

植物学报 ›› 2019, Vol. 54 ›› Issue (4): 503-508.DOI: 10.11983/CBB19087 cstr: 32102.14.CBB19087
收稿日期:2019-05-12
接受日期:2019-06-14
出版日期:2019-07-01
发布日期:2019-07-01
通讯作者:
程祝宽
Xinjie Cheng1,Hengxiu Yu1,Zhukuan Cheng2,*( )
)
Received:2019-05-12
Accepted:2019-06-14
Online:2019-07-01
Published:2019-07-01
Contact:
Zhukuan Cheng
摘要: 减数分裂粗线期染色体研究技术的发展, 很大程度上克服了水稻(Oryza sativa)细胞遗传研究中较小染色体所带来的研究困难。减数分裂染色体的制备与观察已经成为水稻细胞遗传学研究中的常规方法。该文详细描述了水稻中常用的减数分裂染色体制备、荧光原位杂交和免疫荧光染色的实验方法。
程新杰, 于恒秀, 程祝宽. 水稻减数分裂染色体分析方法. 植物学报, 2019, 54(4): 503-508.
Xinjie Cheng, Hengxiu Yu, Zhukuan Cheng. Protocols for Analyzing Rice Meiotic Chromosomes. Chinese Bulletin of Botany, 2019, 54(4): 503-508.
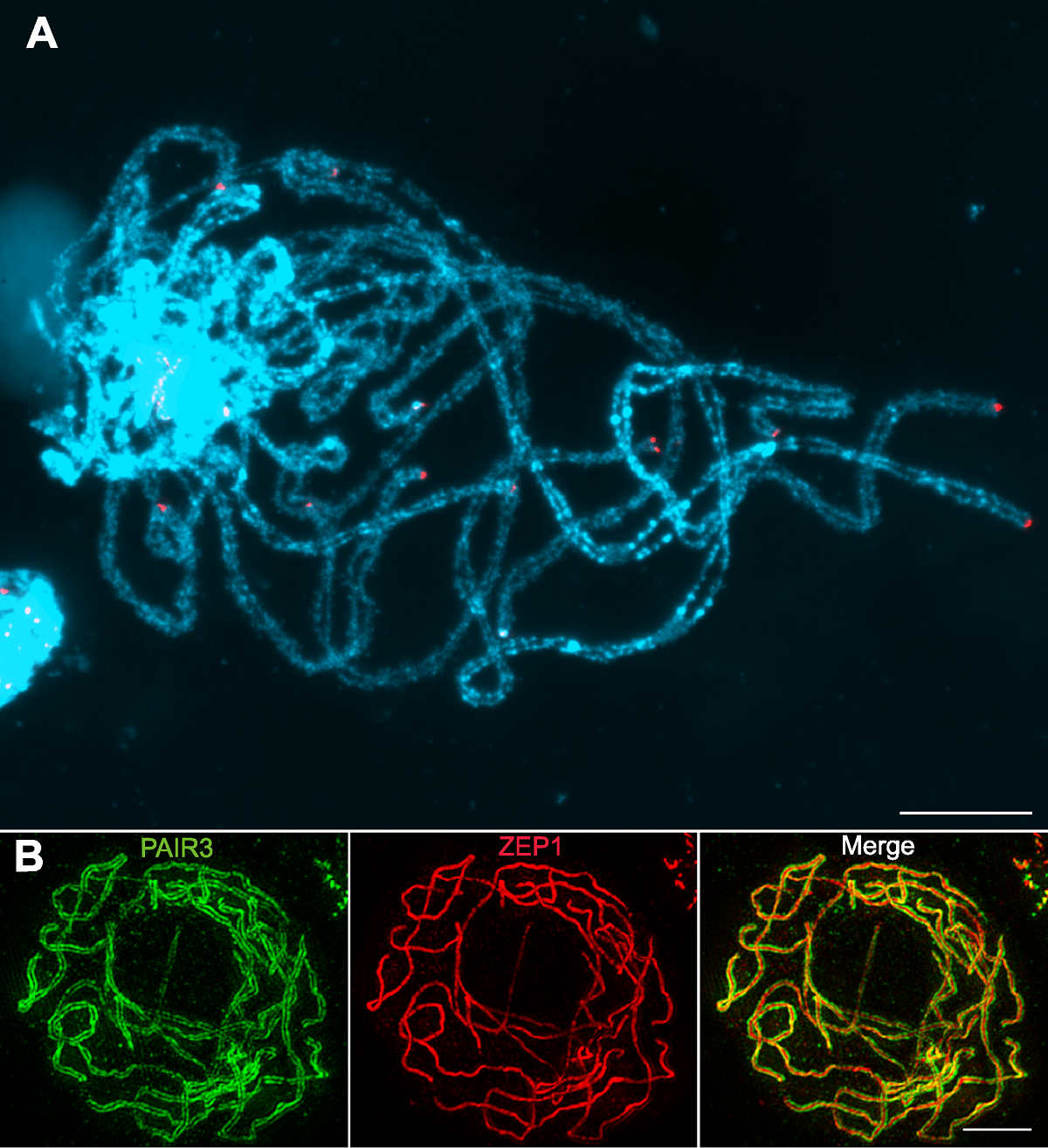
图1 水稻减数分裂粗线期染色体 (A) 水稻花粉母细胞减数分裂粗线期染色体的荧光原位杂交结果, 红色为端粒探针pAtT4的杂交信号, 蓝色为DAPI染色的粗线期染色体(普通荧光显微镜下观察); (B) 水稻花粉母细胞减数分裂粗线期染色体的免疫荧光染色结果, 绿色为PAIR3荧光信号, 红色为ZEP1荧光信号(超分辨荧光显微镜下观察)。Bars=5 µm
Figure 1 Characterization of rice meiotic pachytene chromosomes (A) Fluorescent in situ hybridization using rice pachytene chromosomes probed with the telomeric sequence pAtT4 (red); Chromosomes stained with DAPI (blue) (Images taken with a regular fluorescence microscope); (B) Immunodetection of PAIR3 (green) and ZEP1 (red) in rice pachytene chromosomes (Images taken with a super resolution microscope). Bars=5 µm
| [1] | Arumuganathan K, Earle ED ( 1991). Estimation of nuclear DNA content of plants by flow cytometry. Plant Mol Biol Rep 9, 229-241. |
| [2] | Che L, Tang D, Wang K, Wang M, Zhu K, Yu H, Gu M, Cheng Z ( 2011). OsAM1 is required for leptotene transition in rice. Cell Res 21, 654-665. |
| [3] | Cheng Z, Buell CR, Wing RA, Gu M, Jiang J ( 2001 a). Toward a cytological characterization of the rice genome. Genome Res 11, 2133-2141. |
| [4] | Cheng Z, Dong F, Langdon T, Ouyang S, Buell CR, Gu M, Blattner FR, Jiang J ( 2002). Functional rice centromeres are marked by a satellite repeat and a centromere-specific retrotransposon. Plant Cell 14, 1691-1704. |
