

植物学报 ›› 2022, Vol. 57 ›› Issue (1): 56-68.DOI: 10.11983/CBB21220 cstr: 32102.14.CBB21220
秦怡1, 刘艳爽1,2, 仇柳柳1, 周敏1, 杜小杉1, 戴绍军1,*( ), 孙美红1,*(
), 孙美红1,*( )
)
收稿日期:2021-12-16
接受日期:2022-01-18
出版日期:2022-01-01
发布日期:2022-01-19
通讯作者:
戴绍军,孙美红
作者简介:sunmeihong@shnu.edu.cn基金资助:
Yi Qin1, Yanshuang Liu1,2, Liuliu Qiu1, Min Zhou1, Xiaoshan Du1, Shaojun Dai1,*( ), Meihong Sun1,*(
), Meihong Sun1,*( )
)
Received:2021-12-16
Accepted:2022-01-18
Online:2022-01-01
Published:2022-01-19
Contact:
Shaojun Dai,Meihong Sun
摘要: MBF1是一种进化上高度保守的转录共激活因子, 存在于所有真核生物中, 可通过连接基础转录机器组分与转录因子来激活基因转录。植物MBF1具有多种重要生物学功能, 包括调控植物生长发育和逆境适应等。该文综述了植物MBF1分子结构与调控机制相关研究进展, 重点总结了AtMBF1c参与植物热胁迫应答调控的分子机制。
秦怡, 刘艳爽, 仇柳柳, 周敏, 杜小杉, 戴绍军, 孙美红. MBF1调控植物热应答与生长发育分子机制研究进展. 植物学报, 2022, 57(1): 56-68.
Yi Qin, Yanshuang Liu, Liuliu Qiu, Min Zhou, Xiaoshan Du, Shaojun Dai, Meihong Sun. Advance in Molecular Mechanism of MBF1 Regulating Plant Heat Response and Development. Chinese Bulletin of Botany, 2022, 57(1): 56-68.
| 物种 | 拉丁名 | 蛋白名 | 蛋白编号(JGI数据库) | 参考文献 |
|---|---|---|---|---|
| 拟南芥 | Arabidopsis thaliana | AtMBF1a | AT2G42680.1 | Tsuda et al., |
| AtMBF1b | AT3G58680.1 | |||
| AtMBF1c | AT3G24500.1 | |||
| 番茄 | Solanum lycopersicum | SlMBF1a | Solyc10g007350.3.1 | Zhang et al., |
| SlMBF1b | Solyc12g014290.2.1 | |||
| SlMBF1c | Solyc07g062400.3.1 | |||
| SlMBF1d | Solyc09g055470.1.1 | |||
| SlER24 | Solyc01g104740.3.1 | |||
| 水稻 | Oryza sativa | OsMBF1a | LOC_Os08g27850.1 | Zhang et al., |
| OsMBF1c | LOC_Os06g39240.1 | |||
| 菠菜 | Spinacia oleracea | SoMBF1b | Spov3_C0009.00073 | Xu et al., |
| SoMBF1c | Spov3_C0062.00022 | |||
| 马铃薯 | S. tuberosum | StMBF1a | PGSC0003DMP400012592 | Yu et al., |
| StMBF1b | PGSC0003DMP400026892 | |||
| StMBF1c | PGSC0003DMP400051869 | |||
| StMBF1d | PGSC0003DMP400051868 | |||
| 小麦 | Triticum aestivum | TaMBF1a-1 | Traes_2AL_A9D390619.1 | Qin et al., |
| TaMBF1a-2 | Traes_2BL_4F31B5695.1 | |||
| TaMBF1a-3 | Traes_2DL_D4AB94C53.1 | |||
| TaMBF1a-4 | Traes_3AS_719D37CCA.1 | |||
| TaMBF1a-5 | Traes_3B_EC2B74116.1 | |||
| TaMBF1b | Traes_4DS_F1C77E7B0.1 | |||
| TaMBF1c-7A | Traes_7AL_FA77CC1F41.1 | |||
| TaMBF1c-7B | Traes_7BL_A002364C5.1 | |||
| TaMBF1c-7C | Traes_7DL_D5AD8EB4B.1 | |||
| 葡萄 | Vitis vinifera | VvMBF1a | VIT_212s0028g02020.1 | Yan et al., |
| VvMBF1a-like | VIT_219s0014g01260.1 | |||
| VvMBF1c | VIT_211s0016g04080.1 | |||
| 大豆 | Glycine max | GmMBF1a-1 | Glyma.06G276200.1.p | Tsuda et al., |
| GmMBF1a-2 | Glyma.06G276300.1.p | |||
| GmMBF1a-3 | Glyma.12G129100.1.p | |||
