

植物学报 ›› 2019, Vol. 54 ›› Issue (5): 606-619.DOI: 10.11983/CBB19053 cstr: 32102.14.CBB19053
周纯1,焦然1,胡萍2,林晗1,胡娟1,徐娜1,吴先美2,饶玉春1,*( ),王跃星2,*(
),王跃星2,*( )
)
收稿日期:2019-03-20
接受日期:2019-06-20
出版日期:2019-09-01
发布日期:2020-03-10
通讯作者:
饶玉春,王跃星
基金资助:
Chun Zhou1,Ran Jiao1,Ping Hu2,Han Lin1,Juan Hu1,Na Xu1,Xianmei Wu2,Yuchun Rao1,*( ),Yuexing Wang2,*(
),Yuexing Wang2,*( )
)
Received:2019-03-20
Accepted:2019-06-20
Online:2019-09-01
Published:2020-03-10
Contact:
Yuchun Rao,Yuexing Wang
摘要: 衰老是植物发育末期自主发生且不可逆的适应性反应, 叶片早衰相关分子机制研究对水稻(Oryza sativa)遗传改良以及抗衰老品种培育有重要意义。LS-es1是通过EMS诱变粳稻品种TP309获得的稳定遗传的早衰突变体。对LS-es1及其野生型的表型观察和生理生化分析表明, LS-es1叶片中积累了大量活性氧且细胞死亡更多, 同时LS-es1与产量相关的农艺性状均显著下降, 这也验证了LS-es1早衰的特征。对LS-es1及其野生型幼苗进行外源激素处理, 结果表明LS-es1对水杨酸(SA)、脱落酸(ABA)和茉莉酸甲酯(MeJA)更敏感。用图位克隆方法将LS-es1基因定位在水稻第7号染色体长臂46.2 kb区间内, 该区间共包括8个开放阅读框(ORF)。对该区间内的基因进行生物信息学分析, 结果发现Os07g0275300和Os07g0276000两个候选功能基因与早衰途径相关, 并且这2个基因的表达量在野生型和突变体中差异较大。研究结果为进一步克隆LS-es1基因并深入研究其生物学功能奠定了基础。
周纯,焦然,胡萍,林晗,胡娟,徐娜,吴先美,饶玉春,王跃星. 水稻早衰突变体LS-es1的基因定位及候选基因分析. 植物学报, 2019, 54(5): 606-619.
Chun Zhou,Ran Jiao,Ping Hu,Han Lin,Juan Hu,Na Xu,Xianmei Wu,Yuchun Rao,Yuexing Wang. Gene Mapping and Candidate Gene Analysis of Rice Early Senescence Mutant LS-es1. Chinese Bulletin of Botany, 2019, 54(5): 606-619.
| Primer name | Forward primer (5'-3') | Reverse primer (5'-3') |
|---|---|---|
| InDel-1 | AGCGGGGATGGAGATGATG | CTTGCCTCACACCAGATCTG |
| InDel-2 | GGCGCCTTTGTTCCATAGTT | GAGGAGCCAGTGGTAGCAG |
| InDel-3 | CGTTTTTACAACCAATTTTGGAA | CCATCTTCTACCTCCGGACA |
| InDel-4 | GATTGGATTGGTTGCTCGCT | AACAGCGAATCGAGATGCAC |
| InDel-5 | TTACTGCTGCCGTTGTTTCA | TTGTGGACCTCCAGGATCAG |
| SGR | AGGGGTGGTACAACAAGCTG | GCTCCTTGCGGAAGATGTAG |
| Osh36 | GCACGGAGGCGAACGA | TTGAGCGGTAGCACCCATT |
| Osl85 | GAGCAACGGCGTGGAGA | GCGGCGGTAGAGGAGATG |
| OsNAP | CAAGAAGCCGAACGGTTC | GTTAGAGTGGAGCAGCAT |
| Actin | CAGGCCGTCCTCTCTCTGTA | AAGGATAGCATGGGGGAGAG |
表1 基因定位及qRT-PCR所用引物序列
Table 1 Primers used for gene mapping and qRT-PCR
| Primer name | Forward primer (5'-3') | Reverse primer (5'-3') |
|---|---|---|
| InDel-1 | AGCGGGGATGGAGATGATG | CTTGCCTCACACCAGATCTG |