| [5] | Cheng Z, Stupar RM, Gu M, Jiang J ( 2001 b). A tandemly repeated DNA sequence is associated with both knob-like heterochromatin and a highly decondensed structure in the meiotic pachytene chromosomes of rice. Chromosoma 110, 24-31. |
| [6] | Goff SA ( 1999). Rice as a model for cereal genomics. Curr Opin Plant Biol 2, 86-89. |
| [7] | Ji JH, Tang D, Shen Y, Xue ZH, Wang HJ, Shi WQ, Zhang C, Du GJ, Li YF, Cheng ZK ( 2016). P31 comet, a member of the synaptonemal complex, participates in meiotic DSB formation in rice . Proc Natl Acad Sci USA 113, 10577-10582. |
| [8] | Kurata N, Omura T, Iwata N ( 1981). Studies on centromere, chromomere and nucleolus in pachytene nuclei of rice, Oryza sativa, microsporocytes. Cytologia 46, 791-800. |
| [9] | Li X, Chao D, Wu Y, Huang X, Chen K, Cui L, Su L, Ye W, Chen H, Chen H, Dong N, Guo T, Shi M, Feng Q, Zhang P, Han B, Shan J, Gao J, Lin H ( 2015). Natural alleles of a proteasome α2 subunit gene contribute to thermotolerance and adaptation of African rice. Nat Genet 47, 827-833. |
| [10] | Li YF, Qin BX, Shen Y, Zhang FF, Liu CZ, You HL, Du GJ, Tang D, Cheng ZK ( 2018). HEIP1 regulates crossover formation during meiosis in rice. Proc Natl Acad Sci USA 115, 10810-10815. |
| [11] | Luo Q, Li YF, Shen Y, Cheng ZK ( 2014). Ten years of gene discovery for meiotic event control in rice. J Genet Genomics 41, 125-137. |
| [12] | Ma Y, Dai X, Xu Y, Luo W, Zheng X, Zeng D, Pan Y, Lin X, Liu H, Zhang D, Xiao J, Guo X, Xu S, Niu Y, Jin J, Zhang H, Xu X, Li L, Wang W, Qian Q, Ge S, Chong K ( 2015). COLD1 confers chilling tolerance in rice. Cell 160, 1209-1221. |
| [13] | Miao CB, Tang D, Zhang HG, Wang M, Li YF, Tang SZ, Yu HX, Gu MH, Cheng ZK ( 2013). CENTRAL REGION COMPONENT 1, a novel synaptonemal complex component, is essential for meiotic recombination initiation in rice. Plant Cell 25, 2998-3009. |