| 玉米 | Zea mays | ZmMBF1a | ZmPHB47.01G345100.1.p | Tsuda et al., |
| ZmMBF1b | ZmPHB47.04G054300.1.p | |||
| ZmMBF1c | ZmPHB47.09G119100.1.p | |||
| 蓖麻 | Ricinus communis | RcMBF1b | 27894.m000799 | Tsuda et al., |
| RcMBF1c | 29912.m005549 | |||
| 蒺藜苜蓿 | Medicago truncatula | MtMBF1b-1 | Medtr2g084220.1 | Tsuda et al., |
| MtMBF1b-2 | Medtr4g080090.1 | |||
| MtMBF1b-3 | Medtr6g018330.1 | |||
| MtMBF1c | Medtr6g086280.1 |
表1 不同植物物种中MBF1蛋白家族成员
Table 1 Members of the MBF1 protein family in different plant species
| 物种 | 拉丁名 | 蛋白名 | 蛋白编号(JGI数据库) | 参考文献 |
|---|---|---|---|---|
| 拟南芥 | Arabidopsis thaliana | AtMBF1a | AT2G42680.1 | Tsuda et al., |
| AtMBF1b | AT3G58680.1 | |||
| AtMBF1c | AT3G24500.1 | |||
| 番茄 | Solanum lycopersicum | SlMBF1a | Solyc10g007350.3.1 | Zhang et al., |
| SlMBF1b | Solyc12g014290.2.1 | |||
| SlMBF1c | Solyc07g062400.3.1 | |||
| SlMBF1d | Solyc09g055470.1.1 | |||
| SlER24 | Solyc01g104740.3.1 | |||
| 水稻 | Oryza sativa | OsMBF1a | LOC_Os08g27850.1 | Zhang et al., |
| OsMBF1c | LOC_Os06g39240.1 | |||
| 菠菜 | Spinacia oleracea | SoMBF1b | Spov3_C0009.00073 | Xu et al., |
| SoMBF1c | Spov3_C0062.00022 | |||
| 马铃薯 | S. tuberosum | StMBF1a | PGSC0003DMP400012592 | Yu et al., |
| StMBF1b | PGSC0003DMP400026892 | |||
| StMBF1c | PGSC0003DMP400051869 | |||
| StMBF1d | PGSC0003DMP400051868 | |||
| 小麦 | Triticum aestivum | TaMBF1a-1 | Traes_2AL_A9D390619.1 | Qin et al., |
| TaMBF1a-2 | Traes_2BL_4F31B5695.1 | |||
| TaMBF1a-3 | Traes_2DL_D4AB94C53.1 | |||
| TaMBF1a-4 | Traes_3AS_719D37CCA.1 | |||
| TaMBF1a-5 | Traes_3B_EC2B74116.1 | |||
| TaMBF1b | Traes_4DS_F1C77E7B0.1 | |||
| TaMBF1c-7A | Traes_7AL_FA77CC1F41.1 | |||
| TaMBF1c-7B | Traes_7BL_A002364C5.1 | |||
| TaMBF1c-7C | Traes_7DL_D5AD8EB4B.1 | |||
| 葡萄 | Vitis vinifera | VvMBF1a | VIT_212s0028g02020.1 | Yan et al., |
| VvMBF1a-like | VIT_219s0014g01260.1 | |||
| VvMBF1c | VIT_211s0016g04080.1 | |||
| 大豆 | Glycine max | GmMBF1a-1 | Glyma.06G276200.1.p | Tsuda et al., |
| GmMBF1a-2 | Glyma.06G276300.1.p | |||
| GmMBF1a-3 | Glyma.12G129100.1.p | |||
| 玉米 | Zea mays | ZmMBF1a | ZmPHB47.01G345100.1.p | Tsuda et al., |
| ZmMBF1b | ZmPHB47.04G054300.1.p | |||
| ZmMBF1c | ZmPHB47.09G119100.1.p | |||
| 蓖麻 | Ricinus communis | RcMBF1b | 27894.m000799 | Tsuda et al., |
| RcMBF1c | 29912.m005549 | |||
| 蒺藜苜蓿 | Medicago truncatula | MtMBF1b-1 | Medtr2g084220.1 | Tsuda et al., |
| MtMBF1b-2 | Medtr4g080090.1 | |||
| MtMBF1b-3 | Medtr6g018330.1 | |||
| MtMBF1c | Medtr6g086280.1 |
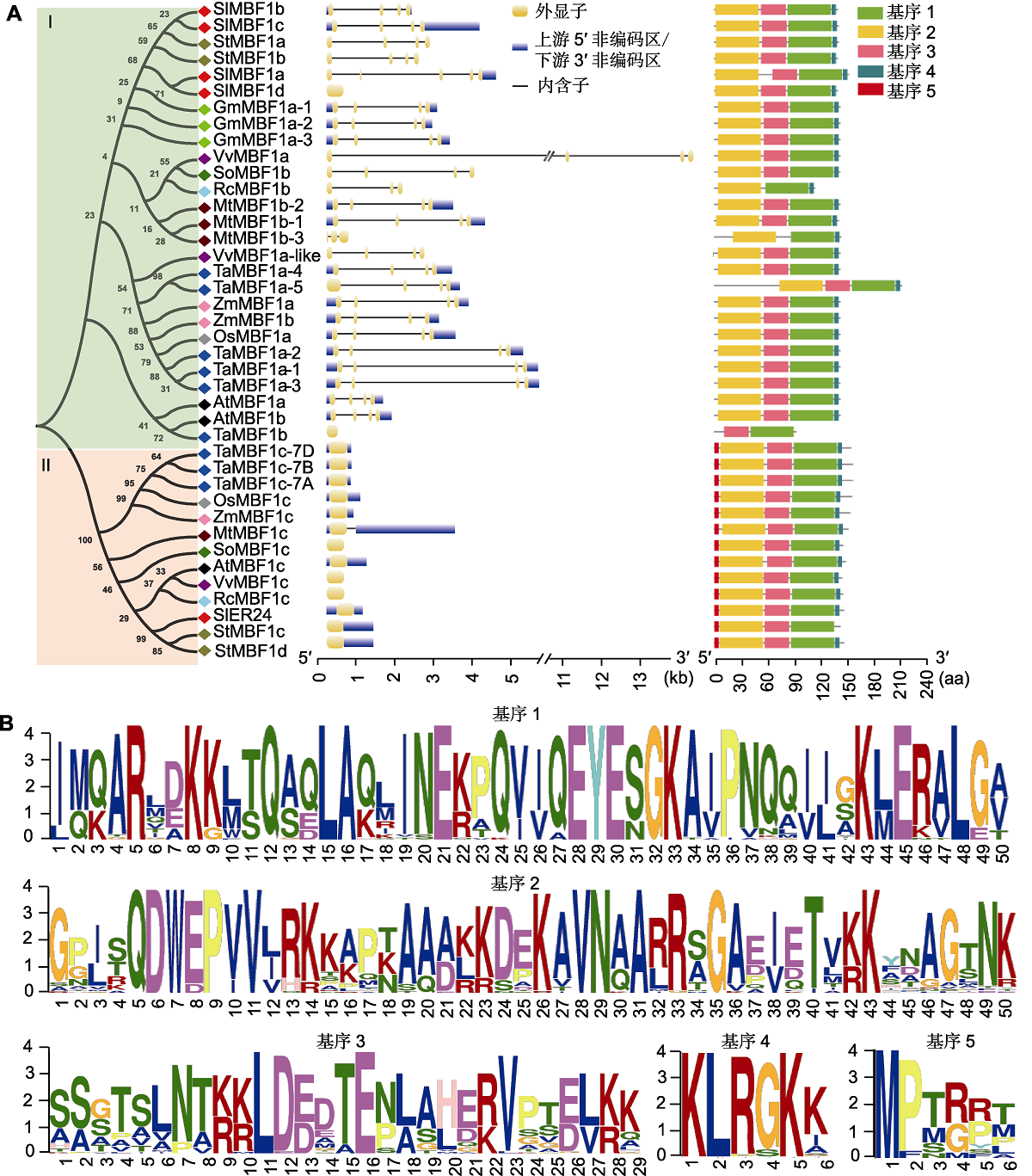
