

植物学报 ›› 2025, Vol. 60 ›› Issue (2): 172-185.DOI: 10.11983/CBB24110 cstr: 32102.14.CBB24110
曹婕1,†, 卢秋连1,†, 翟健平1, 刘宝辉1,2, 方超1,2, 李世晨1,2,*( ), 苏彤1,2,*(
), 苏彤1,2,*( )
)
收稿日期:2024-07-22
接受日期:2024-12-14
出版日期:2025-03-10
发布日期:2024-12-27
通讯作者:
李世晨,苏彤
作者简介:第一联系人:†共同第一作者
基金资助:
Jie Cao1,†, Qiulian Lu1,†, Jianping Zhai1, Baohui Liu1,2, Chao Fang1,2, Shichen Li1,2,*( ), Tong Su1,2,*(
), Tong Su1,2,*( )
)
Received:2024-07-22
Accepted:2024-12-14
Online:2025-03-10
Published:2024-12-27
Contact:
Shichen Li, Tong Su
About author:First author contact:†These authors contributed equally to this paper
摘要: 海藻糖-6-磷酸合酶(trehalose-6-phosphate synthase, TPS)是合成海藻糖的关键酶, 已在多个物种中被报道参与调节光合作用、糖代谢、生长发育和逆境响应等生理过程。目前, TPS在大豆(Glycine max)中的报道极少。该文在大豆全基因组中鉴定了20个TPS基因及其包含的10种重要蛋白保守基序。启动子元件分析显示, 大豆TPS基因的启动子区富含大量胁迫响应元件; 盐胁迫处理后, 17个TPS基因的表达发生变化, 其中12个基因上调表达, 5个基因下调表达。对TPS进行单倍型和选择趋势分析, 发现TPS8、TPS13、TPS15、TPS17和TPS18存在2种主要的等位变异, 其中携带TPS15H2、TPS13H2、TPS17H2和TPS18H2的品种在栽培品种中大量富集, 受到强烈的人工选择。该研究揭示了大豆TPS基因家族的分子特征以及在盐胁迫下的表达模式和进化历史, 旨在为进一步解析大豆TPS基因的功能以及培育耐盐大豆品种提供理论依据和遗传材料。
曹婕, 卢秋连, 翟健平, 刘宝辉, 方超, 李世晨, 苏彤. 大豆TPS基因家族在盐胁迫下的表达变化及单倍型选择规律分析(长英文摘要). 植物学报, 2025, 60(2): 172-185.