| InDel-2 | GGCGCCTTTGTTCCATAGTT | GAGGAGCCAGTGGTAGCAG |
| InDel-3 | CGTTTTTACAACCAATTTTGGAA | CCATCTTCTACCTCCGGACA |
| InDel-4 | GATTGGATTGGTTGCTCGCT | AACAGCGAATCGAGATGCAC |
| InDel-5 | TTACTGCTGCCGTTGTTTCA | TTGTGGACCTCCAGGATCAG |
| SGR | AGGGGTGGTACAACAAGCTG | GCTCCTTGCGGAAGATGTAG |
| Osh36 | GCACGGAGGCGAACGA | TTGAGCGGTAGCACCCATT |
| Osl85 | GAGCAACGGCGTGGAGA | GCGGCGGTAGAGGAGATG |
| OsNAP | CAAGAAGCCGAACGGTTC | GTTAGAGTGGAGCAGCAT |
| Actin | CAGGCCGTCCTCTCTCTGTA | AAGGATAGCATGGGGGAGAG |

图1 水稻野生型(WT) TP309和突变体LS-es1的表型特征 (A) 苗期表型; (B) 分蘖期表型; (C) 成熟期表型。Bars=6 cm
Figure 1 Phenotypes of rice wild type (WT) TP309 and mu- tant LS-es1 (A) Phenotypes at seedling stage; (B) Phenotypes at tillering stage; (C) Phenotypes at maturity stage. Bars=6 cm
| Agronomic traits | TP309 | LS-es1 |
|---|---|---|
| Effective number of panicle | 13.40±5.41 | 8.20±0.84 |
| Flag leaf length (cm) | 39.46±8.16 | 35.68±4.08 |
| Secondary branch number | 29.80±6.87 | 18.00±4.47* |
| Tiller number | 16.40±5.46 | 11.40±2.70 |
| Filled grain number per panicle | 169.60±8.08 | 125.60±24.83** |
| Seed-setting rate (%) | 81.01±6.98 | 64.76±17.23 |
表2 水稻野生型TP309和突变体LS-es1的农艺性状比较
Table 2 The comparison of agronomic traits between rice wild type TP309 and mutant LS-es1
| Agronomic traits | TP309 | LS-es1 |
|---|---|---|
| Effective number of panicle | 13.40±5.41 | 8.20±0.84 |
| Flag leaf length (cm) | 39.46±8.16 | 35.68±4.08 |
| Secondary branch number | 29.80±6.87 | 18.00±4.47* |
| Tiller number | 16.40±5.46 | 11.40±2.70 |
| Filled grain number per panicle | 169.60±8.08 | 125.60±24.83** |
| Seed-setting rate (%) | 81.01±6.98 | 64.76±17.23 |
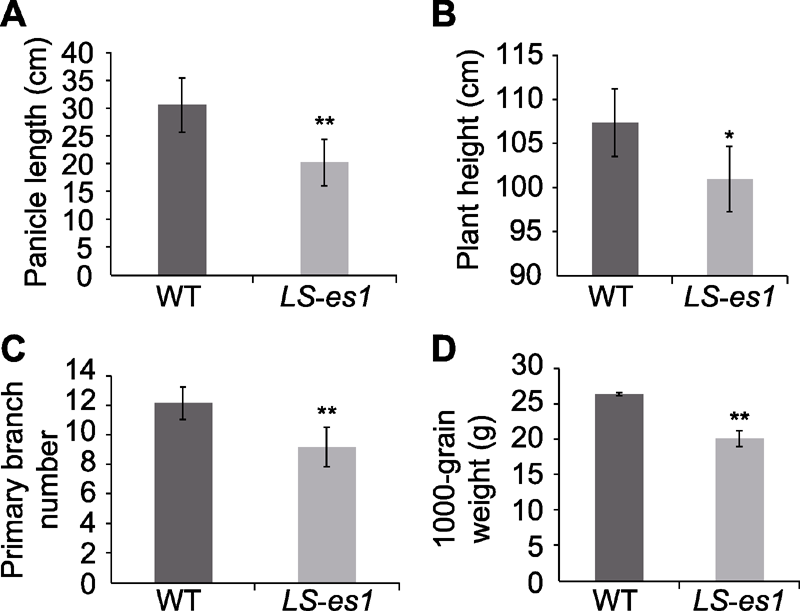
图2 水稻野生型(WT) TP309和突变体LS-es1的农艺性状比较 (A) 穗长; (B) 株高; (C) 一次枝梗数; (D) 千粒重。*和**分别表示TP309与LS-es1在0.05和0.01水平上差异显著。
Figure 2 The comparison of agronomic traits between rice wild type (WT) TP309 and mutant LS-es1 (A) Panicle length; (B) Plant height; (C) Primary branch number; (D) 1000-grain weight. * and ** indicate significant differences between TP309 and LS-es1 at 0.05 and 0.01 level, respectively.
