

植物学报 ›› 2021, Vol. 56 ›› Issue (2): 201-217.DOI: 10.11983/CBB20159 cstr: 32102.14.CBB20159
收稿日期:2020-09-29
接受日期:2020-12-25
出版日期:2021-03-01
发布日期:2021-03-17
通讯作者:
杨超
作者简介:*E-mail: 798916640@qq.com基金资助:
Xibao Li, Minyi Lai, Shan Liang, Xiaojing Wang, Caiji Gao, Chao Yang( )
)
Received:2020-09-29
Accepted:2020-12-25
Online:2021-03-01
Published:2021-03-17
Contact:
Chao Yang
About author:First author contact:† These authors contributed equally to this paper
摘要: 自噬(autophagy)是真核生物长期进化形成的一种高度保守的细胞内物质降解和周转途径, 通过形成双层膜结构的自噬体将包裹其中的待降解大分子物质, 如受损伤的蛋白质、蛋白质复合物和细胞器, 运送至液泡或溶酶体进行降解并产生可循环利用的降解产物。细胞自噬在植物生长发育和环境应答等过程中发挥重要作用。在拟南芥(Arabidopsis thaliana)和水稻(Oryza sativa)等模式植物中已鉴定到40多个自噬基因, 并发现其中多个基因在植物叶片衰老、种子成熟等发育阶段以及营养饥饿、干旱和病原菌侵染等逆境胁迫响应过程中显著上调表达, 但具体的转录激活或抑制机制有待阐明。该文综述了自噬基因在植物生长发育和胁迫应答过程中的功能与转录调控网络。
李喜豹, 赖敏怡, 梁山, 王小菁, 高彩吉, 杨超. 植物细胞自噬基因的功能与转录调控机制. 植物学报, 2021, 56(2): 201-217.
Xibao Li, Minyi Lai, Shan Liang, Xiaojing Wang, Caiji Gao, Chao Yang. Function and Transcriptional Regulation of Autophagy-related Genes in Plants. Chinese Bulletin of Botany, 2021, 56(2): 201-217.
| 物种 | 发育时期及组织 | 检测方法 | 自噬基因表达 | 参考文献 |
|---|---|---|---|---|
| 拟南芥 (Arabidopsis thaliana) | 自然衰老的叶片(10%黄化的叶片) | Microarray | 5个AtATG上调 | Buchanan-Wollaston et al. |
| 衰老期叶片(7-39天的叶片) | 28个AtATG上调 | Breeze et al. | ||
| 授粉后12-30天的种子 | qRT-PCR | 31个AtATG上调 | Di Berardino et al. 2018 | |
| 授粉后的乳突细胞 | RNA-Seq | 9个AtATG上调 | Ye et al. | |
| 玉米 (Zea mays) | 授粉后2-24天的种子 | RNA-Seq | 11个ZmATG上调 | Li et al. |
| 授粉后12-24天的胚乳 | 29个ZmATG上调 | |||
| 授粉后0-20天的叶片 | 30个ZmATG上调 | |||
| 烟草 (Nicotiana tabacum) | 授粉后4-20天的种子 | qRT-PCR | 27个NtATG上调 | Zhou et al. |
| 萌发0.25-4小时的花粉 | 29个NtATG上调 | Zhao et al. | ||
| 蓖麻 (Ricinus communis) | 授粉后20-50天的胚乳 | RNA-Seq, qRT-PCR | 34个RcATG上调 | Han et al. |
| 授粉后20-30天的种皮 | 31个RcATG上调 | |||
| 种子萌发后1-10天的胚乳 | 34个RcATG上调 |
表1 植物生长发育过程中上调表达的自噬基因(ATG genes)
Table 1 Up-regulated autophagy genes (ATG genes) during plant growth and development
| 物种 | 发育时期及组织 | 检测方法 | 自噬基因表达 | 参考文献 |
|---|---|---|---|---|
| 拟南芥 (Arabidopsis thaliana) | 自然衰老的叶片(10%黄化的叶片) | Microarray | 5个AtATG上调 | Buchanan-Wollaston et al. |
| 衰老期叶片(7-39天的叶片) | 28个AtATG上调 | Breeze et al. | ||
| 授粉后12-30天的种子 | qRT-PCR | 31个AtATG上调 | Di Berardino et al. 2018 | |
| 授粉后的乳突细胞 | RNA-Seq | 9个AtATG上调 | Ye et al. | |
| 玉米 (Zea mays) | 授粉后2-24天的种子 | RNA-Seq | 11个ZmATG上调 | Li et al. |
| 授粉后12-24天的胚乳 | 29个ZmATG上调 | |||
| 授粉后0-20天的叶片 | 30个ZmATG上调 | |||
| 烟草 (Nicotiana tabacum) | 授粉后4-20天的种子 | qRT-PCR | 27个NtATG上调 | Zhou et al. |
| 萌发0.25-4小时的花粉 | 29个NtATG上调 | Zhao et al. | ||
| 蓖麻 (Ricinus communis) | 授粉后20-50天的胚乳 | RNA-Seq, qRT-PCR | 34个RcATG上调 | Han et al. |
| 授粉后20-30天的种皮 | 31个RcATG上调 | |||
| 种子萌发后1-10天的胚乳 | 34个RcATG上调 |
| 物种 | 发育时期/组织 | 胁迫类型/时间 | 检测方法 | 自噬基因表达 | 参考文献 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 拟南芥 (Arabidopsis thaliana) | 4周/叶 | 淹水胁迫/24小时 | qRT-PCR | 6个AtATG上调 | Chen et al. | ||||||
| 2周/幼苗 | 缺锌胁迫/14天 | 13个AtATG上调 | Eguchi et al. | ||||||||
| 5天/幼苗 | 42°C热激后/24小时 | 33个AtATG上调 | Sedaghatmehr et al. | ||||||||
| 5天/幼苗 | 光照到黑暗/24小时 | 11个AtATG上调 | Yang et al. | ||||||||
| 30天/茎叶 | 黑暗胁迫/3天 | RNA-Seq | 23个AtATG上调 | Yan et al. | |||||||