Jie Cao, Qiulian Lu, Jianping Zhai, Baohui Liu, Chao Fang, Shichen Li, Tong Su. Changes in the Expression of the Soybean TPS Gene Family Under Salt Stress and Haplotype Selection Analysis. Chinese Bulletin of Botany, 2025, 60(2): 172-185.
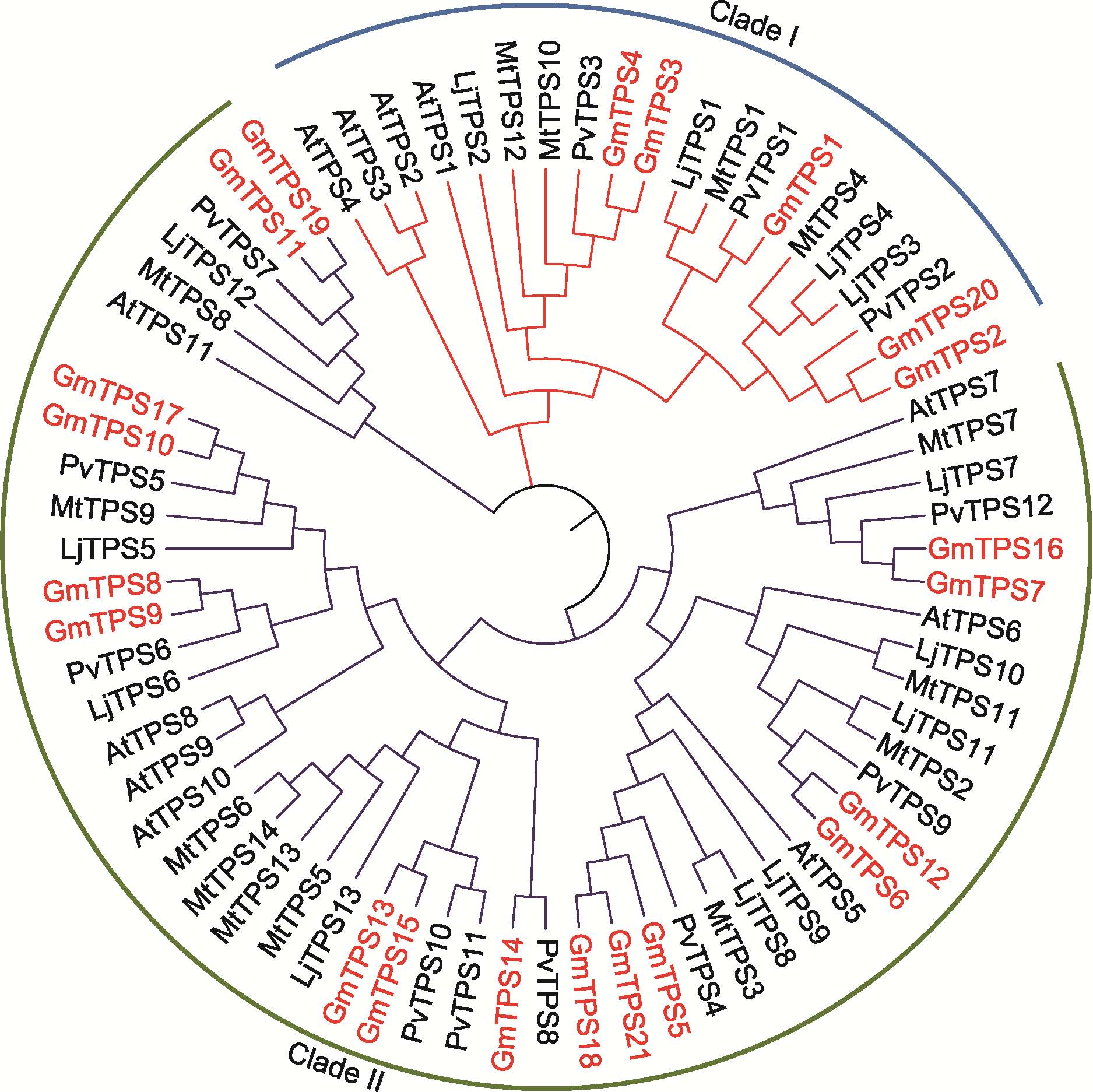
图1 拟南芥和豆科植物海藻糖-6-磷酸合酶(TPS)的系统发育树 拟南芥和豆科植物TPS蛋白分为2类(clade I和clade II)。红色的为大豆TPS蛋白。At: 拟南芥; Gm: 大豆; Pv: 菜豆; Mt: 蒺藜苜蓿; Lj: 百脉根
Figure 1 Phylogenetic tree of trehalose-6-phosphate synthase (TPS) from Arabidopsis and legumes The TPS proteins from Arabidopsis and legumes are divided into two clades (clade I and clade II). The soybean TPS proteins are highlighted in red. At: Arabidopsis thaliana; Gm: Glycine max; Pv: Phaseolus vulgaris; Mt: Medicago truncatula; Lj: Lotus japonicus
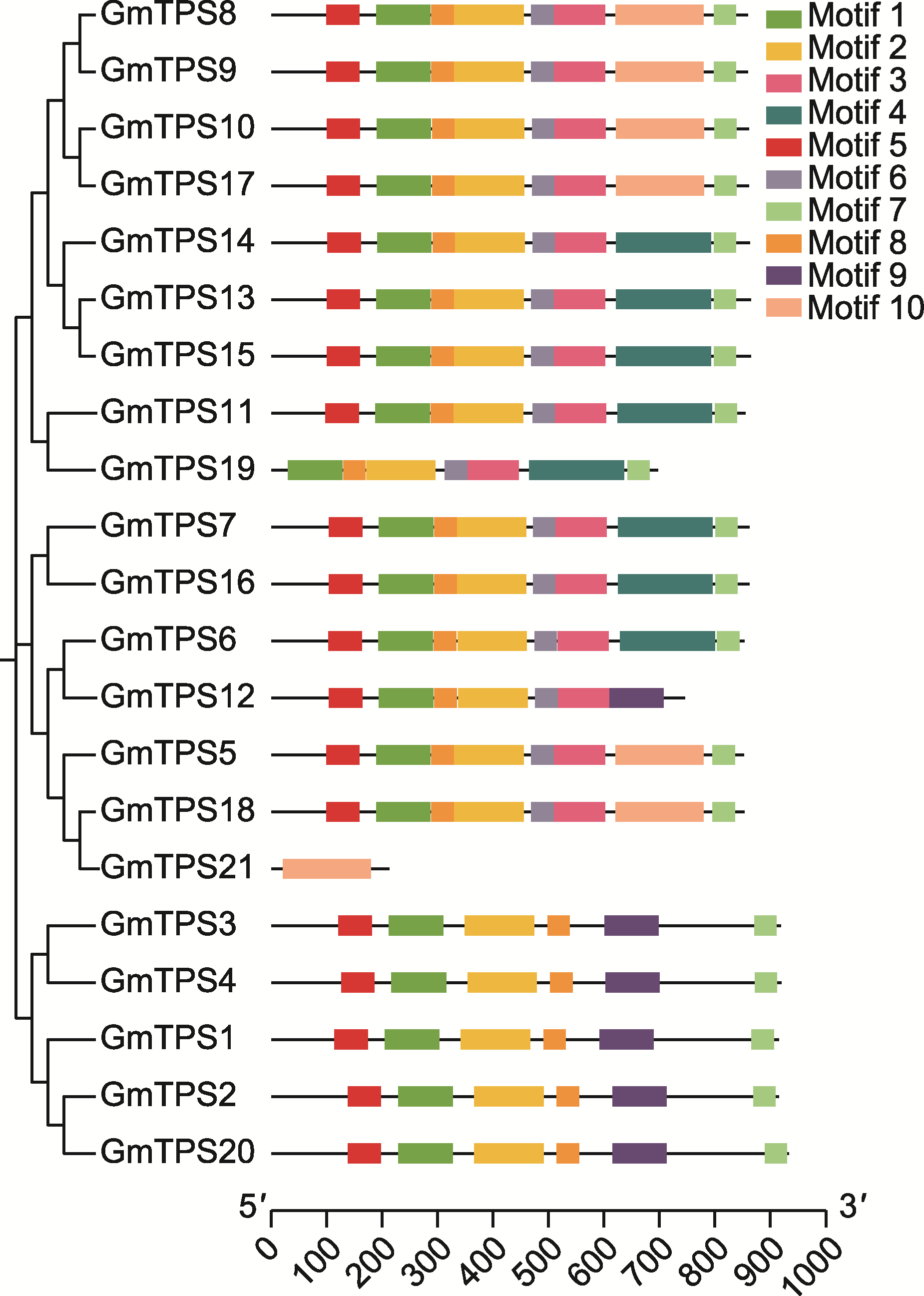
图2 大豆海藻糖-6-磷酸合酶(TPS)的蛋白保守基序 大豆TPS成员之间的系统发育关系与蛋白保守基序的分布, 共鉴定到10个保守基序。底部刻度尺代表氨基酸序列长度(aa)。
Figure 2 Conserved protein motif of trehalose-6-phosphate synthase (TPS) in soybean Phylogenetic relationships among TPS members and distribution of conserved protein motifs in soybean, ten motifs were identified. The bottom scale represents the amino acid sequence length (aa).