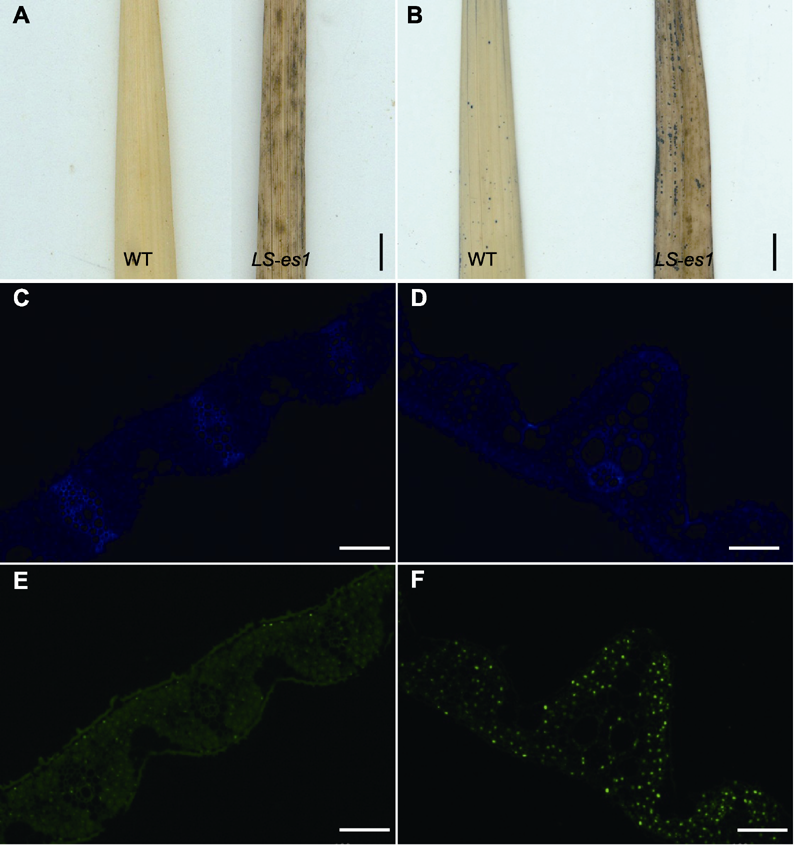
图3 水稻野生型(WT) TP309和突变体LS-es1的叶片组织细胞化学分析 (A) 野生型和突变体叶片的DAB染色(Bar=2 cm); (B) 野生型和突变体叶片的NBT染色(Bar=2 cm); (C), (E) 野生型TP309叶片Tunel检测(Bars=100 μm); (D), (F) 突变体LS-es1叶片Tunel检测(Bars=100 μm)
Figure 3 Histochemical analysis of rice wild type (WT) TP309 and mutant LS-es1 leaves DAB staining of wild-type and LS-es1 leaves (Bar=2 cm);(B) NBT staining of wild-type and LS-es1 leaves (Bar=2 cm); (C), (E) Tunel detection of wild-type leaves (Bars=100 μm); (D), (F) Tunel detection of LS-es1 leaves (Bars=100 μm).
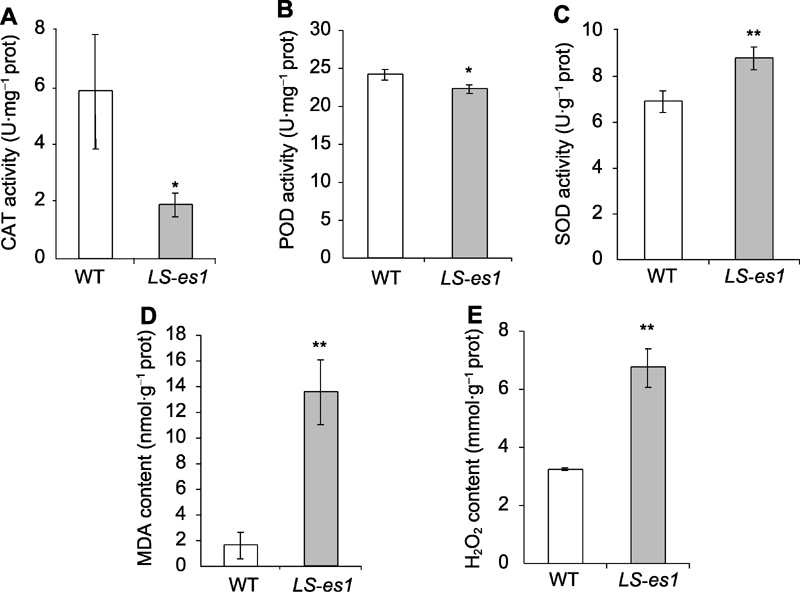
图4 抽穗期水稻突变体LS-es1及其野生型(WT)叶片的过氧化氢酶(CAT) (A)、过氧化物酶(POD) (B)和超氧化物歧化酶(SOD) (C)活性以及丙二醛(MDA) (D)和过氧化氢(H2O2) (E)含量 *和**分别表示TP309与LS-es1在0.05和0.01水平上差异显著。
Figure 4 Catalase (CAT) (A), peroxisome (POD) (B), and superoxide dismutase (SOD) (C) activities and malondialdehyde (MDA) (D) and H2O2 (E) contents of LS-es1 and wild type (WT) at heading stage of rice * and ** indicate significant differences between TP309 and LS-es1 at 0.05 and 0.01 level, respectively.