| 4周/叶 | 黑暗胁迫/6天 | Microarray | 14个AtATG上调 | van der Graaff et al. 2006 | |||||||
| 水稻 (Oryza sativa) | 3周/整株苗 | 250 mmol·L-1 NaCl/4小时 | qRT-PCR | 13个OsATG上调 | Xia et al. | ||||||
| 干旱胁迫/4小时 | 13个OsATG上调 | ||||||||||
| 4°C冷胁迫/4小时 | 21个OsATG上调 | ||||||||||
| 黑暗胁迫/48小时 | 19个OsATG上调 | ||||||||||
| 碳饥饿/48小时 | 17个OsATG上调 | ||||||||||
| 10天/整株苗 | 氮饥饿/7天 | 18个OsATG上调 | |||||||||
| 物种 | 发育时期/组织 | 胁迫类型/时间 | 检测方法 | 自噬基因表达 | 参考文献 | ||||||
| 烟草 (Nicotiana tabacum) | 2周/整株苗 | 碳饥饿/48小时 | qRT-PCR | 17个NtATG上调 | Zhou et al. | ||||||
| 氮饥饿/48小时 | 16个NtATG上调 | ||||||||||
| 黑暗胁迫/48小时 | 16个NtATG上调 | ||||||||||
| 250 mmol·L-1 NaCl/4小时 | 14个NtATG上调 | ||||||||||
| 4°C冷胁迫/4小时 | 14个NtATG上调 | ||||||||||
| 干旱胁迫/4小时 | 7个NtATG上调 | ||||||||||
| 1 μmol ·L-1 NAA/24小时 | 6个NtATG上调 | ||||||||||
| 5 μmol ·L-1 2,4-D/24小时 | 5个NtATG上调 | ||||||||||
| 25 μmol ·L-1 ABA/24小时 | 4个NtATG上调 | ||||||||||
| 5 μmol ·L-1 GA3/24小时 | 7个NtATG下调 | ||||||||||
| 500 μmol ·L-1 SA/24小时 | 4个NtATG上调 | ||||||||||
| 500 μmol ·L-1 JA/24小时 | 11个NtATG上调 | ||||||||||
| 20 μmol ·L-1 Cu2+/24小时 | 12个NtATG上调 | ||||||||||
| 20 μmol ·L-1 Ni2+/24小时 | 15个NtATG上调 | ||||||||||
| 40 μmol ·L-1 Zn2+/24小时 | 13个NtATG上调 | ||||||||||
| 40 μmol ·L-1 Cd2+/24小时 | 11个NtATG上调 | ||||||||||
| 100 μmol ·L-1 Mn2+/24小时 | 14个NtATG上调 | ||||||||||
| 辣椒 (Capsicum annuum) | 5-6叶期/根 | 200 mmol·L-1 NaCl/3小时 | RNA-Seq | 9个CaATG上调 | Zhai et al. | ||||||
| 5-6叶期/茎 | 15个CaATG上调 | ||||||||||
| 5-6叶期/根 | 干旱胁迫/3小时 | 7个CaATG上调 | |||||||||
| 5-6叶期/茎 | 7个CaATG上调 | ||||||||||
| 5-6叶期/整株苗 | 40°C热胁迫/12小时 (耐热品种) | 26个CaATG上调 | |||||||||
| 40°C热胁迫/12小时 (热敏感品种) | 17个CaATG上调 | ||||||||||
| 4°C冷胁迫/3小时 | 14个CaATG上调 | ||||||||||
| 碳饥饿/2天 | 10个CaATG上调 | ||||||||||
| 小麦 (Triticum aestivum) | 10天/叶 | 150 mmol·L-1 NaCl/48小时 | RNA-Seq | 75个TaATG上调 | Yue et al. | ||||||
| 4°C冷胁迫/24小时 | 23个TaATG上调 | ||||||||||
| 42°C热胁迫/6小时 | 36个TaATG上调 | ||||||||||
| 干旱胁迫/24小时 | 23个TaATG上调 | ||||||||||
| 2叶期/根-叶 | 氮饥饿/4天 | qRT-PCR | 9个TaATG8上调 | Zhang et al. | |||||||
| 白粉病真菌侵染/36小时(敏感品种) | 9个TaATG8上调 | ||||||||||
| 白粉病真菌侵染/36小时(抗性品种) | 3个TaATG8上调 | ||||||||||
| 番茄 (Solanum lycopersicum) | 6周/叶 | 干旱胁迫/6天 | qRT-PCR | 20个SlATG上调 | Wang et al. | ||||||
| 6周/叶 | 500 nmol·L-1 BL/12小时 | 8个SlATG上调 | Wang et al. | ||||||||
| 5周/叶 | 4°C冷胁迫/24小时 | 11个SlATG上调 | Chi et al. | ||||||||
| 香蕉 (Musa nana) | 5叶期/根 | 镰刀菌侵染/51小时 | qRT-PCR | 8个MaATG上调 | Wei et al. | ||||||
| 木薯 (Manihot esculenta) | 30天/叶 | 单黄胞菌侵染/6小时 | qRT-PCR | 25个MeATG上调 | Yan et al. | ||||||
| 葡萄 (Vitis vinifera) | 2年/叶 | 400 μmol ·L-1 Cu2+/36小时 | qRT-PCR | 24个VvATG上调 | Shangguan et al. | ||||||
| 线形草沙蚕 (Tripogon loliiformis) | 2月/根-茎 | 脱水处理 | RNA-Seq | 10个TlATG上调 | Williams et al. | ||||||
表2 逆境胁迫诱导表达的植物自噬基因
Table 2 Up-regulated autophagy genes upon stress treatments in plants
| 物种 | 发育时期/组织 | 胁迫类型/时间 | 检测方法 | 自噬基因表达 | 参考文献 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 拟南芥 (Arabidopsis thaliana) | 4周/叶 | 淹水胁迫/24小时 | qRT-PCR | 6个AtATG上调 | Chen et al. | ||||||
| 2周/幼苗 | 缺锌胁迫/14天 | 13个AtATG上调 | Eguchi et al. | ||||||||
| 5天/幼苗 | 42°C热激后/24小时 | 33个AtATG上调 | Sedaghatmehr et al. | ||||||||
| 5天/幼苗 | 光照到黑暗/24小时 | 11个AtATG上调 | Yang et al. | ||||||||
| 30天/茎叶 | 黑暗胁迫/3天 | RNA-Seq | 23个AtATG上调 | Yan et al. | |||||||