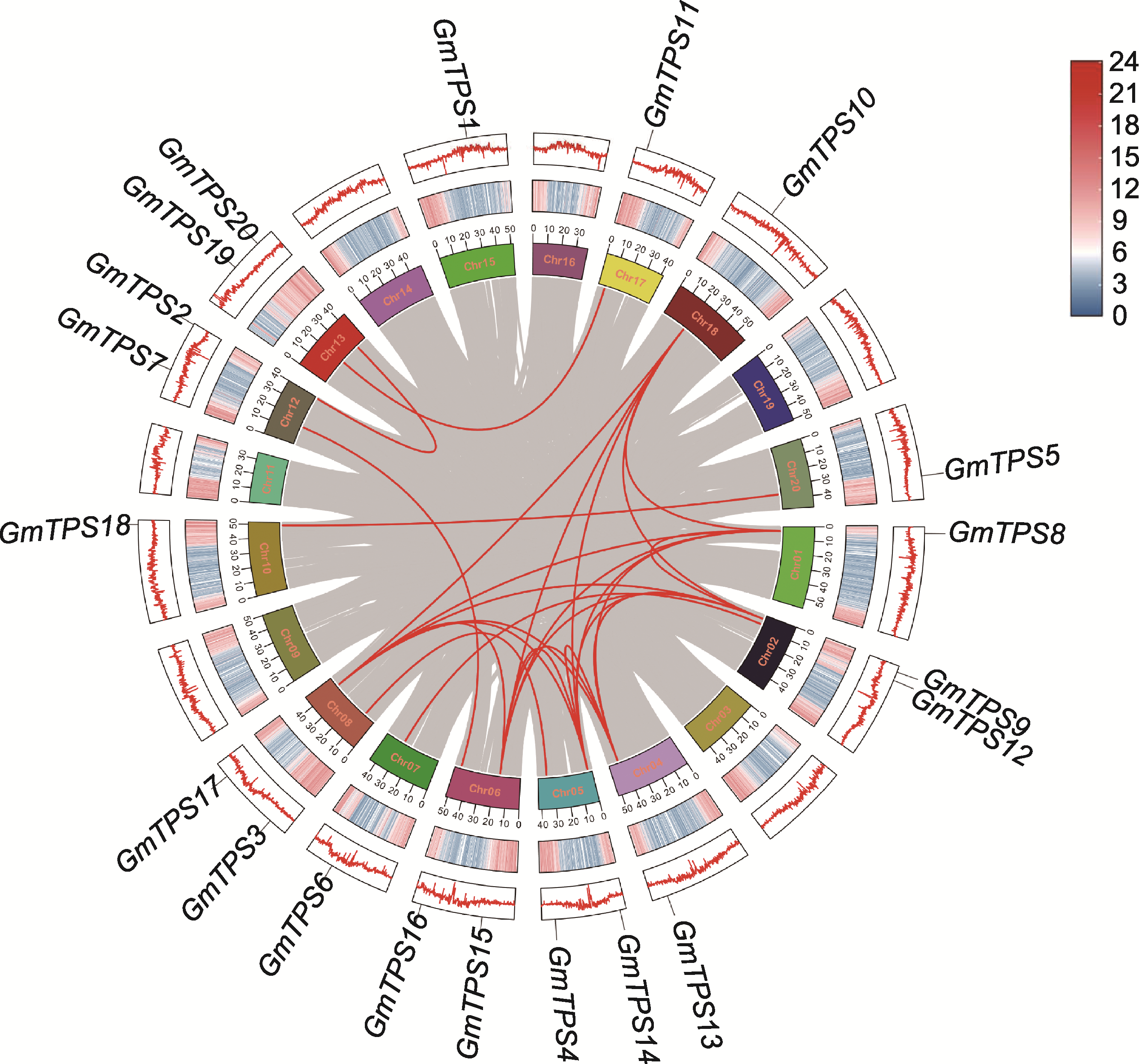
图3 大豆TPS基因的染色体分布及染色体间关系 红色曲线连接片段表示复制的基因对。
Figure 3 Chromosomal distribution and inter-chromosomal relationships of TPS genes in soybean Red curves connecting pairs of genes indicate segmental duplications.
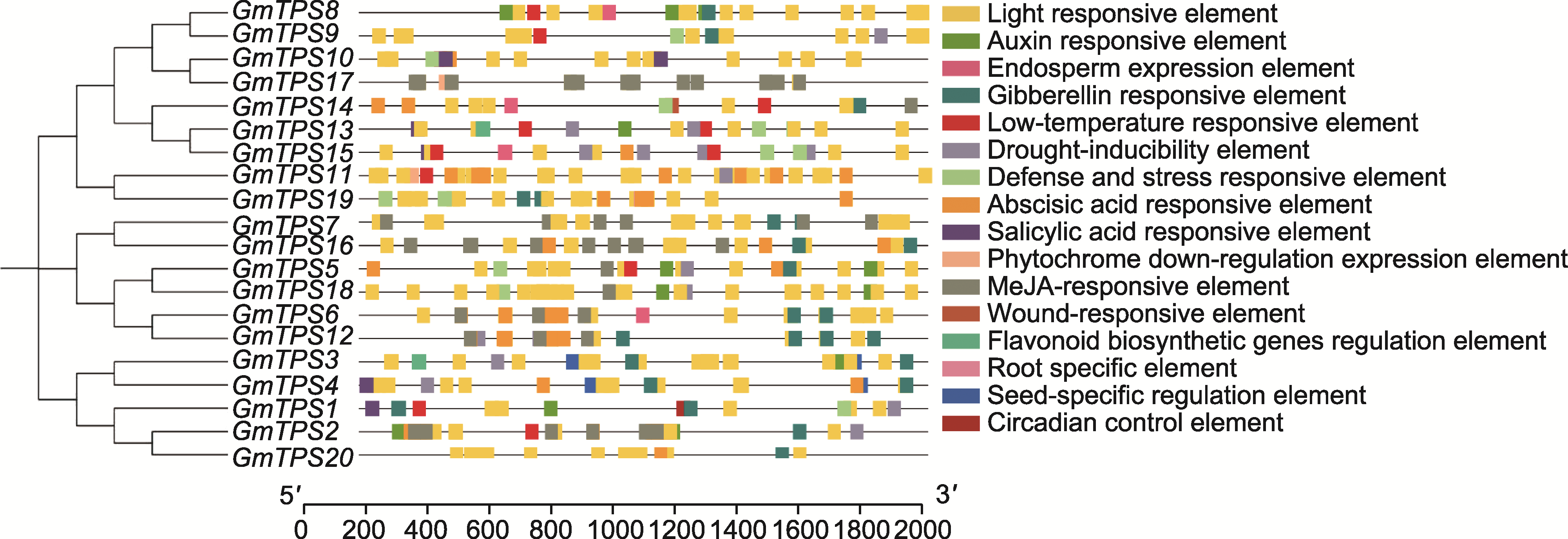
图4 大豆TPS基因的顺式作用元件 基因上游2 Kb启动子区域中的顺式作用元件被映射到每个TPS基因上, 以不同颜色标记不同类型的顺式作用元件, 右侧显示顺式作用元件类型。图中底部的刻度尺表示启动子序列的长度(bp)。
Figure 4 Cis-element analysis of TPS genes in soybean The cis-elements in the 2 Kb upstream promoter regions of each TPS gene are mapped, with different types of cis-elements represented by different colors, as indicated on the right. The scale bars at the bottom of the figure represent the length of the promoter sequence (bp).