图1 植物MBF1系统发育关系、基因结构和保守基序分析 (A) 植物MBF1的系统发育关系、基因结构和保守基序分布图。11个物种MBF1蛋白的全长蛋白质序列来自JGI数据库(https:// phytozome-next.jgi.doe.gov/) (表1)。用MEGA7.0软件基于邻接法构建同源系统进化树, 绿色背景表示I类MBF1蛋白亚家族, 红色背景表示II类MBF1蛋白亚家族。利用GSDS2.0软件(http://gsds.cbi.pku.edu.cn/)在线绘制基因结构图, 上游5'非编码区/下游3'非编码区用蓝色方框表示, 外显子用黄色方框表示, 内含子用黑色直线显示。使用MEME网站(https://meme-suite.org/meme/ tools/meme)在线预测保守基序。通过NCBI网站CDD数据库(https://www.ncbi.nlm.nih.gov/cdd)鉴定蛋白质序列的保守结构域及功能。利用Tbtools软件绘制保守基序分布图, 5个保守基序用不同颜色方框表示。(B) 11个物种MBF1蛋白中预测到的保守基序序列。
Figure 1 The analysis of the phylogenetic relationships, gene structures and conserved motifs of plant MBF1 (A) Phylogenetic relationship, gene structure and conserved motif distribution map of plant MBF1. The full-length protein sequences of MBF1 proteins from 11 species are downloaded from the JGI database (https://phytozome-next.jgi.doe.gov/) (table 1). MEGA7.0 software is used to construct a Neighbor-Joining homologous phylogenetic tree. The green background indicates the type I subfamily, and the red background indicates the type II subfamily. GSDS2.0 software (http://gsds.cbi.pku.edu.cn/) is used to draw a gene structure diagram online. 5'UTR/3'UTR are represented by blue boxes, exons are represented by yellow boxes, and introns are represented by black straight lines. The MEME website (https://meme-suite.org/meme/tools/meme) is used to predict conserved motifs. Conserved domains and functions are identified through the NCBI website CDD database (https://www.ncbi.nlm.nih.gov/cdd). Tbtools software is used to draw a map of the conserved motifs, and the 5 conserved motifs are indicated by boxes with different colors. (B) The predicted conserved motif sequences in MBF1 proteins of 11 species.
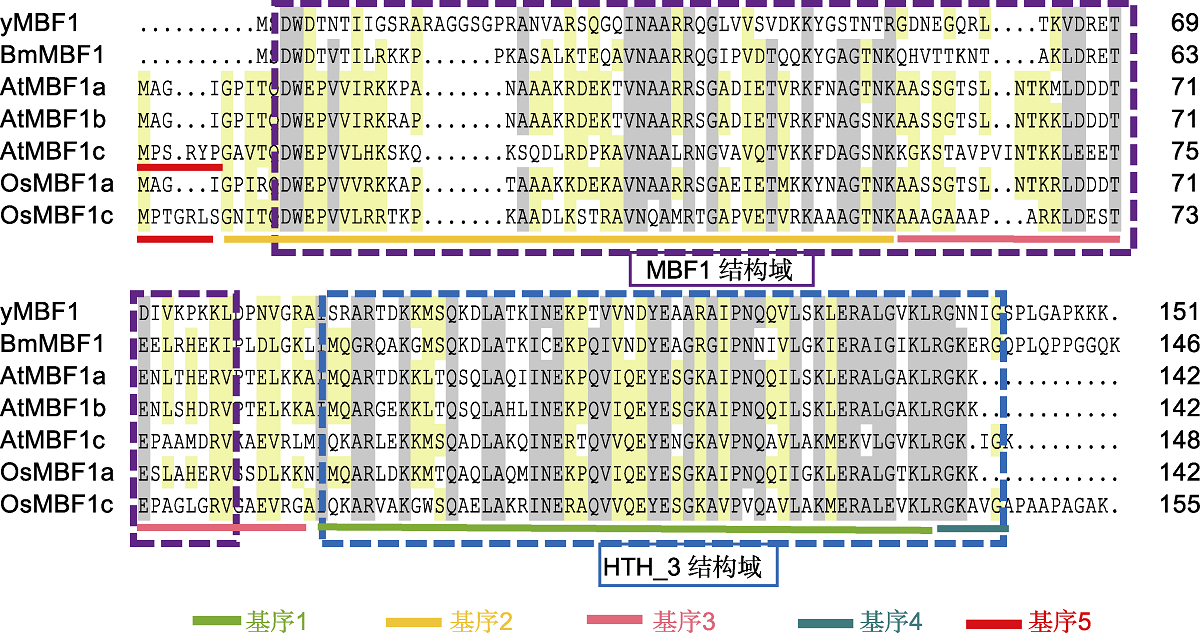
图2 不同物种MBF1s同源序列比对分析 家蚕BmMBF1和酿酒酵母yMBF1蛋白质序列从Ensembl数据库(https://ensemblgenomes.org/)下载, 蛋白编号分别为BGIBMGA 007702和YOR298C-A。利用DNAMAN软件对蛋白序列进行比对。紫色框表示N端MBF1结构域, 蓝色框表示C端HTH_3结构域。拟南芥和水稻MBF1蛋白质序列及蛋白编号见表1。
Figure 2 Homologous sequence alignment analysis of MBF1s of different species The protein sequences of Bombyx mori BmMBF1 and Saccharomyces cerevisiae yMBF1 are downloaded from Ensembl database (https://ensemblgenomes.org/), the corresponding protein accession number is BGIBMGA007702 and YOR298C-A. DNAMAN software is used for sequence alignment. The purple box indicates the N-terminal MBF1 domain, and the blue box indicates the C-terminal HTH_3 domain. The protein sequences and protein accession number of Arabidopsis and rice MBF1 are listed in Table 1.