| 4周/叶 | 黑暗胁迫/6天 | Microarray | 14个AtATG上调 | van der Graaff et al. 2006 | |||||||
| 水稻 (Oryza sativa) | 3周/整株苗 | 250 mmol·L-1 NaCl/4小时 | qRT-PCR | 13个OsATG上调 | Xia et al. | ||||||
| 干旱胁迫/4小时 | 13个OsATG上调 | ||||||||||
| 4°C冷胁迫/4小时 | 21个OsATG上调 | ||||||||||
| 黑暗胁迫/48小时 | 19个OsATG上调 | ||||||||||
| 碳饥饿/48小时 | 17个OsATG上调 | ||||||||||
| 10天/整株苗 | 氮饥饿/7天 | 18个OsATG上调 | |||||||||
| 物种 | 发育时期/组织 | 胁迫类型/时间 | 检测方法 | 自噬基因表达 | 参考文献 | ||||||
| 烟草 (Nicotiana tabacum) | 2周/整株苗 | 碳饥饿/48小时 | qRT-PCR | 17个NtATG上调 | Zhou et al. | ||||||
| 氮饥饿/48小时 | 16个NtATG上调 | ||||||||||
| 黑暗胁迫/48小时 | 16个NtATG上调 | ||||||||||
| 250 mmol·L-1 NaCl/4小时 | 14个NtATG上调 | ||||||||||
| 4°C冷胁迫/4小时 | 14个NtATG上调 | ||||||||||
| 干旱胁迫/4小时 | 7个NtATG上调 | ||||||||||
| 1 μmol ·L-1 NAA/24小时 | 6个NtATG上调 | ||||||||||
| 5 μmol ·L-1 2,4-D/24小时 | 5个NtATG上调 | ||||||||||
| 25 μmol ·L-1 ABA/24小时 | 4个NtATG上调 | ||||||||||
| 5 μmol ·L-1 GA3/24小时 | 7个NtATG下调 | ||||||||||
| 500 μmol ·L-1 SA/24小时 | 4个NtATG上调 | ||||||||||
| 500 μmol ·L-1 JA/24小时 | 11个NtATG上调 | ||||||||||
| 20 μmol ·L-1 Cu2+/24小时 | 12个NtATG上调 | ||||||||||
| 20 μmol ·L-1 Ni2+/24小时 | 15个NtATG上调 | ||||||||||
| 40 μmol ·L-1 Zn2+/24小时 | 13个NtATG上调 | ||||||||||
| 40 μmol ·L-1 Cd2+/24小时 | 11个NtATG上调 | ||||||||||
| 100 μmol ·L-1 Mn2+/24小时 | 14个NtATG上调 | ||||||||||
| 辣椒 (Capsicum annuum) | 5-6叶期/根 | 200 mmol·L-1 NaCl/3小时 | RNA-Seq | 9个CaATG上调 | Zhai et al. | ||||||
| 5-6叶期/茎 | 15个CaATG上调 | ||||||||||
| 5-6叶期/根 | 干旱胁迫/3小时 | 7个CaATG上调 | |||||||||
| 5-6叶期/茎 | 7个CaATG上调 | ||||||||||
| 5-6叶期/整株苗 | 40°C热胁迫/12小时 (耐热品种) | 26个CaATG上调 | |||||||||
| 40°C热胁迫/12小时 (热敏感品种) | 17个CaATG上调 | ||||||||||
| 4°C冷胁迫/3小时 | 14个CaATG上调 | ||||||||||
| 碳饥饿/2天 | 10个CaATG上调 | ||||||||||
| 小麦 (Triticum aestivum) | 10天/叶 | 150 mmol·L-1 NaCl/48小时 | RNA-Seq | 75个TaATG上调 | Yue et al. | ||||||
| 4°C冷胁迫/24小时 | 23个TaATG上调 | ||||||||||
| 42°C热胁迫/6小时 | 36个TaATG上调 | ||||||||||
| 干旱胁迫/24小时 | 23个TaATG上调 | ||||||||||
| 2叶期/根-叶 | 氮饥饿/4天 | qRT-PCR | 9个TaATG8上调 | Zhang et al. | |||||||
| 白粉病真菌侵染/36小时(敏感品种) | 9个TaATG8上调 | ||||||||||
| 白粉病真菌侵染/36小时(抗性品种) | 3个TaATG8上调 | ||||||||||
| 番茄 (Solanum lycopersicum) | 6周/叶 | 干旱胁迫/6天 | qRT-PCR | 20个SlATG上调 | Wang et al. | ||||||
| 6周/叶 | 500 nmol·L-1 BL/12小时 | 8个SlATG上调 | Wang et al. | ||||||||
| 5周/叶 | 4°C冷胁迫/24小时 | 11个SlATG上调 | Chi et al. | ||||||||
| 香蕉 (Musa nana) | 5叶期/根 | 镰刀菌侵染/51小时 | qRT-PCR | 8个MaATG上调 | Wei et al. | ||||||
| 木薯 (Manihot esculenta) | 30天/叶 | 单黄胞菌侵染/6小时 | qRT-PCR | 25个MeATG上调 | Yan et al. | ||||||
| 葡萄 (Vitis vinifera) | 2年/叶 | 400 μmol ·L-1 Cu2+/36小时 | qRT-PCR | 24个VvATG上调 | Shangguan et al. | ||||||
| 线形草沙蚕 (Tripogon loliiformis) | 2月/根-茎 | 脱水处理 | RNA-Seq | 10个TlATG上调 | Williams et al. | ||||||
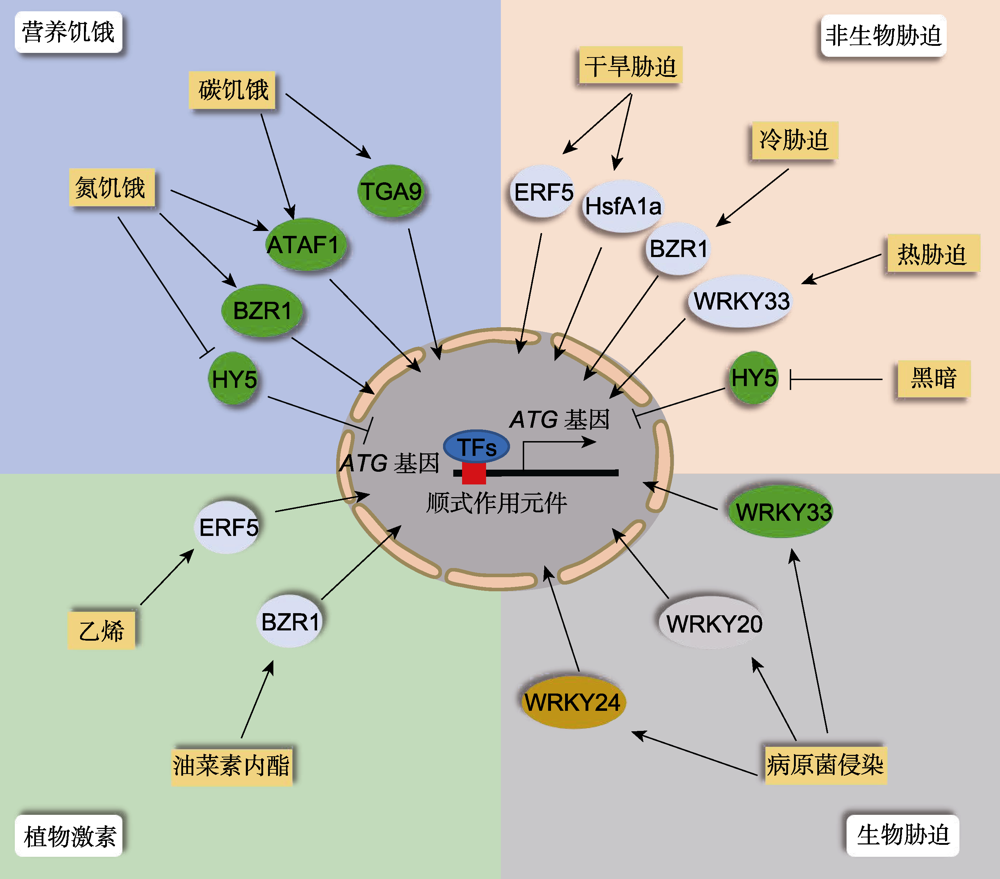
图1 不同逆境胁迫下植物细胞自噬的转录调控网络 当植物遭受逆境胁迫时, 转录因子(TFs)被诱导或抑制表达, 进入细胞核识别特定的顺式作用元件, 结合并调控自噬基因的表达, 从而调控细胞自噬活性。
Figure 1 Transcriptional regulation of plant autophagy upon various stresses When plants encounter adverse environmental conditions, the corresponding transcription factors (TFs) are induced or repressed to activate the gene expression of downstream ATG genes through binding the specific cis-elements in their promoters in the nucleus, thereby controlling the activity of cell autophagy.