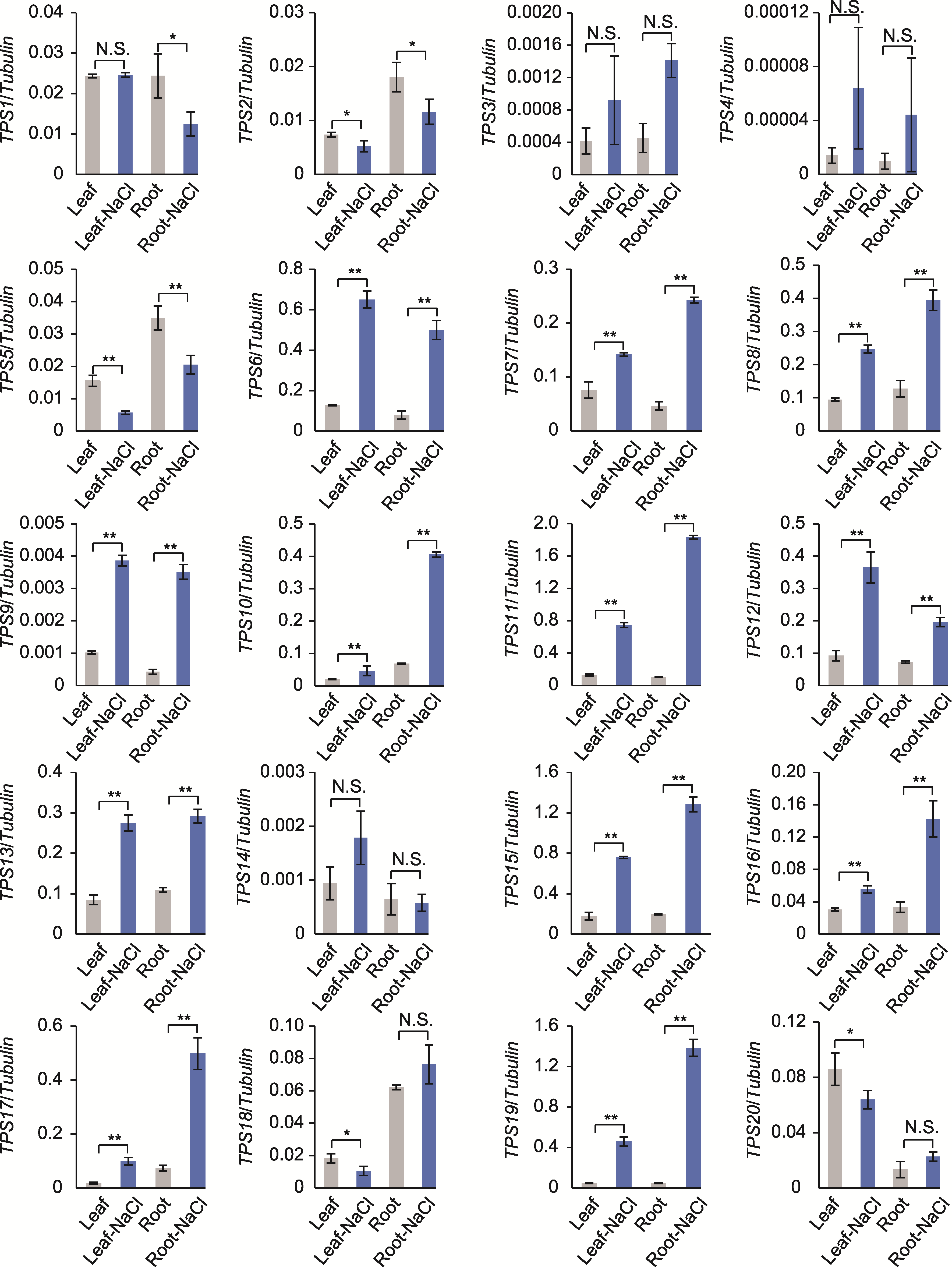
图5 qRT-PCR分析盐胁迫下TPS基因的表达量 图中从左往右, 从上至下依次为TPS1-TPS20。* P<0.05; ** P<0.01; N.S.表示无显著性差异。
Figure 5 qRT-PCR analysis of the TPS genes expression under salt stress From left to right and top to bottom in the picture, they are TPS1 to TPS20 in sequence. * P<0.05; ** P<0.01; N.S. indicate no significant difference.