| [14] | Nonomura K, Morohoshi A, Nakano M, Eiguchi M, Miyao A, Hirochika H, Kurata N ( 2007). A germ cell specific gene of the ARGONAUTE family is essential for the progression of premeiotic mitosis and meiosis during sporogenesis in rice. Plant Cell 19, 2583-2594. |
| [15] | Pawlowski WP, Golubovskaya IN, Timofejeva L, Meeley RB, Sheridan WF, Cande WZ ( 2004). Coordination of meiotic recombination, pairing, and synapsis by PHS1. Science 303, 89-92. |
| [16] | Ren LJ, Tang D, Zhao TT, Zhang FF, Liu CZ, Xue ZH, Shi WQ, Du GJ, Shen Y, Li YF, Cheng ZK ( 2018). OsSPL regulates meiotic fate acquisition in rice. New Phytol 218, 789-803. |
| [17] | Tang X, Bao W, Zhang W, Cheng Z ( 2007). Identification of chromosomes from multiple rice genomes using a universal molecular cytogenetic marker system. J Integr Plant Biol 49, 953-960. |
| [18] | Wang K, Tang D, Wang M, Lu J, Yu H, Liu J, Qian B, Gong Z, Wang X, Chen J, Gu M, Cheng Z ( 2009). MER3 is required for normal meiotic crossover formation, but not for presynaptic alignment in rice. J Cell Sci 122, 2055-2063. |
| [19] | Wang M, Wang K, Tang D, Wei C, Li M, Shen Y, Chi Z, Gu M, Cheng Z ( 2010). The central element protein ZEP1 of the synaptonemal complex regulates the number of crossovers during meiosis in rice. Plant Cell 22, 417-430. |
| [20] | Wang Y, Xiong G, Hu J, Jiang L, Yu H, Xu J, Fang Y, Zeng L, Xu E, Xu J, Ye W, Meng X, Liu R, Chen H, Jing Y, Wang Y, Zhu X, Li J, Qian Q ( 2015). Copy number variation at the GL7 locus contributes to grain size diversity in rice. Nat Genet 47, 944-948. |
| [21] | Wu HK ( 1967). Note on preparing of pachytene chromosomes by double mordant. Sci Agric 15, 40-44. |
| [22] | Yu H, Wang M, Tang D, Wang K, Chen F, Gong Z, Gu M, Cheng Z ( 2010). OsSPO11-1 is essential for both homologous chromosome pairing and crossover formation in rice. Chromosoma 119, 625-636. |