| [1] | 黄晓, 李发强 (2016). 细胞自噬在植物细胞程序性死亡中的作用. 植物学报 51,859-862. |
| [2] | 刘洋, 张静, 王秋玲, 侯岁稳 (2018). 植物细胞自噬研究进展. 植物学报 53,5-16. |
| [3] | 马丹颖, 季东超, 徐勇, 陈彤, 田世平 (2019). 活性氧调控植物细胞自噬的研究进展. 植物学报 54,81-92. |
| [4] | 王燕, 刘玉乐 (2010). 植物细胞自噬研究进展. 中国细胞生物学学报 32,677-689. |
| [5] | Avila-Ospina L, Moison M, Yoshimoto K, Masclaux-Daubresse C (2014). Autophagy, plant senescence, and nutrient recycling. J Exp Bot 65,3799-3811. |
| [6] | Avin-Wittenberg T (2019). Autophagy and its role in plant abiotic stress management. Plant Cell Environ 42, 1045- 1053. |
| [7] | Baumberger N, Tsai CH, Lie M, Havecker E, Baulcombe DC (2007). The polerovirus silencing suppressor P0 targets ARGONAUTE proteins for degradation. Curr Biol 17,1609-1614. |
| [8] | Bewley JD (1997). Seed germination and dormancy. Plant Cell 9,1055-1066. |
| [9] | Blümel M, Dally N, Jung C (2015). Flowering time regula- tion in crops—what did we learn fromArabidopsis? Curr Opin Biotechnol 32,121-129. |
| [10] | Breeze E, Harrison E, McHattie S, Hughes L, Hickman R, Hill C, Kiddle S, Kim YS, Penfold CA, Jenkins D, Zhang C, Morris K, Jenner C, Jackson S, Thomas B, Tabrett A, Legaie R, Moore JD, Wild DL, Ott S, Rand D, Beynon J, Denby K, Mead A, Buchanan-Wollaston V (2011). High-resolution temporal profiling of transcripts during Arabidopsis leaf senescence reveals a distinct chronology of processes and regulation. Plant Cell 23,873-894. |
| [11] | Buchanan-Wollaston V, Page T, Harrison E, Breeze E, Lim PO, Nam HG, Lin JF, Wu SH, Swidzinski J, Ishizaki K, Leaver CJ (2005). Comparative transcriptome analysis reveals significant differences in gene expression and signaling pathways between developmental and dark/ starvation-induced senescence in Arabidopsis. Plant J 42,567-585. |
| [12] | Chen L, Liao B, Qi H, Xie LJ, Huang L, Tan WJ, Zhai N, Yuan LB, Zhou Y, Yu LJ, Chen QF, Shu WS, Xiao S (2015). Autophagy contributes to regulation of the hypoxia response during submergence in Arabidopsis thaliana. Autophagy 11,2233-2246. |
| [13] | Chi C, Li XM, Fang PP, Xia XJ, Shi K, Zhou YH, Zhou J, Yu JQ (2020). Brassinosteroids act as a positive regulator of NBR1-dependent selective autophagy in response to chilling stress in tomato. J Exp Bot 71,1092-1106. |
| [14] | Derrien B, Baumberger N, Schepetilnikov M, Viotti C, De Cillia J, Ziegler-Graff V, Isono E, Schumacher K, Genschik P (2012). Degradation of the antiviral compo- nent ARGONAUTE1 by the autophagy pathway. Proc Natl Acad Sci USA 109,15942-15946. |
| [15] | Deter RL, Baudhuin P, De Duve C (1967). Participation of lysosomes in cellular autophagy induced in rat liver by glucagon. J Cell Biol 35,C11-C16. |
| [16] | Di Bartolomeo S, Nazio F, Cecconi F (2010). The role of autophagy during development in higher eukaryotes. Traffic 11,1280-1289. |
| [17] | Di Berardino J, Marmagne A, Berger A, Yoshimoto K, Cueff G, Chardon F, Masclaux-Daubresse C, Reisdorf- Cren M (2018). Autophagy controls resource allocation and protein storage accumulation in Arabidopsis seeds. J Exp Bot 69,1403-1414. |
| [18] | Doelling JH, Walker JM, Friedman EM, Thompson AR, Vierstra RD (2002). The APG8/12-activating enzyme APG7 is required for proper nutrient recycling and sene- scence in Arabidopsis thaliana. J Biol Chem 277, 33105- 33114. |
| [19] | Dündar G, Shao ZH, Higashitani N, Kikuta M, Izumi M, Higashitani A (2019). Autophagy mitigates high-tem- perature injury in pollen development of Arabidopsis thaliana. Dev Biol 456,190-200. |
| [20] | Eguchi M, Kimura K, Makino A, Ishida H (2017). Autophagy is induced under Zn limitation and contributes to Zn-limited stress tolerance in Arabidopsis (Arabidopsis thaliana). Soil Sci Plant Nutr 63,342-350. |
| [21] | Fan T, Yang W, Zeng X, Xu XL, Xu YL, Fan XR, Luo M, Tian CG, Xia KF, Zhang MY (2020). A rice autophagy gene OsATG8b is involved in nitrogen remobilization and control of grain quality. Front Plant Sci 11,588. |
| [22] | Fujiki Y, Yoshimoto K, Ohsumi Y (2007). An Arabidopsis homolog of yeast ATG6/ VPS30 is essential for pollen germination. Plant Physiol 143,1132-1139. |
| [23] | Gallegos J (2018). Autophagy: both friend and foe in Pseudomonas syringae infection. Plant Cell 30,522-523. |
| [24] | Gangappa SN, Botto JF (2016). The multifaceted roles of HY5 in plant growth and development. Mol Plant 9, 1353-1365. |
| [25] | Gao CJ, Zhuang XH, Shen JB, Jiang LW (2017). Plant ESCRT complexes: moving beyond endosomal sorting. Trends Plant Sci 22,986-998. |
| [26] | Garapati P, Feil R, Lunn JE, Van Dijck P, Balazadeh S, Mueller-Roeber B (2015). Transcription factor Arabidop- sis activating factor 1 integrates carbon starvation res- ponses with trehalose metabolism. Plant Physiol 169,379-390. |
| [27] | Goring DR (2017). Exocyst, exosomes, and autophagy in the regulation of Brassicaceae pollen-stigma interactions. J Exp Bot 69,69-78. |