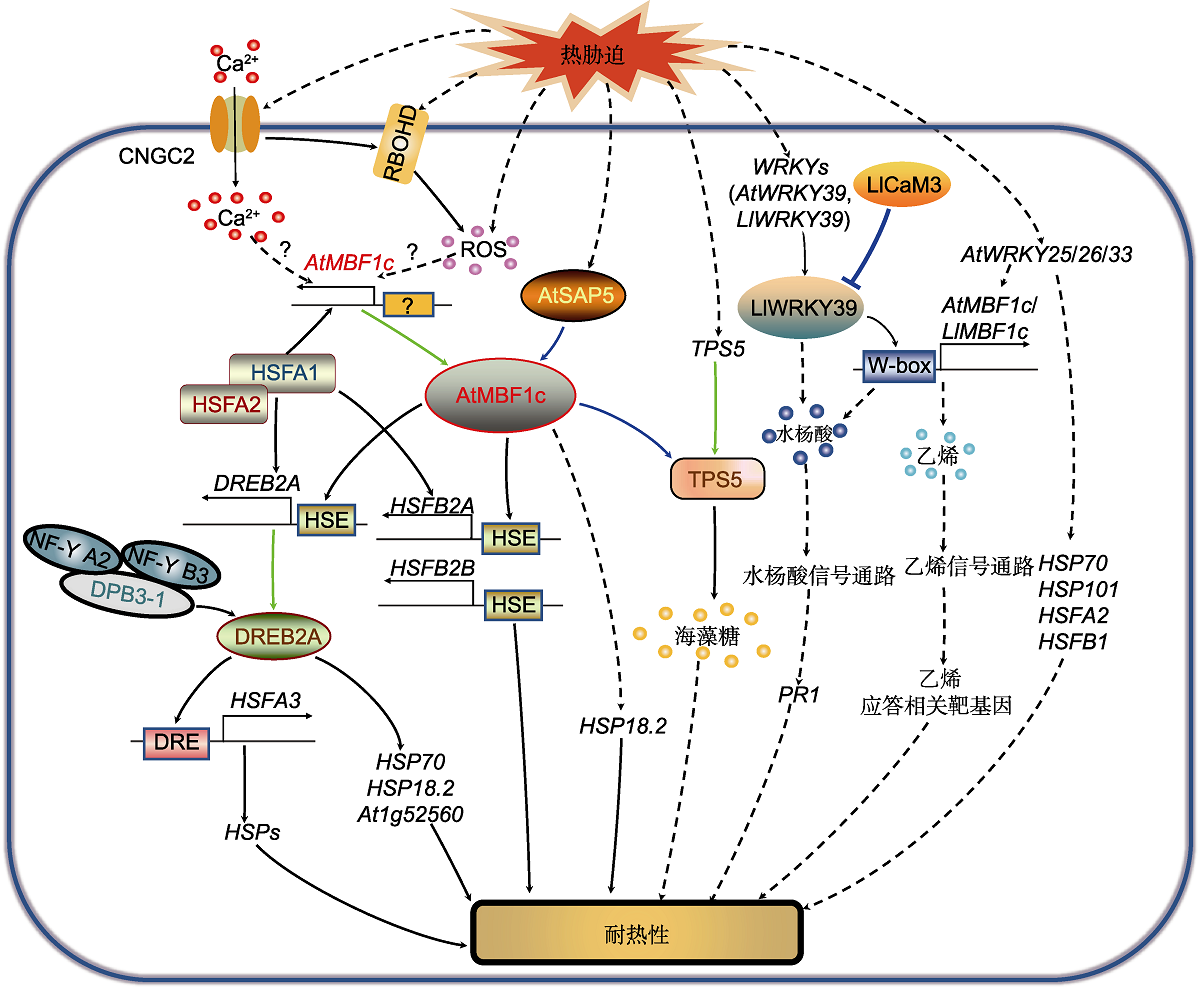
图3 MBF1调控热胁迫应答信号通路 热胁迫通过激活质膜CNGC2引起Ca2+内流, 通过激活质膜结合的RBOHD导致ROS积累。而Ca2+信号和ROS信号通过未知途径激活AtMBF1c及其下游靶基因调节的热胁迫应答。热激转录因子HSFA1与HSFA2相互作用, 直接调控AtMBF1c、HSFA2、HSFBs和DREB2A的表达。同时, AtMBF1c通过与DREB2A、HSFB2A和HSFB2B的启动子HSE元件结合, 调控其基因表达, 提高热胁迫耐受性。DREB2A与DPB3-1、NF-Y A2和NF-Y B3形成的三聚体共激活复合物互作, 增强其对下游靶基因HSFA3的转录激活, 提高植物的耐热性。DREB2A也促进HSP70、HSP18.2和At1g52560的表达, 增强植株耐热性。AtSAP5作为上游调控因子与AtMBF1c发生互作并激活AtMBF1c, 调节细胞核中HSP18.2的表达, 提高植物耐热性。热胁迫诱导TPS5的表达, AtMBF1c与TPS5互作, 通过促进海藻糖的合成和积累提高耐热性。热胁迫诱导AtWRKY39、AtWRKY25、AtWRKY26和AtWRKY33的表达。AtWRKY39通过AtMBF1c调节水杨酸(SA)信号通路下游PR1基因表达, 提高耐热性。AtWRKY25、AtWRKY26和AtWRKY33通过AtMBF1c调节乙烯信号通路下游基因的表达提高耐热性, 同时通过促进HSP70、HSP101、HSFA2和HSFB1的表达提高耐热性。CNGC2: 环核苷酸门控离子通道2; DPB3-1 (NF-YC10): DNA聚合酶II亚基B3-1; DRE: 脱水应答元件; DREB2A: 干旱应答元件结合蛋白2A; HSE: 热休克响应元件; HSFA1/2/3: 热激转录因子A1/2/3; HSFB2A/B: 热激转录因子B2A/B; HSP18.2/70/101: 热激蛋白18.2/70/101; MBF1c: 多蛋白桥梁因子1c; NF-Y A2/B3: 核因子-Y A2/B3; PR1: 病程相关因子1; RBOHD: 呼吸爆发氧化同源蛋白D; ROS: 活性氧; SAP5: 胁迫相关蛋白5; TPS5: 海藻糖磷酸合成酶5。蓝色实线箭头表示蛋白互作; 绿色实线箭头表示基因编码蛋白; 黑色实线箭头表示直接转录激活; 黑色虚线箭头表示间接转录激活。
Figure 3 Signaling pathway of MBF1 regulating heat stress response Heat stress causes Ca2+ influx by activating the plasma membrane-localized protein CNGC2, and leads to the accumulation of ROS by activating the plasma membrane-bound RBOHD. The Ca2+ signal and ROS signal activate the heat stress response by regulating AtMBF1c and its downstream target genes through unknown pathways. The heat stress transcription factor HSFA1 interacts with HSFA2 and directly regulates the expression of AtMBF1c, HSFA2, HSFBs and DREB2A. At the same time, AtMBF1c binds to the HSE elements of DREB2A, HSFB2A and HSFB2B promoters to regulate their gene expression and improve heat stress tolerance. DREB2A interacts with the trimeric co-activation complex formed by DPB3-1, NF-Y A2 and NF-Y B3 to enhance the transcriptional activation of the downstream target gene HSFA3 and improve plant heat tolerance. DREB2A also promotes the expression of HSP70, HSP18.2 and At1g52560 to enhance plant heat tolerance. As the upstream regulator, AtSAP5 interacts with and activates AtMBF1c in the nucleus, regulating the expression of HSP18.2 and improving plant heat tolerance. Heat stress induces the expression of TPS5. AtMBF1c interacts with TPS5 to improve heat tolerance by promoting the synthesis and accumulation of trehalose. Heat stress induces the expression of AtWRKY39, AtWRKY25, AtWRKY26 and AtWRKY33. AtWRKY39 regulates the expression of the downstream gene of salicylic acid (SA) signaling pathway PR1 through AtMBF1c to improve heat tolerance. AtWRKY25, AtWRKY26 and AtWRKY33 regulate the expression of downstream genes in the ethylene (ET) signaling pathway through AtMBF1c to improve heat resistance, and at the same time promote the expression of HSP70, HSP101, HSFA2, and HSFB1. CNGC2: Cyclic nucleotide-gated channel 2; DPB3-1 (NF-YC10): DNA polymerase II subunit B3-1; DRE: Dehydration-responsive element; DREB2A: Dehydration responsive element-binding protein 2A; HSE: Heat shock elements; HSFA1/2/3: Heat stress transcription factor A1/2/3; HSFB2A/B: