wang YC (2020). Identification and characterization of cadmium stress-related lncRNAs from Betula platyphylla. Plant Sci 299, 110601. |
| [82] | Wong-Bajracharya J, Singan VR, Monti R, Plett KL, Ng V, Grigoriev IV, Martin FM, Anderson IC, Plett JM (2022). The ectomycorrhizal fungus Pisolithus microcarpus encodes a microRNA involved in cross-kingdom gene silencing during symbiosis. Proc Natl Acad Sci USA 119, e2103527119. |
| [83] | Wu J, Liu CX, Liu ZG, Li S, Li DD, Liu SY, Huang XQ, Liu SK, Yukawa Y (2019). Pol III-dependent cabbage BoNR8 long ncRNA affects seed germination and growth in Arabidopsis. Plant Cell Physiol 60, 421-435. |
| [84] | Xia KF, Pan XQ, Chen HP, Xu XL, Zhang MY (2023). Rice miR168a-5p regulates seed length, nitrogen allocation and salt tolerance by targeting OsOFP3, OsNPF2.4 and OsAGO1a, respectively. J Plant Physiol 280, 153905. |
| [85] | Xiong XM, Wu Y, Wang Y (2014). Research progresses in the regulation of miRNAs biogenesis and function in plants. Bull Bot Res 34, 282-288. (in Chinese) |
| 熊雪梅, 吴莹, 王洋 (2014). 植物体内调控miRNA合成与功能的机制研究进展. 植物研究 34, 282-288. | |
| [86] | Xu SJ, Dong Q, Deng M, Lin DX, Xiao J, Cheng PL, Xing LJ, Niu YD, Gao CX, Zhang WH, Xu YY, Chong K (2021). The vernalization-induced long non-coding RNA VAS functions with the transcription factor TaRF2b to promote TaVRN1 expression for flowering in hexaploid wheat. Mol Plant 14, 1525-1538. |
| [87] | Xue LZ, Sun MT, Wu Z, Yu L, Yu QH, Tang YP, Jiang FL (2020). LncRNA regulates tomato fruit cracking by coordinating gene expression via a hormone-redox-cell wall network. BMC Plant Biol 20, 162. |
| [88] | Yang SB, Yang T, Tang YP, Aisimutuola P, Zhang GR, Wang BK, Li N, Wang J, Yu QH (2020). Transcriptomic profile analysis of non-coding RNAs involved in Capsicum chinense Jacq. fruit ripening. Sci Hortic 264, 109158. |
| [89] | Yang T, Ma HY, Zhang J, Wu T, Song TT, Tian J, Yao YC (2019). Systematic identification of long noncoding RNAs expressed during light-induced anthocyanin accumulation in apple fruit. Plant J 100, 572-590. |
| [90] | Yang XQ, Fu TT, Yu RE, Zhang LC, Yang YZ, Xiao DD, Wang YY, Wang YL, Wang YW (2023). miR159a modulates poplar resistance against different fungi and bacteria. Plant Physiol Biochem 201, 107899. |
| [91] | Ye XX, Wang S, Zhao XJ, Gao N, Wang Y, Yang YM, Wu E, Jiang C, Cheng YX, Wu WW, Liu SK (2022). Role of lncRNAs in cis- and trans-regulatory responses to salt in Populus trichocarpa. Plant J 110, 978-993. |
| [92] | Yu JX, Qiu KN, Sun WJ, Yang T, Wu T, Song TT, Zhang J, Yao YC, Tian J (2022). A long noncoding RNA functions in high-light-induced anthocyanin accumulation in apple by activating ethylene synthesis. Plant Physiol 189, 66-83. |
| [93] | Yu Y, Zhou YF, Feng YZ, He H, Lian JP, Yang YW, Lei MQ, Zhang YC, Chen YQ (2020). Transcriptional landscape of pathogen-responsive lncRNAs in rice unveils the role of ALEX1 in jasmonate pathway and disease resistance. Plant Biotechnol J 18, 679-690. |