| [28] | Gou WT, Li X, Guo SY, Liu YF, Li FQ, Xie QJ (2019). Autophagy in plant: a new orchestrator in the regulation of the phytohormones homeostasis. Int J Mol Sci 20,2900. |
| [29] | Guiboileau A, Yoshimoto K, Soulay F, Bataillé MP, Avice JC, Masclaux-Daubresse C (2012). Autophagy machi- nery controls nitrogen remobilization at the whole-plant level under both limiting and ample nitrate conditions in Arabidopsis. New Phytol 194,732-740. |
| [30] | Hafrén A, Macia JL, Love AJ, Milner JJ, Drucker M, Hofius D (2017). Selective autophagy limits cauliflower mosaic virus infection by NBR1-mediated targeting of viral capsid protein and particles. Proc Natl Acad Sci USA 114,E2026-E2035. |
| [31] | Hafrén A, Ustün S, Hochmuth A, Svenning S, Johansen T, Hofius D (2018). Turnip Mosaic virus counteracts selective autophagy of the viral silencing suppressor HCpro. Plant Physiol 176,649-662. |
| [32] | Han B, Xu H, Feng YT, Xu W, Cui QH, Liu AZ (2020). Genomic characterization and expressional profiles of autophagy-related genes ( ATGs) in oilseed crop castor bean ( Ricinus communis L.). Int J Mol Sci 21,562. |
| [33] | Hanaoka H, Noda T, Shirano Y, Kato T, Hayashi H, Shibata D, Tabata S, Ohsumi Y (2002). Leaf sene- scence and starvation-induced chlorosis are accelerated by the disruption of an Arabidopsis autophagy gene. Plant Physiol 129,1181-1193. |
| [34] | Harrison-Lowe NJ, Olsen LJ (2008). Autophagy protein 6 (ATG6) is required for pollen germination in Arabidopsis thaliana. Autophagy 4,339-348. |
| [35] | Haxim Y, Ismayil A, Jia Q, Wang Y, Zheng XY, Chen TY, Qian LC, Liu N, Wang YJ, Han SJ, Cheng JX, Qi YJ, Hong YG, Liu YL (2017). Autophagy functions as an antiviral mechanism against geminiviruses in plants. eLife 6,e23897. |
| [36] | Hightower LE (1991). Heat shock, stress proteins, chapero- nes, and proteotoxicity. Cell 66,191-197. |
| [37] | Huang X, Zheng C, Liu F, Yang C, Zheng P, Lu X, Tian J, Chung T, Otegui MS, Xiao S, Gao C, Vierstra RD, Li F (2019). Genetic analyses of the Arabidopsis ATG1 kinase complex reveal both kinase-dependent and independent autophagic routes during fixed-carbon starvation. Plant Cell 31,2973-2995. |
| [38] | Jakoby M, Weisshaar B, Dröge-Laser W, Vicente-Car- bajosa J, Tiedemann J, Kroj T, Parcy F (2002). bZIP transcription factors in Arabidopsis. Trends Plant Sci 7,106-111. |
| [39] | Jiao YL, Lau OS, Deng XW (2007). Light-regulated trans- criptional networks in higher plants. Nat Rev Genet 8,217-230. |
| [40] | Jung H, Lee HN, Marshall RS, Lomax AW, Yoon MJ, Kim J, Kim JH, Vierstra RD, Chung T (2020). Arabidopsis cargo receptor NBR1 mediates selective autophagy of defective proteins. J Exp Bot 71,73-89. |
| [41] | Kurusu T, Koyano T, Hanamata S, Kubo T, Noguchi Y, Yagi C, Nagata N, Yamamoto T, Ohnishi T, Okazaki Y, Kitahata N, Ando D, Ishikawa M, Wada S, Miyao A, Hirochika H, Shimada H, Makino A, Saito K, Ishida H, Kinoshita T, Kurata N, Kuchitsu K (2014). OsATG7 is required for autophagy-dependent lipid metabolism in rice postmeiotic anther development. Autophagy 10,878-888. |
| [42] | Lai ZB, Wang F, Zheng ZY, Fan BF, Chen ZX (2011). A critical role of autophagy in plant resistance to necro- trophic fungal pathogens. Plant J 66,953-968. |
| [43] | Leary AY, Sanguankiattichai N, Duggan C, Tumtas Y, Pandey P, Segretin ME, Linares JS, Savage ZD, Yow RJ, Bozkurt TO (2018). Modulation of plant autophagy during pathogen attack. J Exp Bot 69,1325-1333. |
| [44] | Leng X, Jia HF, Sun X, Shangguan LF, Mu Q, Wang BJ, Fang JG (2015). Comparative transcriptome analysis of grapevine in response to copper stress. Sci Rep 5,17749. |
| [45] | Li FF, Zhang CW, Li YZ, Wu GW, Hou XL, Zhou XP, Wang AM (2018). Beclin1 restricts RNA virus infection in plants through suppression and degradation of the viral poly- merase. Nat Commun 9,1268. |
| [46] | Li FQ, Chung T, Pennington JG, Federico ML, Kaeppler HF, Kaeppler SM, Otegui MS, Vierstra RD (2015). Autophagic recycling plays a central role in maize nitro- gen remobilization. Plant Cell 27,1389-1408. |
| [47] | Li FQ, Chung T, Vierstra RD (2014). AUTOPHAGY-RE- LATED11 plays a critical role in general autophagy- and senescence-induced mitophagy in Arabidopsis. Plant Cell 26,788-807. |
| [48] | Li FQ, Vierstra RD (2012). Autophagy: a multifaceted intracellular system for bulk and selective recycling. Trends Plant Sci 17,526-537. |
| [49] | Li SS, Yan H, Mei WM, Tse YC, Wang H (2020). Boosting autophagy in sexual reproduction: a plant perspective. New Phytol 226,679-689. |
| [50] | Liao CY, Bassham DC (2020). Combating stress: the interplay between hormone signaling and autophagy in plants. J Exp Bot 71,1723-1733. |