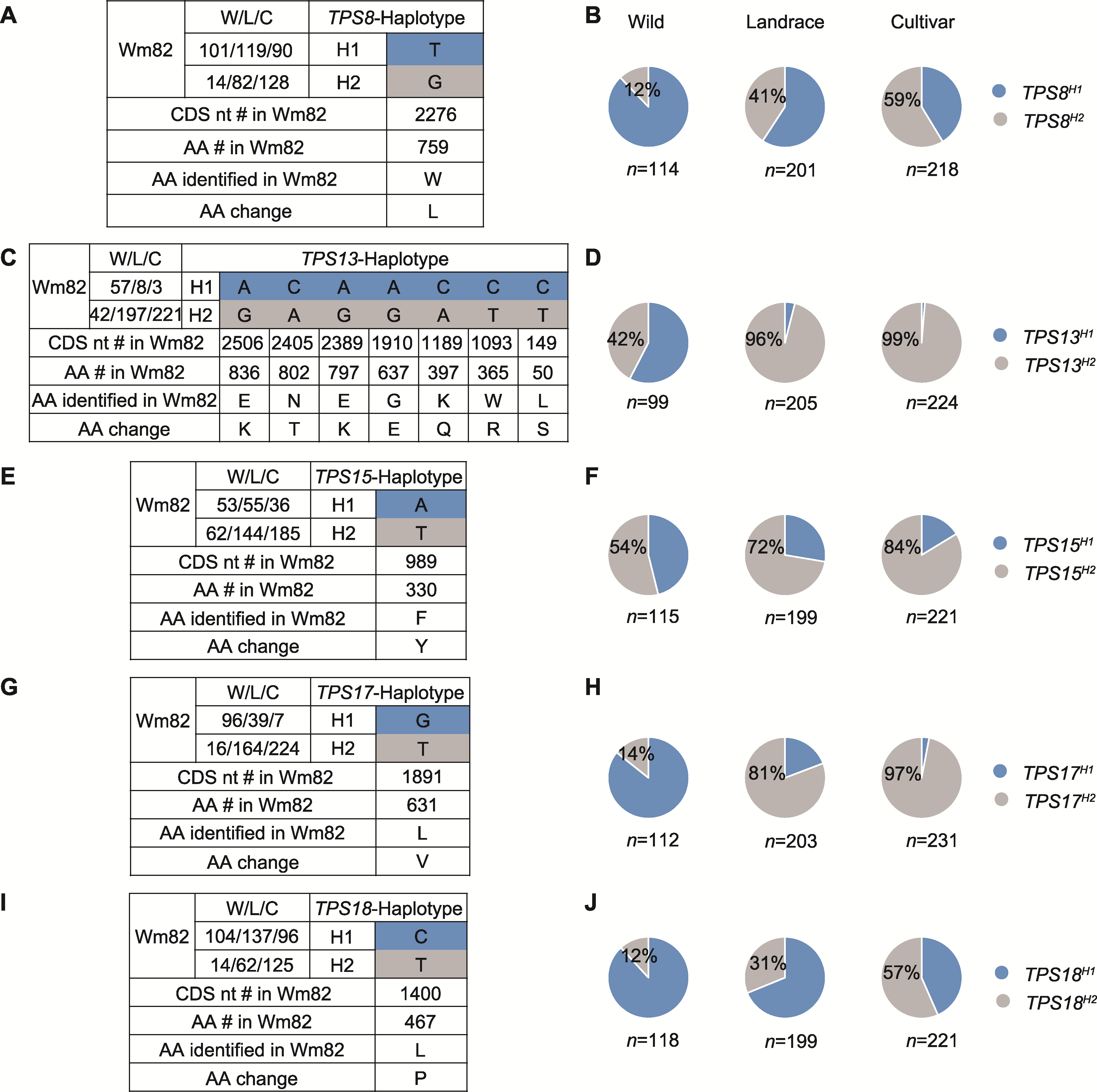
图6 受到人工选择的TPS基因 (A)、(C)、(E)、(G)和(I)分别代表TPS8、TPS13、TPS15、TPS17和TPS18的单倍型。图中所展示的为部分TPS重要位点的单倍型。(B)、(D)、(F)、(H)和(J)分别代表不同等位变异在野生大豆(W)、农家品种(L)和栽培品种(C)中所占的比例。数据来自559份已测序的材料(121份野生大豆、207份农家品种和231份栽培品种)。
Figure 6 TPS genes subjected to intense artificial selection (A), (C), (E), (G), and (I) represent the haplotypes of TPS8, TPS13, TPS15, TPS17, and TPS18, respectively. The figure shows several important TPS haplotypes. (B), (D), (F), (H), and (J) represent the proportions of different alleles in wild (W) soybeans, Landraces (L) and cultivated varieties (C), respectively. Data were obtained from 559 sequenced accessions (121 wild soybeans, 207 landraces, and 231 improved cultivars).
| [1] | Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 37, W202-W208. |
| [2] | Cannon SB, Mitra A, Baumgarten A, Young ND, May G (2004). The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol 4, 10. |
| [3] | Carter TE Jr, Nelson RL, Sneller CH, Cui ZL (2004). Genetic diversity in soybean. In: ShiblesRM, HarperJE, WilsonRF, ShoemakerRC, eds. Soybeans: Improvement, Production, and Uses, Vol. 16, 3rd edn. Madison: American Society of Agronomy. pp. 303-416. |
| [4] |
Chen CJ, Wu Y, Li JW, Wang X, Zeng ZH, Xu J, Liu YL, Feng JT, Chen H, He YH, Xia R (2023). TBtools-II: a "one for all, all for one" bioinformatics platform for biological big-data mining. Mol Plant 16, 1733-1742.
DOI PMID |
| [5] |
Conant GC, Wolfe KH (2008). Turning a hobby into a job: how duplicated genes find new functions. Nat Rev Genet 9, 938-950.
DOI PMID |
| [6] |
Delorge I, Figueroa C, Feil R, Lunn J, Van Dijck P (2015). Trehalose-6-phosphate synthase 1 is not the only active TPS in Arabidopsis thaliana. Biochem J 466, 283-290.