| Net photosynthetic rate | Stomatal conductance | Intercellular CO2 concentration | Transpiration rate | SPAD | |
|---|---|---|---|---|---|
| TP309 | 9.4±0.961 | 0.110±0.01 | 263±3 | 4.63±0.289 | 40.633±1.206 |
| LS-es1 | 2.225±1.407** | 0.0403±0.007** | 312.5±10.606** | 2.3±0.283** | 27.65±2.333** |
表3 水稻野生型TP309和突变体LS-es1的叶绿素相对含量(SPAD值)和光合速率比较
Table 3 The comparison of SPAD value and photosynthetic rate between rice TP309 and LS-es1
| Net photosynthetic rate | Stomatal conductance | Intercellular CO2 concentration | Transpiration rate | SPAD | |
|---|---|---|---|---|---|
| TP309 | 9.4±0.961 | 0.110±0.01 | 263±3 | 4.63±0.289 | 40.633±1.206 |
| LS-es1 | 2.225±1.407** | 0.0403±0.007** | 312.5±10.606** | 2.3±0.283** | 27.65±2.333** |
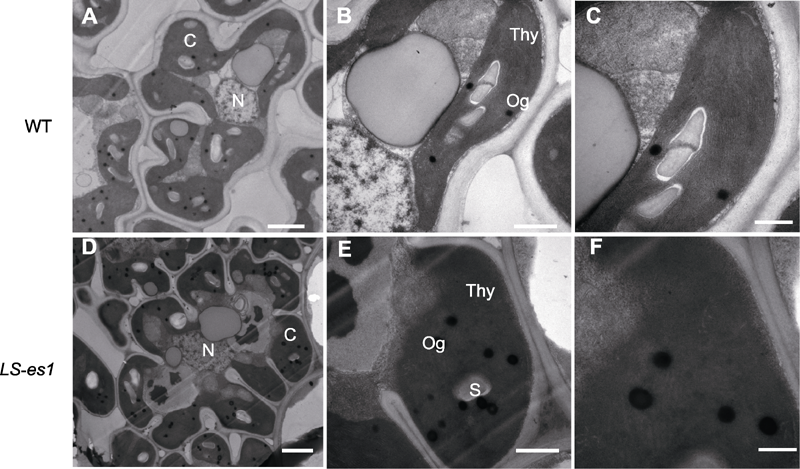
图5 水稻野生型(WT) TP309和突变体LS-es1叶片的透射电镜(TEM)观察 (A) TP309叶片细胞, 6000X; (B) TP309叶片细胞, 25000X; (C) TP309叶片细胞, 40000X; (D) LS-es1叶片细胞, 6000X; (E) LS-es1叶片细胞, 25000X; (F) LS-es1叶片细胞, 40000X。N: 细胞核; C: 叶绿体; Thy: 类囊体; S: 淀粉颗粒; Og: 嗜锇小体。Bars=1 μm
Figure 5 Transmission electron microscopy (TEM) analysis of chloroplast in rice wild type (WT) TP309 and mutant LS-es1 (A) TP309 leaf cells in 6000X; (B) TP309 leaf cells in 25000X; (C) TP309 leaf cells in 40000X; (D) LS-es1 leaf cells in 6000X; (E) LS-es1 leaf cells in 25000X; (F) LS-es1 leaf cells in 40000X. N: Cell nucleus; C: Chloroplast; Thy: Thylakoid; S: Starch granule; Og: Eosinophil. Bars=1 μm
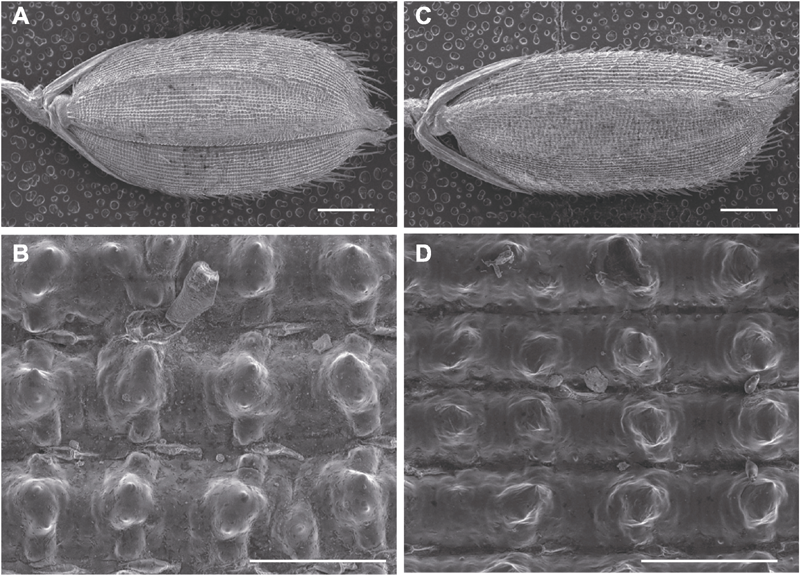
图6 水稻野生型TP309和突变体LS-es1籽粒的扫描电镜(SEM)观察 (A), (B) TP309籽粒; (C), (D) LS-es1籽粒。(A), (C) Bars=1 mm; (B), (D) Bars=100 μm
Figure 6 Scanning electron microscopy (SEM) analysis of seeds in rice wild type TP309 and mutant LS-es1 (A), (B) TP309 seed; (C), (D) LS-es1 seed. (A), (C) Bars=1 mm; (B), (D) Bars=100 μm
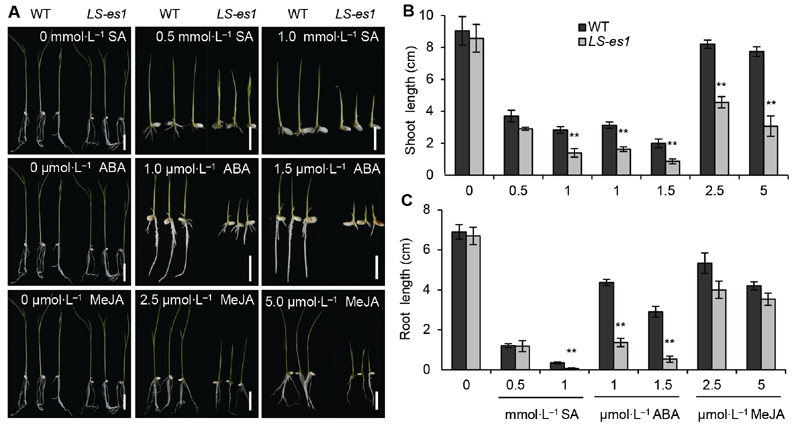
图7 外源激素处理对水稻野生型(WT) TP309和突变体LS-es1幼苗生长的抑制作用 (A) 水杨酸(SA)处理(上), 脱落酸(ABA)处理(中), 茉莉酸甲酯(MeJA)处理(下) (Bars=2 cm); (B) 激素处理后芽长的比较; (C) 激素处理后根长的比较。** 表示TP309与LS-es1在0.01水平上差异显著。
Figure 7 Inhibition of exogenous hormone treatment on the growth of seedlings in rice wild type (WT) TP309 and mutant LS-es1 (A) Salicylic acid (SA) (top), abscisic acid (ABA) (middle), methyl jasmonate (MeJA) (bottom) (Bars=2 cm); (B) Comparison of shoot length of TP309 and LS-es1 seedlings treated with hormones; (C) Comparison of root length of TP309 and LS-es1 seedlings treated with hormones. ** indicate significant differences at 0.01 level between TP309 and LS-es1.