| [51] | Lindemose S, Jensen MK, Van De Velde J, O'Shea C, Heyndrickx KS, Workman CT, Vandepoele K, Skriver K, De Masi F (2014). A DNA-binding-site landscape and regulatory network analysis for NAC transcription factors in Arabidopsis thaliana. Nucleic Acids Res 42, 7681- 7693. |
| [52] | Liu F, Hu WM, Vierstra RD (2018). The vacuolar protein sorting-38 subunit of the Arabidopsis phosphatidylinositol- 3-kinase complex plays critical roles in autophagy, endosome sorting, and gravitropism. Front Plant Sci 9,781. |
| [53] | Liu GY, Zeng HQ, Li X, Wei YX, Shi HT (2019). Functional analysis of MaWRKY24 in transcriptional activation of autophagy-related gene 8f/ g and plant disease suscep- tibility to soil-borne Fusarium oxysporum f. sp. cubense. Pathogens 8,264. |
| [54] | Liu YL, Schiff M, Czymmek K, Tallóczy Z, Levine B, Dinesh-Kumar SP (2005). Autophagy regulates program- med cell death during the plant innate immune response. Cell 121,567-577. |
| [55] | Liu YM, Xiong Y, Bassham DC (2009). Autophagy is required for tolerance of drought and salt stress in plants. Autophagy 5,954-963. |
| [56] | Luo LM, Zhang PP, Zhu RH, Fu J, Su J, Zheng J, Wang ZY, Wang D, Gong QQ (2017). Autophagy is rapidly induced by salt stress and is required for salt tolerance in Arabidopsis. Front Plant Sci 8, 1459. |
| [57] | Maleck K, Levine A, Eulgem T, Morgan A, Schmid J, Lawton KA, Dangl JL, Dietrich RA (2000). The trans- criptome of Arabidopsis thaliana during systemic acquired resistance. Nat Genet 26,403-410. |
| [58] | Marshall RS, Vierstra RD (2018). Autophagy: the master of bulk and selective recycling. Annu Rev Plant Biol 69,173- 208. |
| [59] | Masclaux-Daubresse C, Chen QW, Have M (2017). Regu- lation of nutrient recycling via autophagy. Curr Opin Plant Biol 39,8-17. |
| [60] | Michaeli S, Clavel M, Lechner E, Viotti C, Wu J, Dubois M, Hacquard T, Derrien B, Izquierdo E, Lecorbeiller M, Bouteiller N, De Cilia J, Ziegler-Graff V, Vaucheret H, Galili G, Genschik P (2019). The viral F-box protein P0 induces an ER-derived autophagy degradation pathway for the clearance of membrane-bound AGO1. Proc Natl Acad Sci USA 116,22872-22883. |
| [61] | Minina EA, Moschou PN, Vetukuri RR, Sanchez-Vera V, Cardoso C, Liu QS, Elander PH, Dalman K, Beganovic M, Yilmaz JL, Marmon S, Shabala L, Suarez MF, Ljung K, Novák O, Shabala S, Stymne S, Hofius D, Bozhkov PV (2018). Transcriptional stimulation of rate-limiting com- ponents of the autophagic pathway improves plant fitness. J Exp Bot 69,1415-1432. |
| [62] | Naumann C, Müller J, Sakhonwasee S, Wieghaus A, Hause G, Heisters M, Bürstenbinder K, Abel S (2019). The local phosphate deficiency response activates endo- plasmic reticulum stress-dependent autophagy. Plant Phy- siol 179,460-476. |
| [63] | Osterlund MT, Hardtke CS, Wei N, Deng XW (2000). Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405,462-466. |
| [64] | Pu YT, Luo XJ, Bassham DC (2017). TOR-dependent and -independent pathways regulate autophagy in Arabi- dopsis thaliana. Front Plant Sci 8, 1204. |
| [65] | Qin GJ, Ma ZQ, Zhang L, Xing SF, Hou XH, Deng J, Liu JJ, Chen ZL, Qu LJ, Gu HY (2007). Arabidopsis AtBECLIN1/ AtAtg6/ AtVps30 is essential for pollen germi- nation and plant development. Cell Res 17,249-263. |
| [66] | Ren CX, Liu JF, Gong QQ (2014). Functions of autophagy in plant carbon and nitrogen metabolism. Front Plant Sci 5,301. |
| [67] | Rodrigues J, Inzé D, Nelissen H, Saibo NJM (2019). Source-sink regulation in crops under water deficit. Trends Plant Sci 24,652-663. |
| [68] | Rodriguez E, Chevalier J, Olsen J, Ansbøl J, Kapousidou V, Zuo ZL, Svenning S, Loefke C, Koemeda S, Drozdowskyj PS, Jez J, Durnberger G, Kuenzl F, Schutzbier M, Mechtler K, Ebstrup EN, Lolle S, Dagdas Y, Petersen M (2020). Autophagy mediates temporary reprogramming and dedifferentiation in plant somatic cells. EMBO J 39,e103315. |
| [69] | Rushton PJ, Somssich IE, Ringler P, Shen QJ (2010). WRKY transcription factors. Trends Plant Sci 15,247-258. |
| [70] | Sedaghatmehr M, Thirumalaikumar VP, Kamranfar I, Marmagne A, Masclaux-Daubresse C, Balazadeh S (2019). A regulatory role of autophagy for resetting the memory of heat stress in plants. Plant Cell Environ 42,1054-1064. |
| [71] | Shangguan LF, Fang X, Chen LD, Cui LW, Fang JG (2018). Genome-wide analysis of autophagy-related ge- nes (ARGs) in grapevine and plant tolerance to copper stress. Planta 247,1449-1463. |
| [72] | Shibuya K, Niki T, Ichimura K (2013). Pollination induces autophagy in petunia petals via ethylene. J Exp Bot 64,1111-1120. |
| [73] | Signorelli S, Tarkowski ŁP, Van Den Ende W, Bassham DC (2019). Linking autophagy to abiotic and biotic stress responses. Trends Plant Sci 24,413-430. |
| [74] | Suzuki N, Rivero RM, Shulaev V, Blumwald E, Mittler R (2014). Abiotic and biotic stress combinations. New Phytol 203,32-43. |