| [23] | Zhang F, Tang D, Shen Y, Xue ZH, Shi WQ, Ren LJ, Du GJ, Li Y, Cheng ZK ( 2017). The F-box protein ZYGO1 mediates bouquet formation to promote homologous pairing, synapsis, and recombination in rice meiosis. Plant Cell 29, 2597-2609. |
| [24] | Zhao TT, Ren LJ, Chen XJ, Yu HX, Liu CJ, Shen Y, Shi WQ, Tang D, Du GJ, Li YF, Ma BJ, Cheng ZK ( 2018). The OsRR24/LEPTO1 type-B response regulator is essential for the organization of leptotene chromosomes in rice meiosis. Plant Cell 30, 3024-3037. |
| [1] | 叶灿, 姚林波, 金莹, 高蓉, 谭琪, 李旭映, 张艳军, 陈析丰, 马伯军, 章薇, 张可伟. 水稻水杨酸代谢突变体高通量筛选方法的建立与应用[J]. 植物学报, 2025, 60(4): 1-0. |
| [2] | 赵凌, 管菊, 梁文化, 张勇, 路凯, 赵春芳, 李余生, 张亚东. 基于高密度Bin图谱的水稻苗期耐热性QTL定位[J]. 植物学报, 2025, 60(3): 342-353. |
| [3] | 李新宇, 谷月, 徐非非, 包劲松. 水稻胚乳淀粉合成相关蛋白的翻译后修饰研究进展[J]. 植物学报, 2025, 60(2): 256-270. |
| [4] | 李建国, 张怡, 张文君. 水稻根系铁膜形成及对磷吸收的影响[J]. 植物学报, 2025, 60(1): 132-143. |
| [5] | 姚瑞枫, 谢道昕. 水稻独脚金内酯信号感知的激活和终止[J]. 植物学报, 2024, 59(6): 873-877. |
| [6] | 连锦瑾, 唐璐瑶, 张伊诺, 郑佳兴, 朱超宇, 叶语涵, 王跃星, 商文楠, 傅正浩, 徐昕璇, 吴日成, 路梅, 王长春, 饶玉春. 水稻抗氧化性状遗传位点挖掘及候选基因分析[J]. 植物学报, 2024, 59(5): 738-751. |
| [7] | 黄佳慧, 杨惠敏, 陈欣雨, 朱超宇, 江亚楠, 胡程翔, 连锦瑾, 芦涛, 路梅, 张维林, 饶玉春. 水稻突变体pe-1对弱光胁迫的响应机制[J]. 植物学报, 2024, 59(4): 574-584. |
| [8] | 周俭民. 收放自如的明星战车[J]. 植物学报, 2024, 59(3): 343-346. |
| [9] | 朱超宇, 胡程翔, 朱哲楠, 张芷宁, 汪理海, 陈钧, 李三峰, 连锦瑾, 唐璐瑶, 钟芊芊, 殷文晶, 王跃星, 饶玉春. 水稻穗部性状QTL定位及候选基因分析[J]. 植物学报, 2024, 59(2): 217-230. |
| [10] | 夏婧, 饶玉春, 曹丹芸, 王逸, 柳林昕, 徐雅婷, 牟望舒, 薛大伟. 水稻中乙烯生物合成关键酶OsACS和OsACO调控机制研究进展[J]. 植物学报, 2024, 59(2): 291-301. |
| [11] | 罗兰莎, 宋雯佩, 化青珠, 李大卫, 梁红, 张宪智. 植物性别决定基因及其表观遗传调控研究进展[J]. 植物学报, 2024, 59(2): 278-290. |
| [12] | 朱宝, 赵江哲, 张可伟, 黄鹏. 水稻细胞分裂素氧化酶9参与调控水稻叶夹角发育[J]. 植物学报, 2024, 59(1): 10-21. |
| [13] | 方妍力, 田传玉, 苏如意, 刘亚培, 王春连, 陈析丰, 郭威, 纪志远. 水稻抗细菌性条斑病基因挖掘与初定位[J]. 植物学报, 2024, 59(1): 1-9. |
| [14] | 贾绮玮, 钟芊芊, 顾育嘉, 陆天麒, 李玮, 杨帅, 朱超宇, 胡程翔, 李三峰, 王跃星, 饶玉春. 水稻茎秆细胞壁相关组分含量QTL定位及候选基因分析[J]. 植物学报, 2023, 58(6): 882-892. |
| [15] | 田传玉, 方妍力, 沈晴, 王宏杰, 陈析丰, 郭威, 赵开军, 王春连, 纪志远. 2019-2021年我国南方稻区白叶枯病菌的毒力与遗传多样性调查研究[J]. 植物学报, 2023, 58(5): 743-749. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||