DOI PMID |
| [7] | Dong LD, Hou ZH, Li HY, Li ZB, Fang C, Kong LP, Li YL, Du H, Li T, Wang LS, He ML, Zhao XH, Cheng Q, Kong FJ, Liu BH (2022). Agronomical selection on loss-of- function of GIGANTEA simultaneously facilitates soybean salt tolerance and early maturity. J Integr Plant Biol 64, 1866-1882. |
| [8] | Du JL, Lin XL, Ma YW, Chen JR, Chen HX, Li YF (2023). Research progress in plant Trehalose-6-phosphate synthase genes. Plant Sci J 41, 411-420. (in Chinese) |
| 杜姣林, 蔺新兰, 马豫皖, 陈己任, 陈海霞, 李玉帆 (2023). 植物海藻糖-6-磷酸合成酶基因研究进展. 植物科学学报 41, 411-420. | |
| [9] | Du LS, Qi SY, Ma JJ, Xing LB, Fan S, Zhang SW, Li YM, Shen YW, Zhang D, Han MY (2017). Identification of TPS family members in apple (Malus × domestica Borkh.) and the effect of sucrose sprays on TPS expression and floral induction. Plant Physiol Bioch 120, 10-23. |
| [10] | El-Bashiti T, Hamamcı H, Öktem HA, Yücel M (2005). Biochemical analysis of trehalose and its metabolizing enzymes in wheat under abiotic stress conditions. Plant Sci 169, 47-54. |
| [11] | Essah PA, Davenport R, Tester M (2003). Sodium influx and accumulation in Arabidopsis. Plant Physiol 133, 307-318. |
| [12] | Fehr WR, Caviness CE, Burmood DT, Pennington JS (1971). Stage of development descriptions for soybeans, Glycine max (L.) Merrill. Crop Sci 11, 929-931. |
| [13] | Gao CS, Yuan JZ, Zhi JY, Cheng YH, Dong LD, Cheng Q (2024). Identification and domestication analysis of ERF salt tolerance genes in soybean. J Plant Genet Resour 25, 30-38. (in Chinese) |
|
高超升, 袁嘉志, 植健怡, 程玉汉, 董利东, 程群 (2024). 大豆ERF耐盐基因的鉴定和驯化分析. 植物遗传资源学报 25, 30-38.
DOI |
|
| [14] | Hammer K (1984). Das domestikationssyndrom. Die Kulturpflanze 32, 11-34. |
| [15] |
Hayes S, Pantazopoulou CK, Van Gelderen K, Reinen E, Tween AL, Sharma A, De Vries M, Prat S, Schuurink RC, Testerink C, Pierik R (2019). Soil salinity limits plant shade avoidance. Curr Biol 29, 1669-1676.
DOI PMID |
| [16] | Hou ZH, Li YL, Cheng YH, Li WW, Li T, Du H, Kong FJ, Dong LD, Zheng DF, Feng NJ, Liu BH, Cheng Q (2022). Genome-wide analysis of DREB genes identifies a novel salt tolerance gene in wild soybean (Glycine soja). Front Plant Sci 13, 821647. |
| [17] | Hu Y, Yang F, Yang N, Jia W, Cui Y (2023). Analysis and prospects of saline-alkali land in China from the perspective of utilization. Chin J Soil Sci 54, 489-494. (in Chinese) |
| 胡炎, 杨帆, 杨宁, 贾伟, 崔勇 (2023). 盐碱地资源分析及利用研究展望. 土壤通报 54, 489-494. | |
| [18] |
Huang XH, Huang SW, Han B, Li JY (2022). The integrated genomics of crop domestication and breeding. Cell 185, 2828-2839.
DOI PMID |
| [19] | Hymowitz T (1970). On the domestication of the soybean. Econ Bot 24, 408-421. |
| [20] |
Hyten DL, Song QJ, Zhu YL, Choi IY, Nelson RL, Costa JM, Specht JE, Shoemaker RC, Cregan PB (2006). Impacts of genetic bottlenecks on soybean genome diversity. Proc Natl Acad Sci USA 103, 16666-16671.
DOI PMID |
| [21] | Jiang T, Zhai H, Wang FB, Zhou HN, Si ZZ, He SZ, Liu QC (2014). Cloning and characterization of a salt tolerance- associated gene encoding trehalose-6-phosphate synthase in sweetpotato. J Integr Plant Biol 13, 1651-1661. |
| [22] | Kelley L, Mezulis S, Yates CM, Wass MN, Sternberg MJE (2015). The Phyre2 web portal for protein modeling, prediction and analysis. Nature 10, 845-858. |
| [23] | Kondrashov FA, Rogozin IB, Wolf YI, Koonin EV (2002). Selection in the evolution of gene duplications. Genome Biol 3, research0008. |
| [24] | Kosar F, Akram NA, Sadiq M, Al-Qurainy F, Ashraf M (2019). Trehalose: a key organic osmolyte effectively involved in plant abiotic stress tolerance. J Plant Growth Regul 38, 606-618. |
| [25] |
Krzywinski M, Schein J, Birol İ, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA (2009). Circos: an information aesthetic for comparative genomics. Genome Res 19, 1639-1645.
DOI PMID |
| [26] | Lam HM, Xu X, Liu X, Chen WB, Yang GH, Wong FL, Li MW, He W, Qin N, Wang B, Li J, Jian M, Wang J, Shao GH, Wang J, Sun SSM, Zhang GY (2010). Resequencing of 31 wild and cultivated soybean genomes identifies patterns of genetic diversity and selection. Nat Genet 42, 1053-1059. |
| [27] | Letunic I, Bork P (2021). Interactive tree of life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res 49, W293-W296. |
| [28] | Li HW, Zang BS, Deng XW, Wang XP (2011). Overexpression of the trehalose-6-phosphate synthase gene OsTPS1 enhances abiotic stress tolerance in rice. Planta 234, 1007-1018. |
| [29] | Li ML, Xue M, Ma HY, Feng P, Chen T, Sun XH, Li Q, Ding XD, Zhang SZ, Xiao JL (2024). Wild soybean (Glycine soja) transcription factor GsWRKY40 plays positive roles in plant salt tolerance. Crop J 12, 766-775. |
| [30] | Li S, Wang N, Ji DD, Zhang WX, Wang Y, Yu YC, Zhao SZ, Lyu M, You JJ, Zhang YY, Wang LL, Wang XF, Liu ZH, Tong JH, Xiao LT, Bai MY, Xiang FN (2019). A GmSIN1/GmNCED3s/GmRbohBs feed-forward loop acts as a signal amplifier that regulates root growth in soybean exposed to salt stress. Plant Cell 31, 2107-2130. |
| [31] |
Livak KJ, Schmittgen TD (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCTmethod. Methods 25, 402-408.