| [75] | Takahashi S, Murata N (2008). How do environmental stresses accelerate photoinhibition? Trends Plant Sci 13,178-182. |
| [76] | Takeshige K, Baba M, Tsuboi S, Noda T, Ohsumi Y (1992). Autophagy in yeast demonstrated with proteinase- deficient mutants and conditions for its induction. J Cell Biol 119,301-311. |
| [77] | Tasaki M, Asatsuma S, Matsuoka K (2014). Monitoring protein turnover during phosphate starvation-dependent autophagic degradation using a photoconvertible fluores- cent protein aggregate in tobacco BY-2 cells. Front Plant Sci 5,172. |
| [78] | Tsukada M, Ohsumi Y (1993). Isolation and characteri- zation of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett 333,169-174. |
| [79] | Ustün S, Hafrén A, Liu QS, Marshall RS, Minina EA, Bozhkov PV, Vierstra RD, Hofius D (2018). Bacteria exploit autophagy for proteasome degradation and en- hanced virulence in plants. Plant Cell 30,668-685. |
| [80] | Van Der Graaff E, Schwacke R, Schneider A, Desimone M, Flügge UI, Kunze R (2006). Transcription analysis of Arabidopsis membrane transporters and hormone path- ways during developmental and induced leaf senescence. Plant Physiol 141,776-792. |
| [81] | Von Koskull-Doring P, Scharf KD, Nover L (2007). The diversity of plant heat stress transcription factors. Trends Plant Sci 12,452-457. |
| [82] | Wada S, Hayashida Y, Izumi M, Kurusu T, Hanamata S, Kanno K, Kojima S, Yamaya T, Kuchitsu K, Makino A, Ishida H (2015). Autophagy supports biomass production and nitrogen use efficiency at the vegetative stage in rice. Plant Physiol 168,60-73. |
| [83] | Wang P, Nolan TM, Yin YH, Bassham DC (2020). Identi- fication of transcription factors that regulate ATG8 expres- sion and autophagy in Arabidopsis. Autophagy 16, 123- 139. |
| [84] | Wang P, Sun X, Wang N, Jia X, Ma FW (2017). Ectopic expression of an autophagy-associated MdATG7b gene from apple alters growth and tolerance to nutrient stress in Arabidopsis thaliana. Plant Cell Tissue Organ Cult 128,9-23. |
| [85] | Wang Y, Cai SY, Yin LL, Shi K, Xia XJ, Zhou YH, Yu JQ, Zhou J (2015). Tomato HsfA1a plays a critical role in plant drought tolerance by activating ATG genes and inducing autophagy. Autophagy 11,2033-2047. |
| [86] | Wang Y, Cao JJ, Wang KX, Xia XJ, Shi K, Zhou YH, Yu JQ, Zhou J (2019). BZR1 mediates brassinosteroid- induced autophagy and nitrogen starvation in tomato. Plant Physiol 179,671-685. |
| [87] | Wang Y, Yu BJ, Zhao JP, Guo JB, Li Y, Han SJ, Huang L, Du YM, Hong YG, Tang D, Liu YL (2013). Autophagy contributes to leaf starch degradation. Plant Cell 25, 1383- 1399. |
| [88] | Wei YX, Liu W, Hu W, Liu GY, Wu CJ, Liu W, Zeng HQ, He CZ, Shi HT (2017). Genome-wide analysis of autophagy- related genes in banana highlights MaATG8s in cell death and autophagy in immune response to Fusarium wilt. Plant Cell Rep 36,1237-1250. |
| [89] | Williams B, Njaci I, Moghaddam L, Long H, Dickman MB, Zhang XR, Mundree S (2015). Trehalose accumulation triggers autophagy during plant desiccation. PLoS Genet 11,e1005705. |
| [90] | Woo HR, Kim HJ, Lim PO, Nam HG (2019). Leaf senes- cence: systems and dynamics aspects. Annu Rev Plant Biol 70,347-376. |
| [91] | Wu YR, Deng ZY, Lai JB, Zhang YY, Yang CP, Yin BJ, Zhao QZ, Zhang L, Li Y, Yang CW, Xie Q (2009). Dual function of Arabidopsis ATAF1 in abiotic and biotic stress responses. Cell Res 19,1279-1290. |
| [92] | Xia KF, Liu T, Ouyang J, Wang R, Fan T, Zhang MY (2011). Genome-wide identification, classification, and expression analysis of autophagy-associated gene homo- logues in rice ( Oryza sativa L.). DNA Res 18,363-377. |
| [93] | Xia XJ, Fang PP, Guo X, Qian XJ, Zhou J, Shi K, Zhou YH, Yu JQ (2018). Brassinosteroid-mediated apoplastic H 2O 2-glutaredoxin 12/14 cascade regulates antioxidant capacity in response to chilling in tomato. Plant Cell En- viron 41,1052-1064. |
| [94] | Xiong Y, Contento AL, Nguyen PQ, Bassham DC (2007). Degradation of oxidized proteins by autophagy during oxidative stress in Arabidopsis. Plant Physiol 143, 291- 299. |
| [95] | Yamauchi S, Mano S, Oikawa K, Hikino K, Teshima KM, Kimori Y, Nishimura M, Shimazaki KI, Takemiya A (2019). Autophagy controls reactive oxygen species homeostasis in guard cells that is essential for stomatal opening. Proc Natl Acad Sci USA 116,19187-19192. |
| [96] | Yan HY, Liu Y, Zhang K, Song J, Xu WY, Su Z (2019). Chromatin state-based analysis of epigenetic H3K4me3 marks of Arabidopsis in response to dark stress. Front Genet 10,306. |
| [97] | Yan Y, Wang P, He CZ, Shi HT (2017). MeWRKY20 and its interacting and activating autophagy-related protein 8 (MeATG8) regulate plant disease resistance in cassava. Biochem Biophys Res Commun 494,20-26. |