Heat stress transcription factor B2A/B; HSP18.2/ 70/101: Heat shock protein 18.2/70/101; MBF1c: Multiprotein bridging factor 1c; NF-Y A2/B3: Nuclear factor Y A2/B3; PR1: Pathogenesis-related factor 1; RBOHD: Respiratory burst oxidase homologue D; ROS: Reactive oxygen species; SAP5: Stress-associated protein 5; TPS5: Trehalose phosphate synthetase 5. The blue solid arrow indicates protein interaction; the green solid arrow indicates gene encoding protein; the black solid arrow indicates direct transcription activation; the black dashed arrow indicates indirect transcription activation.
| [1] | 李思佳, 张咏雪, 贾明生, 李莹, 戴绍军 (2020). 植物类LORELEI糖基磷脂酰肌醇锚定蛋白研究进展. 植物学报 55, 541-550. |
| [2] |
邱丽丽, 赵琪, 张玉红, 戴绍军 (2017). 植物质膜蛋白质组的逆境应答研究进展. 植物学报 52, 128-147.
DOI |
| [3] |
张洵, 喻娟娟, 王思竹, 李莹, 戴绍军 (2019). 植物DREPP基因家族研究进展. 植物学报 54, 582-595.
DOI |
| [4] | Arc E, Sechet J, Corbineau F, Rajjou L, Marion-Poll A (2013). ABA crosstalk with ethylene and nitric oxide in seed dormancy and germination. Front Plant Sci 4, 63. |
| [5] |
Arce DP, Godoy AV, Tsuda K, Yamazaki KI, Valle EM, Iglesias MJ, Di Mauro MF, Casalongué CA (2010). The analysis of an Arabidopsis triple knock-down mutant reveals functions for MBF1 genes under oxidative stress conditions. J Plant Physiol 167, 194-200.
DOI URL |
| [6] |
Avonce N, Leyman B, Mascorro-Gallardo JO, Van Dijck P, Thevelein JM, Iturriaga G (2004). The Arabidopsis trehalose-6-P synthase AtTPS1 gene is a regulator of glucose, abscisic acid, and stress signaling. Plant Physiol 136, 3649-3659.
DOI URL |
| [7] | Bita CE, Gerats T (2013). Plant tolerance to high temperature in a changing environment: scientific fundamentals and production of heat stress-tolerant crops. Front Plant Sci 4, 273. |
| [8] |
Blombach F, Launay H, Snijders APL, Zorraquino V, Wu H, de Koning B, Brouns SJJ, Ettema TJG, Camilloni C, Cavalli A, Vendruscolo M, Dickman MJ, Cabrita LD, La Teana A, Benelli D, Londei P, Christodoulou J, van der Oost J (2014). Archaeal MBF1 binds to 30S and 70S ribosomes via its helix-turn-helix domain. Biochem J 462, 373-384.
DOI PMID |
| [9] |
Busk PK, Wulf-Andersen L, Strøm CC, Enevoldsen M, Thirstrup K, Haunsø S, Sheikh SØP (2003). Multiprotein bridging factor 1 cooperates with c-jun and is necessary for cardiac hypertrophy in vitro. Exp Cell Res 286, 102-114.
DOI URL |
| [10] |
Clarke SM, Mur LAJ, Wood JE, Scott IM (2004). Salicylic acid dependent signaling promotes basal thermotolerance but is not essential for acquired thermotolerance in Arabidopsis thaliana. Plant J 38, 432-447.
DOI URL |
| [11] | Corbineau F, Xia Q, Bailly C, El-Maarouf-Bouteau H (2014). Ethylene, a key factor in the regulation of seed dormancy. Front Plant Sci 5, 539. |
| [12] |
De Boeck HJ, Bassin S, Verlinden M, Zeiter M, Hiltbrunner E (2016). Simulated heat waves affected alpine grassland only in combination with drought. New Phytol 209, 531-541.
DOI PMID |
| [13] |
Di Mauro MF, Iglesias MJ, Arce DP, Valle EM, Arnold RB, Tsuda K, Yamazaki KI, Casalongué CA, Godoy AV (2012). MBF1s regulate ABA-dependent germination of Arabidopsis seeds. Plant Signal Behav 7, 188-192.