| Hybrid combination (male/female) | F1 pheno- type | Namber of F2 normal phenotype | Namber of F2 mutant phenotype | Number of F2 population | χ2 (3:1) |
|---|---|---|---|---|---|
| LS-es1/TN1 | Normal | 1381 | 471 | 1852 | 0.1842 |
| LS-es1/ZF802 | Normal | 526 | 178 | 704 | 0.0303 |
表4 水稻早衰突变体LS-es1的遗传分析
Table 4 Genetic analysis of rice early senescence phenotypes of LS-es1
| Hybrid combination (male/female) | F1 pheno- type | Namber of F2 normal phenotype | Namber of F2 mutant phenotype | Number of F2 population | χ2 (3:1) |
|---|---|---|---|---|---|
| LS-es1/TN1 | Normal | 1381 | 471 | 1852 | 0.1842 |
| LS-es1/ZF802 | Normal | 526 | 178 | 704 | 0.0303 |
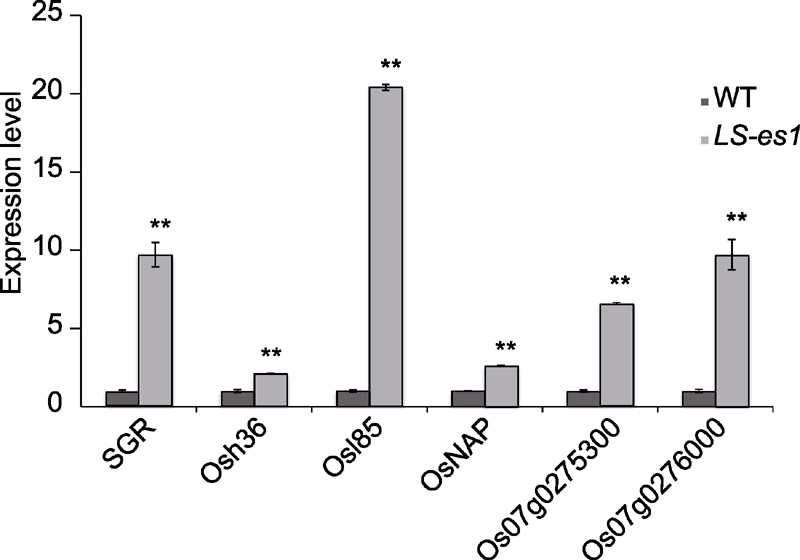
图9 水稻早衰突变体LS-es1候选基因及衰老相关基因的表达量 ** 表示TP309和LS-es1在0.01水平上差异显著。
Figure 9 Expression of LS-es1 candidate genes and senescence-related genes in rice ** indicate significant differences at 0.01 level.
| 1 | 陈昆松, 李方, 徐昌杰, 张上隆, 傅承新 (2004). 改良CTAB法用于多年生植物组织基因组DNA的大量提取. 遗传 26, 529-531. |
| 2 | 邓接楼, 王艾平, 何长水, 王爱斌, 徐芬芬 (2011). 硅肥对水稻生长发育及产量品质的影响. 广东农业科学 38(12), 58-61. |
| 3 | 段俊, 梁承邺, 黄毓文 (1997). 杂交水稻开花结实期间叶片衰老. 植物生理学报 23, 139-144. |
| 4 | 华春, 王仁雷 (2003). 杂交稻及其三系叶片衰老过程中SOD、CAT活性和MDA含量的变化. 西北植物学报 23, 406-409. |
| 5 | 冷语佳 (2013). 水稻早衰基因ES10的遗传分析与基因定位. 硕士论文. 北京: 中国农业科学院. pp. 16-17. |
| 6 | 刘翔 (2014). EMS诱变技术在植物育种中的研究进展. 激光生物学报 23, 197-201. |
| 7 | 孙玉莹 (2013). 水稻叶片早衰基因PSL2的图位克隆及功能初步分析. 硕士论文. 北京: 中国农业科学院. pp. 17-65. |
| 8 | 徐娜, 徐江民, 蒋玲欢, 饶玉春 (2017). 水稻叶片早衰成因及分子机理研究进展. 植物学报 52, 102-112. |
| 9 | 张丽霞 (2000). 水稻叶片衰老相关基因的分离. 硕士论文. 福州: 福建农林大学. pp. 20-54. |
| 10 | Ansari MI, Lee RH, Chen SCG (2005). A novel senescence-associated gene encoding γ-aminobutyric acid (GABA): pyruvate transaminase is upregulated during rice leaf senescence. Physiol Plant 123, 1-8. |
| 11 | Chen HL, Li CR, Liu LP, Zhao JY, Cheng XZ, Jiang GH, Zhai WX (2016). The Fd-GOGAT1 mutant gene lc7 confers resistance to Xanthomonas oryzae pv. oryzae in rice. Sci Rep 6, 26411. |
| 12 | Chen LJ, Wuriyanghan H, Zhang YQ, Duan KX, Chen HW, Li QT, Lu X, He SJ, Ma B, Zhang WK, Lin Q, Chen SY, Zhang JS (2013a). An S-domain receptor-like kinase, OsSIK2, confers abiotic stress tolerance and delays dark- induced leaf senescence in rice. Plant Physiol 163, 1752-1765. |
| 13 | Chen Y, Xu YY, Luo W, Li WX, Chen N, Zhang DJ, Chong K (2013b). The F-box protein OsFBK12 targets OsSAMS1 for degradation and affects pleiotropic phenotypes including leaf senescence in rice. Plant Physiol 163, 1673-1685. |
| 14 | Fanata WID, Lee KH, Son BH, Yoo JY, Harmoko R, Ko KS, Ramasamy NK, Kim KH, Oh DB, Jung HS, Kim JY, Lee SY, Lee KO (2013). N-glycan maturation is crucial for cytokinin-mediated development and cellulose synthesis in Oryza sativa. Plant J 73, 966-979. |
| 15 | Gan S, Amasino RM (1997). Making sense of senescence: molecular genetic regulation and manipulation of leaf senescence. Plant Physiol 113, 313-319. |
| 16 | Gan SS, Hörtensteiner S (2013). Frontiers in plant senescence research: from bench to bank. Plant Mol Biol 82, 503-504. |