DOI PMID |
| [32] |
Lu SJ, Fang C, Abe J, Kong FJ, Liu BH (2022). Current overview on the genetic basis of key genes involved in soybean domestication. aBIOTECH 3, 126-139.
DOI PMID |
| [33] |
Lunn JE (2007). Gene families and evolution of trehalose metabolism in plants. Funct Plant Biol 34, 550-563.
DOI PMID |
| [34] | Lunn JE, Delorge I, Figueroa CM, Van Dijck P, Stitt M (2014). Trehalose metabolism in plants. Plant J 79, 544-567. |
| [35] |
Mu M, Lu XK, Wang JJ, Wang DL, Yin ZJ, Wang S, Fan WL, Ye WW (2016). Genome-wide identification and analysis of the stress-resistance function of the TPS (trehalose-6-phosphate synthase) gene family in cotton. BMC Genet 17, 54.
DOI PMID |
| [36] |
Munns R, Tester M (2008). Mechanisms of salinity tolerance. Annu Rev Plant Biol 59, 651-681.
DOI PMID |
| [37] | Papiernik SK, Grieve CM, Lesch SM, Yates SR (2005). Effects of salinity, imazethapyr, and chlorimuron application on soybean growth and yield. Commun Soil Sci Plant Anal 36, 951-967. |
| [38] | Paul MJ, Watson A, Griffiths CA (2020). Trehalose 6- phosphate signaling and impact on crop yield. Biochem Soc Trans 48, 2127-2137. |
| [39] | Ramon M, De Smet I, Vandesteene L, Naudts M, Leyman B, Van Dijck P, Rolland F, Beeckman T, Thevelein JM (2009). Extensive expression regulation and lack of heterologous enzymatic activity of the Class II trehalose metabolism proteins from Arabidopsis thaliana. Plant Cell Environ 32, 1015-1032. |
| [40] | Ramsey J, Schemske DW (1998). Pathways, mechanisms, and rates of polyploid formation in flowering plants. Annu Rev Ecol Syst 29, 467-501. |
| [41] |
Singh M, Nara U, Kumar A, Choudhary A, Singh H, Thapa S (2021). Salinity tolerance mechanisms and their breeding implications. J Genet Eng Biotechnol 19, 173.
DOI PMID |
| [42] | Song JB, Mao HY, Cheng J, Zhou Y, Chen RR, Zeng LM, Li H, Wang YH (2021). Identification of the trehalose- 6-phosphate synthase gene family in Medicago truncatula and expression analysis under abiotic stresses. Gene 787, 145641. |
| [43] | Su T, Liu H, Wu YC, Wang JH, He FL, Li HY, Li SC, Wang LS, Li LX, Cao J, Lu QL, Zhao XH, Xiang HT, Lin C, Lu SJ, Liu BH, Kong FJ, Fang C (2024). Soybean hypocotyl elongation is regulated by a MYB33-SWEET11/21-GA2ox8c module involving long-distance sucrose transport. Plant Biotechnol J 22, 2859-2872. |
| [44] | Sun XP, Xiang YL, Dou NN, Zhang H, Pei SR, Franco AV, Menon M, Monier B, Ferebee T, Liu T, Liu SY, Gao YC, Wang JB, Terzaghi W, Yan JB, Hearne S, Li L, Li F, Dai MQ (2023). The role of transposon inverted repeats in balancing drought tolerance and yield-related traits in maize. Nat Biotechnol 41, 120-127. |
| [45] |
Tamura K, Stecher G, Kumar S (2021). MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol 38, 3022-3027.
DOI PMID |
| [46] |
Van Dijken AJH, Schluepmann H, Smeekens SCM (2004). Arabidopsis trehalose-6-phosphate synthase 1 is essential for normal vegetative growth and transition to flowering. Plant Physiol 135, 969-977.
DOI PMID |
| [47] |
Vandesteene L, Ramon M, Le Roy K, Van Dijck P, Rolland F (2010). A single active trehalose-6-P synthase (TPS) and a family of putative regulatory TPS-like proteins in Arabidopsis. Mol Plant 3, 406-419.
DOI PMID |
| [48] | Wang JH, Sun ZH, Liu H, Yue L, Wang F, Liu SR, Su BH, Liu BH, Kong FJ, Fang C (2023). Genome-wide identification and characterization of the soybean Snf2 gene family and expression response to rhizobia. Int J Mol Sci 24, 7250. |
| [49] | Wang Y, Tang H, Debarry JD, Tan X, Li J, Wang X, Lee TH, Jin H, Marler B, Guo H, Kissinger JC, Paterson AH (2012). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res 40, e49. |
| [50] |
Wendel JF (2000). Genome evolution in polyploids. Plant Mol Biol 42, 225-249.
PMID |
| [51] | Xia ZJ (2017). Research progress in whole-genome analysis and cloning of genes underlying important agronomic traits in soybean. Chin Bull Bot 52,148-158. (in Chinese) |
|
夏正俊 (2017). 大豆基因组解析与重要农艺性状基因克隆研究进展. 植物学报 52, 148-158.