DOI URL |
| [14] |
Ding LP, Wu Z, Teng RD, Xu SJ, Cao X, Yuan GZ, Zhang DH, Teng NJ (2021). LlWRKY39 is involved in thermotolerance by activating LlMBF1c and interacting with LlCaM3 in lily (Lilium longiflorum). Hortic Res 8, 36.
DOI URL |
| [15] |
Eulgem T, Somssich IE (2007). Networks of WRKY transcription factors in defense signaling. Curr Opin Plant Biol 10, 366-371.
DOI URL |
| [16] |
Finka A, Cuendet AFH, Maathuis FJM, Saidi Y, Goloubinoff P (2012). Plasma membrane cyclic nucleotide gated calcium channels control land plant thermal sensing and acquired thermotolerance. Plant Cell 24, 3333-3348.
DOI URL |
| [17] |
Gao F, Han XW, Wu JH, Zheng SZ, Shang ZL, Sun DY, Zhou RG, Li B (2012). A heat-activated calcium-permeable channel-Arabidopsis cyclic nucleotide-gated ion channel 6-is involved in heat shock responses. Plant J 70, 1056-1069.
DOI URL |
| [18] | Godoy AV, Zanetti ME, San Segundo B, Casalongué CA (2001). Identification of a putative Solanum tuberosum transcriptional coactivator up-regulated in potato tubers by Fusarium solani f. sp. eumartii infection and wounding. Physiol Plant 112, 217-222. |
| [19] |
Gong ZZ, Xiong LM, Shi HZ, Yang SH, Herrera-Estrella LR, Xu GH, Chao DY, Li JR, Wang PY, Qin F, Li JJ, Ding YL, Shi YT, Wang Y, Yang YQ, Guo Y, Zhu JK (2020). Plant abiotic stress response and nutrient use efficiency. Sci China Life Sci 63, 635-674.
DOI URL |
| [20] |
Grennan AK (2007). The role of trehalose biosynthesis in plants. Plant Physiol 144, 3-5.
PMID |
| [21] |
Higashi Y, Ohama N, Ishikawa T, Katori T, Shimura A, Kusakabe K, Yamaguchi-Shinozaki K, Ishida J, Tanaka M, Seki M, Shinozaki K, Sakata Y, Hayashi T, Taji T (2013). HsfA1d, a protein identified via FOX hunting using Thellungiella salsuginea cDNAs improves heat tolerance by regulating heat-stress-responsive gene expression. Mol Plant 6, 411-422.
DOI PMID |
| [22] |
Hommel M, Khalil-Ahmad Q, Jaimes-Miranda F, Mila I, Pouzet C, Latché A, Pech JC, Bouzayen M, Regad F (2008). Over-expression of a chimeric gene of the transcriptional co-activator MBF1 fused to the EAR repressor motif causes developmental alteration in Arabidopsis and tomato. Plant Sci 175, 168-177.
DOI URL |
| [23] |
Hozain M, Abdelmageed H, Lee J, Kang M, Fokar M, Allen RD, Holaday AS (2012). Expression of AtSAP5 in cotton up-regulates putative stress-responsive genes and improves the tolerance to rapidly developing water deficit and moderate heat stress. J Plant Physiol 169, 1261-1270.
DOI URL |
| [24] |
Jaimes-Miranda F, Montes RAC (2020). The plant MBF1 protein family: a bridge between stress and transcription. J Exp Bot 71, 1782-1791.
DOI PMID |
| [25] |
Jegadeesan S, Beery A, Altahan L, Meir S, Pressman E, Firon N (2018). Ethylene production and signaling in tomato (Solanum lycopersicum) pollen grains is responsive to heat stress conditions. Plant Reprod 31, 367-383.
DOI PMID |
| [26] |
Katano K, Honda K, Suzuki N (2018a). Integration between ROS regulatory systems and other signals in the regulation of various types of heat responses in plants. Int J Mol Sci 19, 3370.
DOI URL |
| [27] |
Katano K, Kataoka R, Fujii M, Suzuki N (2018b). Differences between seedlings and flowers in anti-ROS based heat responses of Arabidopsis plants deficient in cyclic nucleotide gated channel 2. Plant Physiol Biochem 123, 288-296.
DOI URL |
| [28] |
Kim GD, Cho YH, Yoo SD (2015). Regulatory functions of evolutionarily conserved AN1/A20-like Zinc finger family proteins in Arabidopsis stress responses under high temperature. Biochem Biophys Res Commun 457, 213-220.
DOI URL |
| [29] |
Kotak S, Larkindale J, Lee U, von Koskull-Döring P, Vierling E, Scharf KD (2007). Complexity of the heat stress response in plants. Curr Opin Plant Biol 10, 310-316.
DOI URL |
| [30] |
Larkindale J, Hall JD, Knight MR, Vierling E (2005). Heat stress phenotypes of Arabidopsis mutants implicate multiple signaling pathways in the acquisition of thermotolerance. Plant Physiol 138, 882-897.
PMID |
| [31] |
Li BJ, Gao K, Ren HM, Tang WQ (2018). Molecular mechanisms governing plant responses to high temperatures. J Integr Plant Biol 60, 757-779.
DOI URL |
| [32] |
Li SJ, Fu QT, Chen LG, Huang WD, Yu DQ (2011). Arabidopsis thaliana WRKY25, WRKY26, and WRKY33 coordinate induction of plant thermotolerance. Planta 233, 1237-1252.
DOI URL |
| [33] |
Li SJ, Zhou X, Chen LG, Huang WD, Yu DQ (2010). Functional characterization of Arabidopsis thaliana WRKY39 in heat stress. Mol Cells 29, 475-483.
DOI URL |
| [34] | Miller G, Schlauch K, Tam R, Cortes D, Torres MA, Shulaev V, Dangl JL, Mittler R (2009). The plant NADPH oxidase RBOHD mediates rapid systemic signaling in response to diverse stimuli. Sci Signal 2, ra45. |
| [35] |
Mittler R (2006). Abiotic stress, the field environment and stress combination. Trends Plant Sci 11, 15-19.
PMID |
| [36] |
Mittler R, Finka A, Goloubinoff P (2012). How do plants feel the heat? Trends Biochem Sci 37, 118-125.
DOI PMID |
| [37] |
Müller M, Munné-Bosch S (2015). Ethylene response factors: a key regulatory hub in hormone and stress signaling. Plant Physiol 169, 32-41.