| 17 | Hideg E, Kálai T, Kós PB, Asada K, Hideg K (2006). Singlet oxygen in plants—its significance and possible detection with double (fluorescent and spin) indicator reagents. Photochem Photobiol 82, 1211-1218. |
| 18 | Huang LM, Sun QW, Qin FJ, Li C, Zhao Y, Zhou DX (2007). Down-regulation of a SILENT INFORMATION REGULATOR 2-related histone deacetylase gene, OsS- RT1, induces DNA fragmentation and cell death in rice. Plant Physiol 144, 1508-1519. |
| 19 | Huang QN, Shi YF, Zhang XB, Song LX, Feng BH, Wang HM, Xu X, Li XH, Guo D, Wu JL (2016). Single base substitution in OsCDC48 is responsible for premature senescence and death phenotype in rice. J Integr Plant Biol 58, 12-28. |
| 20 | Jiang HW, Li MR, Liang NT, Yan HB, Wei YB, Xu XL, Liu J, Xu ZF, Chen F, Wu GJ (2007). Molecular cloning and function analysis of the stay green gene in rice. Plant J 52, 197-209. |
| 21 | Jiao BB, Wang JJ, Zhu XD, Zeng LJ, Li Q, He ZH (2012). A novel protein RLS1 with NB-ARM domains is involved in chloroplast degradation during leaf senescence in rice. Mol Plant 5, 205-217. |
| 22 | Kariola T, Brader G, Li J, Palva ET (2005). Chlorophyllase 1, a damage control enzyme, affects the balance between defense pathways in plants. Plant Cell 17, 282-294. |
| 23 | Kong ZS, Li MN, Yang WY, Xu WY, Xue YB (2006). A novel nuclear-localized CCCH-type zinc finger protein, OsDOS, is involved in delaying leaf senescence in rice. Plant Physiol 141, 1376-1388. |
| 24 | Kusaba M, Ito H, Morita R, Iida S, Sato Y, Fujimoto M, Kawasaki S, Tanaka R, Hirochika H, Nishimura M, Tanaka A (2007). Rice NON-YELLOW COLORING1 is involved in light-harvesting complex II and grana degradation during leaf senescence. Plant Cell 19, 1362-1375. |
| 25 | Lee RH, Lin MC, Chen SC (2004). A novel alkaline α-galactosidase gene is involved in rice leaf senescence. Plant Mol Biol 55, 281-295. |
| 26 | Lee RH, Wang CH, Huang LT, Chen SCG (2001). Leaf senescence in rice plants: cloning and characterization of senescence up-regulated genes. J Exp Bot 52, 1117-1121. |
| 27 | Leng YJ, Yang YL, Ren DY, Huang LC, Dai LP, Wang YQ, Chen L, Tu ZJ, Gao YH, Li XY, Zhu L, Hu J, Zhang GH, Gao ZY, Guo LB, Kong ZS, Lin YJ, Qian Q, Zeng DL (2017). A rice PECTATE LYASE-LIKE gene is required for plant growth and leaf senescence. Plant Physiol 174, 1151-1166. |
| 28 | Liang CZ, Wang YQ, Zhu YN, Tang JY, Hu B, Liu LC, Ou SJ, Wu HK, Sun XH, Chu JF, Chu CC (2014). OsNAP connects abscisic acid and leaf senescence by fine-tuning abscisic acid biosynthesis and directly targeting senescence-associated genes in rice. Proc Natl Acad Sci USA 111, 10013-10018. |
| 29 | Lim PO, Kim HJ, Nam HG (2007). Leaf senescence. Annu Rev Plant Biol 58, 115-136. |
| 30 | Lin AH, Wang YQ, Tang JY, Xue P, Li CL, Liu LC, Hu B, Yang FQ, Loake GJ, Chu CC (2012). Nitric oxide and protein S-nitrosylation are integral to hydrogen peroxide-induced leaf cell death in rice. Plant Physiol 158, 451-464. |
| 31 | Lin YH, Tan LB, Zhao L, Sun XY, Sun CQ (2016). RLS3, a protein with AAA+ domain localized in chloroplast, sustains leaf longevity in rice. J Integr Plant Biol 58, 971-982. |
| 32 | Livak KJ, Schmittgen TD (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2 -ΔΔCt method . Methods 25, 402-408. |