DOI |
|
| [52] |
Xie DW, Wang XN, Fu LS, Sun J, Zheng W, Li ZF (2015). Identification of the trehalose-6-phosphate synthase gene family in winter wheat and expression analysis under conditions of freezing stress. J Genet 94, 55-65.
PMID |
| [53] | Xie L, Wang ZX, Huang B (2014). Genome-wide identification classification and expression of TPS family genes in soybean. Chin J Oil Crop Sci 36, 160-167. (in Chinese) |
|
谢翎, 汪章勋, 黄勃 (2014). 大豆TPS基因家族全基因组鉴定、分类与表达分析. 中国油料作物学报 36, 160-167.
DOI |
|
| [54] | Xu CJ, Shan JM, Liu TM, Wang Q, Ji YJ, Zhang YT, Wang MY, Xia N, Zhao L (2023). CONSTANS-LIKE 1a positively regulates salt and drought tolerance in soybean. Plant Physiol 191, 2427-2446. |
| [55] | Xu YC, Wang YJ, Mattson N, Yang L, Jin QJ (2017). Genome-wide analysis of the Solanum tuberosum (potato) trehalose-6-phosphate synthase (TPS) gene family: evolution and differential expression during development and stress. BMC Genomics 18, 926. |
| [56] | Yang HL, Liu YJ, Wang CL, Zeng QY (2012). Molecular evolution of trehalose-6-phosphate synthase (TPS) gene family in Populus, Arabidopsis and rice. PLoS One 7, e42438. |
| [57] | Yuan GP, Liu JP, An GL, Li WH, Si WJ, Sun DX, Zhu YC (2021). Genome-wide identification and characterization of the trehalose-6-phosphate synthetase (TPS) gene family in watermelon (Citrullus lanatus) and their transcriptional responses to salt stress. Int J Mol Sci 23, 276. |
| [58] |
Zentella R, Mascorro-Gallardo JO, Van Dijck P, Folch- Mallol J, Bonini B, Van Vaeck C, Gaxiola R, Covarrubias AA, Nieto-Sotelo J, Thevelein JM, Iturriaga G (1999). A Selaginella lepidophylla trehalose-6-phosphate synthase complements growth and stress-tolerance defects in a yeast tps1 mutant. Plant Physiol 119, 1473-1482.
PMID |
| [59] | Zhuang YB, Wang XT, Li XC, Hu JM, Fan LC, Landis JB, Cannon SB, Grimwood J, Schmutz J, Jackson SA, Doyle JJ, Zhang XS, Zhang DJ, Ma JX (2022). Phylogenomics of the genus Glycine sheds light on polyploid evolution and life-strategy transition. Nat Plants 8, 233-244. |
| [1] | 郑立媛, 徐茜竹, 尹嘉淇, 孙小雯, 王艳. 沈阳城郊近河农田退耕地野大豆群落生态位和种间联结研究[J]. 植物生态学报, 2025, 49(濒危植物的保护与恢复): 1-. |
| [2] | 范惠玲, 路妍, 金文海, 王慧, 彭小星, 武学霞, 刘玉皎. 基于根系表型性状的蚕豆耐盐碱性鉴定与综合评价(长英文摘要)[J]. 植物学报, 2025, 60(2): 204-217. |
| [3] | 段政勇, 丁敏, 王宇卓, 丁艺冰, 陈凌, 王瑞云, 乔治军. 糜子SBP基因家族全基因组鉴定及表达分析[J]. 植物学报, 2024, 59(2): 231-244. |
| [4] | 陈佳欣, 梅浩, 黄彩翔, 梁宗原, 全依桐, 李东鹏, 布威麦尔耶姆·赛麦提, 李欣欣, 廖红. 利用转基因毛状根高效培育大豆嵌合植株的方法[J]. 植物学报, 2024, 59(1): 89-98. |
| [5] | 孙福辉, 方慧仪, 温小蕙, 张亮生. 马银花MADS-box基因家族系统进化与表达分析[J]. 植物学报, 2023, 58(3): 404-416. |
| [6] | 王韫慧, 王一帆, 蔺佳雨, 李金红, 姚士恩, 冯湘池, 曹振林, 王俊, 李美娜. 植物驱动蛋白: 从微管阵列到生理活动调控[J]. 植物学报, 2022, 57(3): 358-374. |
| [7] | 杜梦柯, 连文婷, 张晓, 李欣欣. 氮处理对大豆根瘤固氮能力及GmLbs基因表达的影响[J]. 植物学报, 2021, 56(4): 391-403. |
| [8] | 王研, 贾博为, 孙明哲, 孙晓丽. 野生大豆耐逆分子调控机制研究进展[J]. 植物学报, 2021, 56(1): 104-115. |
| [9] | 夏正俊, 李玉卓, 朱金龙, 吴红艳, 徐坤, 翟红. 快速、无损大豆种子连续取样技术及其DNA制备[J]. 植物学报, 2021, 56(1): 56-61. |
| [10] | 祝光涛,黄三文. 360度群体遗传变异扫描——大豆泛基因组研究[J]. 植物学报, 2020, 55(4): 403-406. |
| [11] | 冯锋,战勇,田志喜. 新疆地区发展大豆生产的可行性和初步建议[J]. 植物学报, 2020, 55(2): 199-204. |
| [12] | 范业赓,丘立杭,黄杏,周慧文,甘崇琨,李杨瑞,杨荣仲,吴建明,陈荣发. 甘蔗节间伸长过程赤霉素生物合成关键基因的表达及相关植物激素动态变化[J]. 植物学报, 2019, 54(4): 486-496. |
| [13] | 唐康,杨若林. 大豆蛋白编码基因起源与进化[J]. 植物学报, 2019, 54(3): 316-327. |
| [14] | 王甜甜, 郝怀庆, 冯雪, 景海春. 植物HKT蛋白耐盐机制研究进展[J]. 植物学报, 2018, 53(5): 710-725. |
| [15] | 叶子飘, 段世华, 安婷, 康华靖. 最大电子传递速率的确定及其对电子流分配的影响[J]. 植物生态学报, 2018, 42(4): 498-507. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||