DOI PMID |
| [38] |
Nakashima K, Yamaguchi-Shinozaki K (2013). ABA signaling in stress-response and seed development. Plant Cell Rep 32, 959-970.
DOI PMID |
| [39] |
Ohama N, Sato H, Shinozaki K, Yamaguchi-Shinozaki K (2017). Transcriptional regulatory network of plant heat stress response. Trends Plant Sci 22, 53-65.
DOI URL |
| [40] |
Ozaki J, Takemaru KI, Ikegami T, Mishima M, Ueda H, Hirose S, Kabe Y, Handa H, Shirakawa M (1999). Identification of the core domain and the secondary structure of the transcriptional coactivator MBF1. Genes Cells 4, 415-424.
PMID |
| [41] |
Pandey GK, Grant JJ, Cheong YH, Kim BG, Li LG, Luan S (2005). ABR1, an APETALA2-domain transcription factor that functions as a repressor of ABA response in Arabidopsis. Plant Physiol 139, 1185-1193.
DOI URL |
| [42] |
Qin DD, Wang F, Geng XL, Zhang LY, Yao YY, Ni ZF, Peng HR, Sun QX (2015). Overexpression of heat stress-responsive TaMBF1c, a wheat (Triticum aestivum L.) multiprotein bridging factor, confers heat tolerance in both yeast and rice. Plant Mol Biol 87, 31-45.
DOI URL |
| [43] |
Sajid M, Rashid B, Ali Q, Husnain T (2018). Mechanisms of heat sensing and responses in plants. It is not all about Ca2+ ions. Biol Plant 62, 409-420.
DOI URL |
| [44] |
Sakuma Y, Maruyama K, Qin F, Osakabe Y, Shinozaki K, Yamaguchi-Shinozaki K (2006). Dual function of an Arabidopsis transcription factor DREB2A in water-stress- responsive and heat-stress-responsive gene expression. Proc Natl Acad Sci USA 103, 18822-18827.
DOI URL |
| [45] |
Sato H, Mizoi J, Tanaka H, Maruyama K, Qin F, Osakabe Y, Morimoto K, Ohori T, Kusakabe K, Nagata M, Shinozaki K, Yamaguchi-Shinozaki K (2014). Arabidopsis DPB3-1, a DREB2A interactor, specifically enhances heat stress-induced gene expression by forming a heat stress-specific transcriptional complex with NF-Y subunits. Plant Cell 26, 4954-4973.
DOI URL |
| [46] |
Schramm F, Larkindale J, Kiehlmann E, Ganguli A, Englich G, Vierling E, Von Koskull-Döring P (2008). A cascade of transcription factor DREB2A and heat stress transcription factor HsfA3 regulates the heat stress response of Arabidopsis. Plant J 53, 264-274.
DOI URL |
| [47] |
Song C, Ortiz-Urquiza A, Ying SH, Zhang JX, Keyhani NO (2015). Interaction between TATA-binding protein (TBP) and multiprotein bridging factor-1 (MBF1) from the filamentous insect pathogenic fungus Beauveria bassiana. PLoS One 10, e0140538.
DOI URL |
| [48] |
Suzuki N, Bajad S, Shuman J, Shulaev V, Mittler R (2008). The transcriptional co-activator MBF1c is a key regulator of thermotolerance in Arabidopsis thaliana. J Biol Chem 283, 9269-9275.
DOI URL |
| [49] |
Suzuki N, Bassil E, Hamilton JS, Inupakutika MA, Zandalinas SI, Tripathy D, Luo YT, Dion E, Fukui G, Kumazaki A, Nakano R, Rivero RM, Verbeck GF, Azad RK, Blumwald E, Mittler R (2016). ABA is required for plant acclimation to a combination of salt and heat stress. PLoS One 11, e0147625.
DOI URL |
| [50] |
Suzuki N, Rizhsky L, Liang HJ, Shuman J, Shulaev V, Mittler R (2005). Enhanced tolerance to environmental stress in transgenic plants expressing the transcriptional coactivator multiprotein bridging factor 1c. Plant Physiol 139, 1313-1322.
DOI URL |
| [51] |
Suzuki N, Sejima H, Tam R, Schlauch K, Mittler R (2011). Identification of the MBF1 heat-response regulon of Arabidopsis thaliana. Plant J 66, 844-851.
DOI URL |
| [52] |
Takemaru KI, Harashima S, Ueda H, Hirose S (1998). Yeast coactivator MBF1 mediates GCN4-dependent transcriptional activation. Mol Cell Biol 18, 4971-4976.
DOI PMID |
| [53] |
Takemaru KI, Li FQ, Ueda H, Hirose S (1997). Multiprotein bridging factor 1 (MBF1) is an evolutionarily conserved transcriptional coactivator that connects a regulatory factor and TATA element-binding protein. Proc Natl Acad Sci USA 94, 7251-7256.
DOI URL |
| [54] |
Tian X, Qin Z, Zhao Y, Wen J, Lan T, Zhang L, Wang F, Qin D, Yu K, Zhao A, Hu Z, Yao Y, Ni Z, Sun Q, De Smet I, Peng H, Xin M (2021). Stress granule associated TaMBF1c confers thermotolerance through regulating specific mRNA translation in wheat (Triticum aestivum). New Phytol 233, 1719-1731.
DOI URL |
| [55] |
Tsuda K, Tsuji T, Hirose S, Yamazaki KI (2004). Three Arabidopsis MBF1 homologs with distinct expression profiles play roles as transcriptional co-activators. Plant Cell Physiol 45, 225-231.
DOI URL |
| [56] | Tsuda K, Yamazaki KI (2004). Structure and expression analysis of three subtypes of Arabidopsis MBF1 genes. Biochim Biophys Acta 1680, 1-10. |
| [57] |
Wang XL, Du Y, Yu DQ (2019). Trehalose phosphate synthase 5-dependent trehalose metabolism modulates basal defense responses in Arabidopsis thaliana. J Integr Plant Biol 61, 509-527.
DOI URL |
| [58] |
Wang YY, Wei XL, Huang J, Wei JC (2017). Modification and functional adaptation of the MBF1 gene family in the lichenized fungus Endocarpon pusillum under environmental stress. Sci Rep 7, 16333.