| [94] | Yuan ZD, Pan JH, Chen CP, Tang YL, Zhang HS, Guo J, Yang XR, Chen LF, Li CY, Zhao K, Wang Q, Yang B, Sun CH, Deng XJ, Wang PR (2022). DRB2 modulates leaf rolling by regulating accumulation of microRNAs related to leaf development in rice. Int J Mol Sci 23, 11147. |
| [95] | Zhang L, Liu JL, Cheng JR, Sun Q, Zhang Y, Liu JG, Li HM, Zhang Z, Wang P, Cai CW, Chu ZY, Zhang X, Yuan YL, Shi YZ, Cai YF (2022). lncRNA7 and lncRNA2 modulate cell wall defense genes to regulate cotton resistance to Verticillium wilt. Plant Physiol 189, 264-284. |
| [96] | Zhang LL, Lin T, Zhu GN, Wu B, Zhang CJ, Zhu HL (2023). LncRNAs exert indispensable roles in orchestrating the interaction among diverse noncoding RNAs and enrich the regulatory network of plant growth and its adaptive environmental stress response. Hortic Res 10, uhad234. |
| [97] | Zhang N, Liu ZG, Sun SC, Liu SY, Lin JH, Peng YF, Zhang XX, Yang H, Cen X, Wu J (2020). Response of AtR8 lncRNA to salt stress and its regulation on seed germination in Arabidopsis. Chin Bull Bot 55, 421-429. (in Chinese) |
| 张楠, 刘自广, 孙世臣, 刘圣怡, 林建辉, 彭疑芳, 张晓旭, 杨贺, 岑曦, 吴娟 (2020). 拟南芥AtR8 lncRNA对盐胁迫响应及其对种子萌发的调节作用. 植物学报 55, 421-429. | |
| [98] | Zhang P, Dai MQ (2022). CircRNA: a rising star in plant biology. J Genet Genomics 49, 1081-1092. |
| [99] | Zhang P, Fan Y, Sun XP, Chen L, Terzaghi W, Bucher E, Li L, Dai MQ (2019a). A large-scale circular RNA profiling reveals universal molecular mechanisms responsive to drought stress in maize and Arabidopsis. Plant J 98, 697-713. |
| [100] | Zhang QL, Li Y, Zhang Y, Wu CB, Wang SN, Hao L, Wang SY, Li TZ (2017). Md-miR156ab and Md-miR395 target WRKY transcription factors to influence apple resistance to leaf spot disease. Front Plant Sci 8, 526. |
| [101] | Zhang XP, Dong J, Deng FN, Wang W, Cheng YY, Song LR, Hu MJ, Shen J, Xu QJ, Shen FF (2019b). The long non-coding RNA lncRNA973 is involved in cotton response to salt stress. BMC Plant Biol 19, 459. |
| [102] | Zhang XP, Shen J, Xu QJ, Dong J, Song LR, Wang W, Shen FF (2021). Long noncoding RNA lncRNA354 functions as a competing endogenous RNA of miR160b to regulate ARF genes in response to salt stress in upland cotton. Plant Cell Environ 44, 3302-3321. |
| [103] | Zhao XY, Li JR, Lian B, Gu HQ, Li Y, Qi YJ (2018). Global identification of Arabidopsis lncRNAs reveals the regulation of MAF4 by a natural antisense RNA. Nat Commun 9, 5056. |
| [104] | Zheng XM, Chen YJ, Zhou YF, Shi KK, Hu X, Li DY, Ye HZ, Zhou Y, Wang K (2021). Full-length annotation with multistrategy RNA-seq uncovers transcriptional regulation of lncRNAs in cotton. Plant Physiol 185, 179-195. |
| [105] | Zhou BY, Luo Q, Shen YH, Wei L, Song X, Liao HQ, Ni L, Shen T, Du XL, Han JY, Jiang MY, Feng SJ, Wu G (2023a). Coordinated regulation of vegetative phase change by brassinosteroids and the age pathway in Arabidopsis. Nat Commun 14, 2608. |
| [106] | Zhou J, Yang LY, Jia CL, Shi WG, Deng SR, Luo ZB (2022). Identification and functional prediction of poplar root circRNAs involved in treatment with different forms of nitrogen. Front Plant Sci 13, 941380. |