| 33 | Luan WJ, Shen A, Jin ZP, Song SS, Li ZL, Sha AH (2013). Knockdown of OsHox33, a member of the class III homeodomain-leucine zipper gene family, accelerates leaf senescence in rice. Sci China Life Sci 56, 1113-1123. |
| 34 | Mahalingam R, Jambunathan N, Gunjan SK, Faustin E, Weng H, Ayoubi P (2006). Analysis of oxidative signaling induced by ozone in Arabidopsis thaliana. Plant Cell Environ 29, 1357-1371. |
| 35 | McCabe MS, Garratt LC, Schepers F, Jordi WJRM, Stoopen GM, Davelaar E, van Rhijn JHA, Power JB, Davey MR (2001). Effects of PSAG12- IPT gene expression on development and senescence in transgenic lettuce. Plant Physiol 127, 505-516. |
| 36 | Morita R, Sato Y, Masuda Y, Nishimura M, Kusaba M (2010). Defect in non-yellow coloring 3, an α/β hydrolase-fold family protein, causes a stay-green phenotype during leaf senescence in rice. Plant J 59, 940-952. |
| 37 | Navabpour S, Morris K, Allen R, Harrison E, A-H- Mackerness S, Buchanan-Wollaston V (2003). Expression of senescence-enhanced genes in response to oxidative stress. J Exp Bot 54, 2285-2292. |
| 38 | Qiao YL, Jiang WZ, Lee J, Park BS, Choi MS, Piao RH, Woo MO, Roh JH, Han LZ, Paek NC, Seo HS, Koh HJ (2010). SPL28 encodes a clathrin-associated adaptor protein complex 1, medium subunit μ1 (AP1M1) and is responsible for spotted leaf and early senescence in rice (Oryza sativa). New Phytol 185, 258-274. |
| 39 | Rao YC, Yang YL, Xu J, Li XJ, Leng YJ, Dai LP, Huang LC, Shao GS, Ren DY, Hu J, Guo LB, Pan JW, Zeng DL (2015). EARLY SENESCENCE1 encodes a SCAR-LIKE PROTEIN2 that affects water loss in rice. Plant Physiol 169, 1225-1239. |
| 40 | Schippers JH, Schmidt R, Wagstaff C, Jing HC (2015). Living to die and dying to live: the survival strategy behind leaf senescence. Plant Physiol 169, 914-930. |
| 41 | Singh S, Giri MK, Singh PK, Siddiqui A, Nandi AK (2013). Down-regulation of OsSAG12-1 results in enhanced senescence and pathogen-induced cell death in transgenic rice plants. J Biosci 38, 583-592. |
| 42 | Sun LT, Wang YH, Liu LL, Wang CM, Gan T, Zhang ZY, Wang YL, Wang D, Niu M, Long WH, Li XH, Zheng M, Jiang L, Wan JM (2017). Isolation and characterization of a spotted leaf 32 mutant with early leaf senescence and enhanced defense response in rice. Sci Rep 7, 41846. |
| 43 | Tamiru M, Takagi H, Abe A, Yokota T, Kanzaki H, Okamoto H, Saitoh H, Takahashi H, Fujisaki K, Oikawa K, Uemura A, Natsume S, Jikumaru Y, Matsuura H, Umemura K, Terry MJ, Terauchi R (2016). A chloroplast-localized protein LESION AND LAMINA BENDING affects defence and growth responses in rice. New Phytol 210, 1282-1297. |
| 44 | Tang YY, Li MR, Chen YP, Wu PZ, Wu GJ, Jiang HW (2011). Knockdown of Os PAO and OsRCCR1 cause different plant death phenotypes in rice. J Plant Physiol 168, 1952-1959. |
| 45 | Undan JR, Tamiru M, Abe A, Yoshida K, Kosugi S, Takagi H, Yoshida K, Kanzaki H, Saitoh H, Fekih R, Sharma S, Undan J, Yano M, Terauchi R (2012). Mutation in OsLMS, a gene encoding a protein with two double-stranded RNA binding motifs, causes lesion mimic phenotype and early senescence in rice(Oryza sativa L.). Genes Genet Syst 87, 169-179. |