DOI URL |
| [59] |
Xu CX, Jiao C, Sun HH, Cai XF, Wang XL, Ge CH, Zheng Y, Liu WL, Sun XP, Xu YM, Deng J, Zhang ZH, Huang SW, Dai SJ, Mou BQ, Wang QX, Fei ZJ, Wang QH (2017). Draft genome of spinach and transcriptome diversity of 120 Spinacia accessions. Nat Commun 8, 15275.
DOI URL |
| [60] |
Yan Q, Hou HM, Singer SD, Yan XX, Guo RR, Wang XP (2014). The grape VvMBF1 gene improves drought stress tolerance in transgenic Arabidopsis thaliana. Plant Cell Tiss Org 118, 571-582.
DOI URL |
| [61] |
Yoshida T, Sakuma Y, Todaka D, Maruyama K, Qin F, Mizoi J, Kidokoro S, Fujita Y, Shinozaki K, Yamaguchi-Shinozaki K (2008). Functional analysis of an Arabidopsis heat-shock transcription factor HsfA3 in the transcriptional cascade downstream of the DREB2A stress-regulatory system. Biochem Biophys Res Commun 368, 515-521.
DOI URL |
| [62] |
Yu RM, Suo YY, Yang R, Chang YN, Tian T, Song YJ, Wang HJ, Wang C, Yang RJ, Liu HL, Gao G (2021). StMBF1c positively regulates disease resistance to Ralstonia solanacearum via its primary and secondary upregulation combining expression of StTPS5 and resistance marker genes in potato. Plant Sci 307, 110877.
DOI URL |
| [63] |
Zandalinas SI, Balfagón D, Arbona V, Gómez-Cadenas A, Inupakutika MA, Mittler R (2016). ABA is required for the accumulation of APX1 and MBF1c during a combination of water deficit and heat stress. J Exp Bot 67, 5381-5390.
PMID |
| [64] |
Zanetti ME, Blanco FA, Daleo GR, Casalongué CA (2003). Phosphorylation of a member of the MBF1 transcriptional co-activator family, StMBF1, is stimulated in potato cell suspensions upon fungal elicitor challenge. J Exp Bot 54, 623-632.
DOI URL |
| [65] |
Zhang X, Xu ZX, Chen LC, Ren ZH (2019). Comprehensive analysis of multiprotein bridging factor 1 family genes and SlMBF1c negatively regulate the resistance to Botrytis cinerea in tomato. BMC Plant Biol 19, 437.
DOI PMID |
| [66] |
Zou LF, Yu BW, Ma XL, Cao BH, Chen GJ, Chen CM, Lei JJ (2019). Cloning and expression analysis of the BocMBF1c gene involved in heat tolerance in Chinese kale. Int J Mol Sci 20, 5637.
DOI URL |
| [1] | 惠城阳, 章巧依, 刘腾腾, 刘维勇, 周丽娜, 金鑫杰, 张永华, 刘金亮. 温州大罗山主要植被类型及物种组成特征[J]. 植物生态学报, 2025, 49(植被): 1-. |
| [2] | 曹毅 张松林 王旭峰 杨安昌 任敏慧 杨浩 韩超. 兰州市南北两山植物群落数据集[J]. 植物生态学报, 2025, 49(植被): 1-0. |
| [3] | 陈龙 郭柯 勾晓华 赵秀海 马泓若. 祁连圆柏林群落组成及特征[J]. 植物生态学报, 2025, 49(植被): 0-0. |
| [4] | 童金莲, 张博纳, 汤璐瑶, 叶琳峰, 李姝雯, 谢江波, 李彦, 王忠媛. C4植物狗尾草功能性状网络沿降水梯度带的区域分异规律[J]. 植物生态学报, 2025, 49(预发表): 1-. |
| [5] | 闫小红 胡文海. 亚热带地区3种常绿阔叶植物冬季光保护机制的差异[J]. 植物生态学报, 2025, 49(预发表): 0-0. |
| [6] | 赵常明 熊高明 申国珍 葛结林 徐文婷 徐凯 武元帅 谢宗强. 神农架常绿落叶阔叶混交林和亚高山针叶林植物群落特征数据集[J]. 植物生态学报, 2025, 49(典型生态系统数据集): 0-0. |
| [7] | 赵珮杉 高广磊 丁国栋 张英. 林龄和生态位对樟子松人工林地下真菌群落构建的影响[J]. 植物生态学报, 2025, 49(地上地下生态过程关联): 1-0. |
| [8] | 张子睿, 周静, 胡艳萍, 梁爽, 马永鹏, 陈伟乐. 极度濒危植物巧家五针松的根内和根际真菌群落特征[J]. , 2025, 49(濒危植物的保护与恢复): 0-. |
| [9] | 逯子佳, 王天瑞, 郑斯斯, 孟宏虎, 曹建国, Gregor Kozlowski, 宋以刚. 孑遗植物湖北枫杨的环境适应性遗传变异与遗传脆弱性[J]. , 2025, 49(濒危植物的保护与恢复): 0-. |
| [10] | 黄承玲, 黎荣瀚, 覃红玲, 杨胜雄, 田晓玲, 夏国威, 陈正仁, 周玮. 基于SNP分子标记的极小种群野生植物荔波杜鹃保护遗传学研究[J]. , 2025, 49(濒危植物的保护与恢复): 0-. |
| [11] | 周鑫宇, 刘会良, 高贝, 卢妤婷, 陶玲庆, 文晓虎, 张岚, 张元明. 新疆特有濒危植物雪白睡莲繁殖生物学研究[J]. , 2025, 49(濒危植物的保护与恢复): 0-. |
| [12] | 高雨轩, 苏艳军, 冯育才, 张军, 汪小全, 刘玲莉. 珍稀濒危孑遗植物银杉的研究与保护现状[J]. , 2025, 49(濒危植物的保护与恢复): 0-. |
| [13] | 朱润铖, 蔡锡安, 黄娟. 植物防御相关挥发性有机物排放及对氮沉降的响应[J]. 植物生态学报, 2025, 49(5): 681-696. |
| [14] | 平晓燕, 杜毅倩, 赖仕蓉, 孔梦桥, 余国杰. 植物应对食草动物采食的化学防御策略研究进展[J]. 植物生态学报, 2025, 49(5): 667-680. |
| [15] | 贾妍妍, 柳华清, 解欣然, 王博, 张维, 杨允菲. 珍稀濒危植物天山梣林龄结构及种群动态[J]. 植物生态学报, 2025, 49(5): 760-772. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||