| [107] | Zhou JP, Yuan MZ, Zhao YX, Quan Q, Yu D, Yang H, Tang X, Xin XH, Cai GZ, Qian Q, Qi YP, Zhang Y (2021a). Efficient deletion of multiple circle RNA loci by CRISPR-Cas9 reveals Os06circ02797 as a putative sponge for OsMIR408 in rice. Plant Biotechnol J 19, 1240-1252. |
| [108] | Zhou LK, Zhou Z, Jiang XM, Zheng YS, Chen X, Fu Z, Xiao GF, Zhang CY, Zhang LK, Yi YX (2020). Absorbed plant MIR2911 in honeysuckle decoction inhibits SARS- CoV-2 replication and accelerates the negative conversion of infected patients. Cell Discov 6, 54. |
| [109] | Zhou R, Dong YH, Wang CX, Liu JN, Liang Q, Meng XY, Lang XY, Xu SY, Liu WJ, Zhang SH, Wang N, Yang KQ, Fang HC (2023b). LncRNA109897-JrCCR4-JrTLP1b forms a positive feedback loop to regulate walnut resistance against anthracnose caused by Colletotrichum gloeosporioides. Hortic Res 10, uhad086. |
| [110] | Zhou R, Xu LP, Zhao LP, Wang YL, Zhao TM (2018). Genome-wide identification of circRNAs involved in tomato fruit coloration. Biochem Biophys Res Commun 499, 466- 469. |
| [111] | Zhou YF, Zhang YC, Sun YM, Yu Y, Lei MQ, Yang YW, Lian JP, Feng YZ, Zhang Z, Yang L, He RR, Huang JH, Cheng Y, Liu YW, Chen YQ (2021b). The parent-of-origin lncRNA MISSEN regulates rice endosperm development. Nat Commun 12, 6525. |
| [112] | Zhu TD, Yang CW, Xie Y, Huang S, Li L (2023). Shade- induced lncRNA PUAR promotes shade response by repressing PHYA expression. EMBO Rep 24, e56105. |
| [113] | Zhu YX, Jia JH, Yang L, Xia YC, Zhang HL, Jia JB, Zhou R, Nie PY, Yin JL, Ma DF, Liu LC (2019). Identification of cucumber circular RNAs responsive to salt stress. BMC Plant Biol 19, 164. |
| [114] | Zou XY, Ali F, Jin SX, Li FG, Wang Z (2022). RNA-Seq with a novel glabrous-ZM24fl reveals some key lncRNAs and the associated targets in fiber initiation of cotton. BMC Plant Biol 22, 61. |
| [1] | 李青洋, 刘翠, 何李, 彭姗, 马嘉吟, 胡子祎, 刘宏波. 甘蓝型油菜BnaA02.CPSF6基因的克隆及功能分析(长英文摘要)[J]. 植物学报, 2025, 60(1): 62-73. |
| [2] | 王亚萍, 包文泉, 白玉娥. 单细胞转录组学在植物生长发育及胁迫响应中的应用进展[J]. 植物学报, 2025, 60(1): 101-113. |
| [3] | 王涛, 冯敬磊, 张翠. 高温胁迫影响玉米生长发育的分子机制研究进展[J]. 植物学报, 2024, 59(6): 963-977. |
| [4] | 闫恒宇, 李朝霞, 李玉斌. 高温对玉米生长的影响及中国耐高温玉米筛选研究进展[J]. 植物学报, 2024, 59(6): 1007-1023. |
| [5] | 杜庆国, 李文学. lncRNA调控玉米生长发育和非生物胁迫研究进展[J]. 植物学报, 2024, 59(6): 950-962. |
| [6] | 曾鑫海, 陈锐, 师宇, 盖超越, 范凯, 李兆伟. 植物SPL转录因子的生物功能研究进展[J]. 植物学报, 2023, 58(6): 982-997. |
| [7] | 许亚楠, 闫家榕, 孙鑫, 王晓梅, 刘玉凤, 孙周平, 齐明芳, 李天来, 王峰. 红光和远红光在调控植物生长发育及应答非生物胁迫中的作用[J]. 植物学报, 2023, 58(4): 622-637. |
| [8] | 张嘉, 李启东, 李翠, 王庆海, 侯新村, 赵春桥, 李树和, 郭强. 植物MATE转运蛋白研究进展[J]. 植物学报, 2023, 58(3): 461-474. |
| [9] | 王琪, 吴允哲, 刘学英, 孙丽莉, 廖红, 傅向东. 类受体激酶调控水稻生长发育和环境适应研究进展[J]. 植物学报, 2023, 58(2): 199-213. |
| [10] | 刘德帅, 姚磊, 徐伟荣, 冯美, 姚文孔. 褪黑素参与植物抗逆功能研究进展[J]. 植物学报, 2022, 57(1): 111-126. |
| [11] | 岳剑茹, 赫云建, 邱天麒, 郭南南, 韩雪萍, 王显玲. 植物微管骨架参与下胚轴伸长调节机制研究进展[J]. 植物学报, 2021, 56(3): 363-371. |
| [12] | 俞启璐, 赵江哲, 朱晓仙, 张可伟. 水稻根分泌激素调节生长速度[J]. 植物学报, 2021, 56(2): 175-182. |
| [13] | 李喜豹, 赖敏怡, 梁山, 王小菁, 高彩吉, 杨超. 植物细胞自噬基因的功能与转录调控机制[J]. 植物学报, 2021, 56(2): 201-217. |
| [14] | 王益军, 王亚丽, 陈煜东. 转座子来源的植物长链非编码RNA[J]. 植物学报, 2020, 55(6): 768-776. |
| [15] | 张楠,刘自广,孙世臣,刘圣怡,林建辉,彭疑芳,张晓旭,杨贺,岑曦,吴娟. 拟南芥AtR8 lncRNA对盐胁迫响应及其对种子萌发的调节作用[J]. 植物学报, 2020, 55(4): 421-429. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||