| 46 | Wang S, Lei CL, Wang JL, Ma J, Tang S, Wang CL, Zhao KJ, Tian P, Zhang H, Qi CY, Cheng ZJ, Zhang X, Guo XP, Liu LL, Wu CY, Wan JM (2017). SPL33, encoding an eEF1A-like protein, negatively regulates cell death and defense responses in rice. J Exp Bot 68, 899-913. |
| 47 | Wu HB, Wang B, Chen YL, Liu YG, Chen LT (2013). Characterization and fine mapping of the rice premature senescence mutant ospse1. Theor Appl Genet 126, 1897-1907. |
| 48 | Wu ZM, Zhang X, He B, Diao LP, Sheng SL, Wang JL, Guo XP, Su N, Wang LF, Jiang L, Wang CM, Zhai HQ, Wan JM (2007). A chlorophyll-deficient rice mutant with impaired chlorophyllide esterification in chlorophyll biosynthesis. Plant Physiol 145, 29-40. |
| 49 | Yamatani H, Sato Y, Masuda Y, Kato Y, Morita R, Fukunaga K, Nagamura Y, Nishimura M, Sakamoto W, Tanaka A, Kusaba M (2013). Nyc4, the rice ortholog of Arabidopsis THF1, is involved in the degradation of chlorophyll-protein complexes during leaf senescence. Plant J 74, 652-662. |
| 50 | Yoshida S (2003). Molecular regulation of leaf senescence. Curr Opin Plant Biol 6, 79-84. |
| 51 | Zhou Y, Huang WF, Liu L, Chen TY, Zhou F, Lin YJ (2013). Identification and functional characterization of a rice NAC gene involved in the regulation of leaf senescence. BMC Plant Biol 13, 132. |
| 52 | Zhou Y, Liu L, Huang WF, Yuan M, Zhou F, Li XH, Lin YJ (2014). Overexpression of OsSWEET5 in rice causes growth retardation and precocious senescence. PLoS One 9, e94210. |
| [1] | 叶灿, 姚林波, 金莹, 高蓉, 谭琪, 李旭映, 张艳军, 陈析丰, 马伯军, 章薇, 张可伟. 水稻水杨酸代谢突变体高通量筛选方法的建立与应用[J]. 植物学报, 2025, 60(4): 1-0. |
| [2] | 赵凌, 管菊, 梁文化, 张勇, 路凯, 赵春芳, 李余生, 张亚东. 基于高密度Bin图谱的水稻苗期耐热性QTL定位[J]. 植物学报, 2025, 60(3): 342-353. |
| [3] | 李新宇, 谷月, 徐非非, 包劲松. 水稻胚乳淀粉合成相关蛋白的翻译后修饰研究进展[J]. 植物学报, 2025, 60(2): 256-270. |
| [4] | 李建国, 张怡, 张文君. 水稻根系铁膜形成及对磷吸收的影响[J]. 植物学报, 2025, 60(1): 132-143. |
| [5] | 姚瑞枫, 谢道昕. 水稻独脚金内酯信号感知的激活和终止[J]. 植物学报, 2024, 59(6): 873-877. |
| [6] | 连锦瑾, 唐璐瑶, 张伊诺, 郑佳兴, 朱超宇, 叶语涵, 王跃星, 商文楠, 傅正浩, 徐昕璇, 吴日成, 路梅, 王长春, 饶玉春. 水稻抗氧化性状遗传位点挖掘及候选基因分析[J]. 植物学报, 2024, 59(5): 738-751. |
| [7] | 黄佳慧, 杨惠敏, 陈欣雨, 朱超宇, 江亚楠, 胡程翔, 连锦瑾, 芦涛, 路梅, 张维林, 饶玉春. 水稻突变体pe-1对弱光胁迫的响应机制[J]. 植物学报, 2024, 59(4): 574-584. |
| [8] | 周俭民. 收放自如的明星战车[J]. 植物学报, 2024, 59(3): 343-346. |
| [9] | 朱超宇, 胡程翔, 朱哲楠, 张芷宁, 汪理海, 陈钧, 李三峰, 连锦瑾, 唐璐瑶, 钟芊芊, 殷文晶, 王跃星, 饶玉春. 水稻穗部性状QTL定位及候选基因分析[J]. 植物学报, 2024, 59(2): 217-230. |
| [10] | 夏婧, 饶玉春, 曹丹芸, 王逸, 柳林昕, 徐雅婷, 牟望舒, 薛大伟. 水稻中乙烯生物合成关键酶OsACS和OsACO调控机制研究进展[J]. 植物学报, 2024, 59(2): 291-301. |
| [11] | 方妍力, 田传玉, 苏如意, 刘亚培, 王春连, 陈析丰, 郭威, 纪志远. 水稻抗细菌性条斑病基因挖掘与初定位[J]. 植物学报, 2024, 59(1): 1-9. |
| [12] | 朱宝, 赵江哲, 张可伟, 黄鹏. 水稻细胞分裂素氧化酶9参与调控水稻叶夹角发育[J]. 植物学报, 2024, 59(1): 10-21. |
| [13] | 贾绮玮, 钟芊芊, 顾育嘉, 陆天麒, 李玮, 杨帅, 朱超宇, 胡程翔, 李三峰, 王跃星, 饶玉春. 水稻茎秆细胞壁相关组分含量QTL定位及候选基因分析[J]. 植物学报, 2023, 58(6): 882-892. |
| [14] | 戴若惠, 钱心妤, 孙静蕾, 芦涛, 贾绮玮, 陆天麒, 路梅, 饶玉春. 水稻叶色调控机制及相关基因研究进展[J]. 植物学报, 2023, 58(5): 799-812. |
| [15] | 田传玉, 方妍力, 沈晴, 王宏杰, 陈析丰, 郭威, 赵开军, 王春连, 纪志远. 2019-2021年我国南方稻区白叶枯病菌的毒力与遗传多样性调查研究[J]. 植物学报, 2023, 58(5): 743-749. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||