| [98] | Yang C, Shen WJ, Yang LM, Sun Y, Li XB, Lai MY, Wei J, Wang CJ, Xu YC, Li FQ, Liang S, Yang CW, Zhong SW, Luo M, Gao CJ (2020a). HY5-HDA9 module transcrip- tionally regulates plant autophagy in response to light-to- dark conversion and nitrogen starvation. Mol Plant 13,515-531. |
| [99] | Yang M, Ismayil A, Liu YL (2020b). Autophagy in plant- virus interactions. Annu Rev Virol 7,403-419. |
| [100] | Ye H, Ren F, Guo HY, Guo LP, Bai JF, Wang YK (2020). Identification of key genes and transcription factors in ageing Arabidopsis papilla cells by transcriptome analy- sis. Plant Phys Biochem 147,1-9. |
| [101] | Yoshida T, Ohama N, Nakajima J, Kidokoro S, Mizoi J, Nakashima K, Maruyama K, Kim JM, Seki M, Todaka D, Osakabe Y, Sakuma Y, Schöffl F, Shinozaki K, Yamaguchi-Shinozaki K (2011). Arabidopsis HsfA1 transcription factors function as the main positive regu- lators in heat shock-responsive gene expression. Mol Genet Genom 286,321-332. |
| [102] | Yu JL, Zhen XX, Li X, Li N, Xu F (2019). Increased auto- phagy of rice can increase yield and nitrogen use ef- ficiency (NUE). Front Plant Sci 10,584. |
| [103] | Yue WJ, Nie XJ, Cui LC, Zhi YQ, Zhang T, Du XL, Song WN (2018). Genome-wide sequence and expressional analysis of autophagy gene family in bread wheat ( Tri- ticum aestivum L.). J Plant Physiol 229,7-21. |
| [104] | Zhai YF, Guo M, Wang H, Lu JP, Liu JH, Zhang C, Gong ZH, Lu MH (2016). Autophagy, a conserved mechanism for protein degradation, responds to heat, and other abiotic stresses in Capsicum annuum L. Front Plant Sci 7,131. |
| [105] | Zhang JZ, Yang WW, Yue JY, Liu YN, Pei D, Wang HZ (2020). The responses of wheat autophagy and ATG8 family genes to biotic and abiotic stresses. J Plant Growth Regul 39,867-876. |
| [106] | Zhang Y, Li S, Zhou LZ, Fox E, Pao J, Sun W, Zhou C, McCormick S (2011). Overexpression of Arabidopsis thaliana PTEN caused accumulation of autophagic bodies in pollen tubes by disrupting phosphatidylinositol 3-phos- phate dynamics. Plant J 68,1081-1092. |
| [107] | Zhao P, Zhou XM, Zhao LL, Cheung AY, Sun MX (2020). Autophagy-mediated compartmental cytoplasmic deletion is essential for tobacco pollen germination and male fertility. Autophagy 16,2180-2192. |
| [108] | Zhen XX, Xu F, Zhang WZ, Li N, Li X (2019). Overex- pression of rice gene OsATG8b confers tolerance to nitrogen starvation and increases yield and nitrogen use efficiency (NUE) in Arabidopsis. PLoS One 14, e0223011. |
| [109] | Zhou J, Wang J, Yu JQ, Chen ZX (2014). Role and regulation of autophagy in heat stress responses of to- mato plants. Front Plant Sci 5,174. |
| [110] | Zhou XM, Zhao P, Wang W, Zou J, Cheng TH, Peng XB, Sun MX (2015). A comprehensive, genome-wide analysis of autophagy-related genes identified in tobacco suggests a central role of autophagy in plant response to various environmental cues. DNA Res 22,245-257. |
| [111] | Zhu T, Zou LJ, Li Y, Yao XH, Xu F, Deng XG, Zhang DW, Lin HH (2018). Mitochondrial alternative oxidase-depen- dent autophagy involved in ethylene-mediated drought tolerance in Solanum lycopersicum. Plant Biotechnol J 16, 2063-2076. |
| [112] | Zhuang XH, Wang H, Lam SK, Gao CJ, Wang XF, Cai Y, Jiang LW (2013). A BAR-domain protein SH3P2, which binds to phosphatidylinositol 3-phosphate and ATG8, regulates autophagosome formation in Arabidopsis. Plant Cell 25,4596-4615. |
| [1] | 王亚萍, 包文泉, 白玉娥. 单细胞转录组学在植物生长发育及胁迫响应中的应用进展[J]. 植物学报, 2025, 60(1): 101-113. |
| [2] | 李青洋, 刘翠, 何李, 彭姗, 马嘉吟, 胡子祎, 刘宏波. 甘蓝型油菜BnaA02.CPSF6基因的克隆及功能分析(长英文摘要)[J]. 植物学报, 2025, 60(1): 62-73. |
| [3] | 王涛, 冯敬磊, 张翠. 高温胁迫影响玉米生长发育的分子机制研究进展[J]. 植物学报, 2024, 59(6): 963-977. |
| [4] | 杜庆国, 李文学. lncRNA调控玉米生长发育和非生物胁迫研究进展[J]. 植物学报, 2024, 59(6): 950-962. |
| [5] | 闫恒宇, 李朝霞, 李玉斌. 高温对玉米生长的影响及中国耐高温玉米筛选研究进展[J]. 植物学报, 2024, 59(6): 1007-1023. |
| [6] | 路笃贤, 张严妍, 刘艳, 李岩竣, 左新秀, 林金星, 崔亚宁. 非编码RNA在植物生长发育及逆境响应中的研究进展[J]. 植物学报, 2024, 59(5): 709-725. |
| [7] | 曾鑫海, 陈锐, 师宇, 盖超越, 范凯, 李兆伟. 植物SPL转录因子的生物功能研究进展[J]. 植物学报, 2023, 58(6): 982-997. |
| [8] | 许亚楠, 闫家榕, 孙鑫, 王晓梅, 刘玉凤, 孙周平, 齐明芳, 李天来, 王峰. 红光和远红光在调控植物生长发育及应答非生物胁迫中的作用[J]. 植物学报, 2023, 58(4): 622-637. |
| [9] | 张嘉, 李启东, 李翠, 王庆海, 侯新村, 赵春桥, 李树和, 郭强. 植物MATE转运蛋白研究进展[J]. 植物学报, 2023, 58(3): 461-474. |
| [10] | 王琪, 吴允哲, 刘学英, 孙丽莉, 廖红, 傅向东. 类受体激酶调控水稻生长发育和环境适应研究进展[J]. 植物学报, 2023, 58(2): 199-213. |
| [11] | 秦怡, 刘艳爽, 仇柳柳, 周敏, 杜小杉, 戴绍军, 孙美红. MBF1调控植物热应答与生长发育分子机制研究进展[J]. 植物学报, 2022, 57(1): 56-68. |
| [12] | 刘德帅, 姚磊, 徐伟荣, 冯美, 姚文孔. 褪黑素参与植物抗逆功能研究进展[J]. 植物学报, 2022, 57(1): 111-126. |
| [13] | 岳剑茹, 赫云建, 邱天麒, 郭南南, 韩雪萍, 王显玲. 植物微管骨架参与下胚轴伸长调节机制研究进展[J]. 植物学报, 2021, 56(3): 363-371. |
| [14] | 俞启璐, 赵江哲, 朱晓仙, 张可伟. 水稻根分泌激素调节生长速度[J]. 植物学报, 2021, 56(2): 175-182. |
| [15] | 董燕梅, 张文颖, 凌正一, 李靖锐, 白红彤, 李慧, 石雷. 转录因子调控植物萜类化合物生物合成研究进展[J]. 植物学报, 2020, 55(3): 340-350. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||