

植物学报 ›› 2019, Vol. 54 ›› Issue (3): 316-327.DOI: 10.11983/CBB18176 cstr: 32102.14.CBB18176
收稿日期:2018-08-14
接受日期:2018-12-10
出版日期:2019-05-01
发布日期:2019-11-24
通讯作者:
杨若林
基金资助:Received:2018-08-14
Accepted:2018-12-10
Online:2019-05-01
Published:2019-11-24
Contact:
Ruolin Yang
摘要: 物种基因组成是一个高度动态的进化过程, 其中相对较近起源的种系和物种特异性基因会持续整合到包含古老基因的原始基因网络中。新基因在塑造基因组结构中发挥重要作用, 能提高物种适应性。基因复制和新基因的从头起源是产生新基因及改变基因家族大小的2种方式。目前, 大豆(Glycine max)基因起源时间与进化模式的相互联系很大程度上还未被探索。该研究选择19种具有代表性的被子植物基因组, 分析基因含量动态性与大豆基因起源之间的潜在联系。采用基因出现法, 研究显示约58.7%的大豆基因能追溯到大约1.5亿年前, 同时有21.7%的基因为最近起源的orphan基因。研究结果表明, 与新基因相比, 古老基因受到更强的负选择压并且更加保守。此外, 古老基因的表达水平更高且更可能发生选择性剪切。此外, 具有不同拷贝数的基因在上述特征中也具有明显差异。研究结果有助于认识不同年龄基因的进化模式。
唐康,杨若林. 大豆蛋白编码基因起源与进化. 植物学报, 2019, 54(3): 316-327.
Kang Tang,Ruolin Yang. Origin and Evolution of Soybean Protein-coding Genes. Chinese Bulletin of Botany, 2019, 54(3): 316-327.
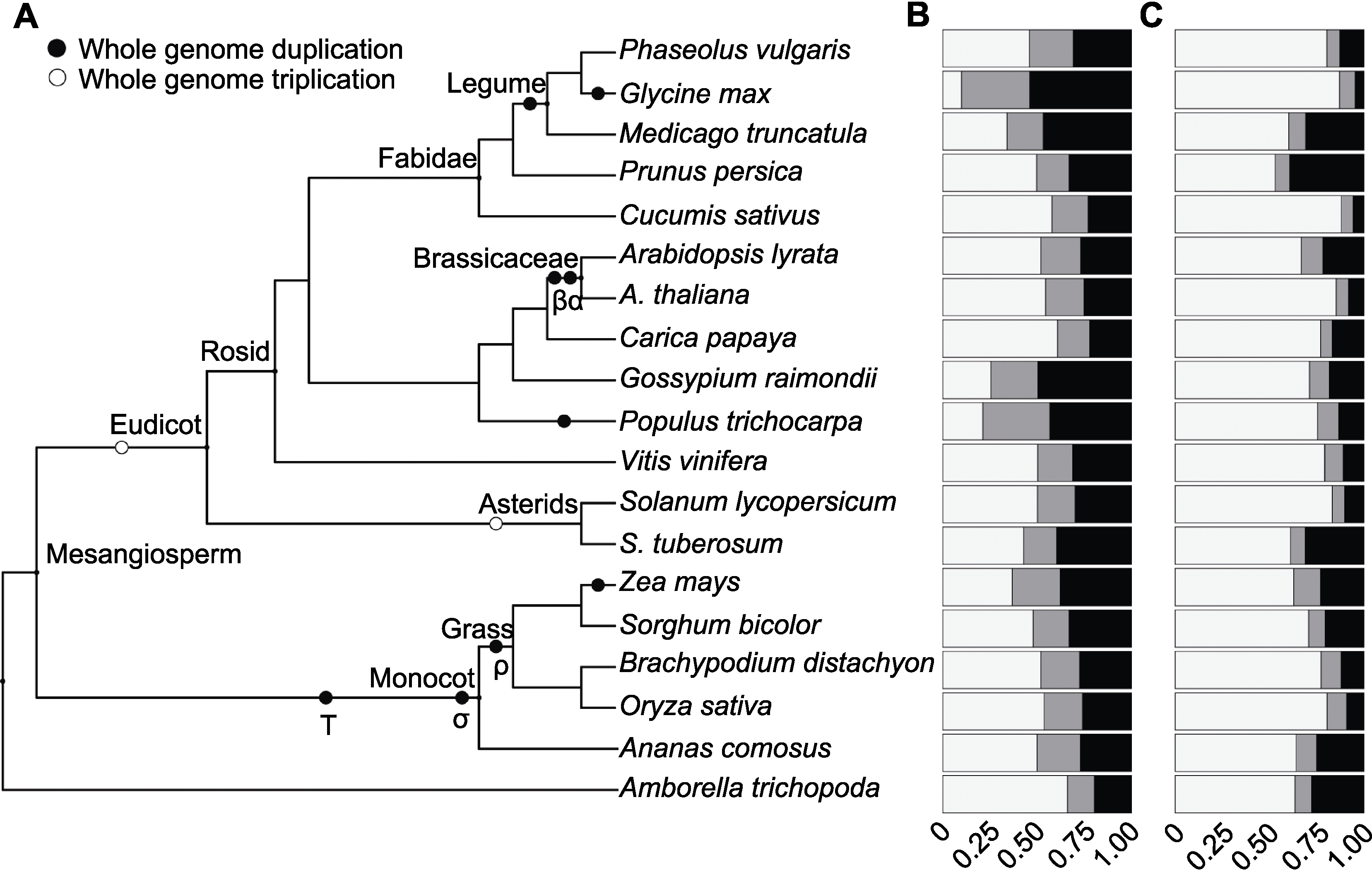
图1 19种被子植物基因家族大小分布(A) 系统发育树代表19种被子植物的进化关系; (B) 直系同源基因家族大小; (C) orphan基因家族大小。白、灰、黑分别代表单拷贝、两拷贝和多拷贝基因所占比例。
Figure 1 Gene family size distribution of 19 angiosperm species(A) Phylogenetic tree showing the relationships between the 19 angiosperm species used in this study; (B) Homologous gene family sizes; (C) Gene family sizes of orphan genes. The colors indicate the proportions of genes, white for singletons, grey for two-genes and black for multigenes.
| Species | Singletons | Two-gene families | Multigene families | Total gene families | Maximum gene family size |
|---|---|---|---|---|---|
| Amborella trichopoda | 9823 | 1061(2122) | 523(2935) | 11407 | 207 |
| Ananas comosus | 9059 | 2087(4174) | 1007(4916) | 12153 | 124 |
| Oryza sativa | 11966 | 2269(4538) | 1167(5805) | 15402 | 64 |
| Brachypodium distachyon | 11455 | 2264(4528) | 1209(6066) | 14928 | 50 |
| Sorghum bicolor | 12663 | 2529(5058) | 1399(8749) | 16591 | 416 |
| Zea mays | 10277 | 3568(7136) | 1964(10539) | 15809 | 297 |
| Solanum tuberosum | 11592 | 2390(4780) | 1399(10741) | 15381 | 1051 |
| S. lycopersicum | 12210 | 2448(4896) | 1371(7277) | 16029 | 72 |
| Vitis vinifera | 10408 | 1931(3862) | 1104(6483) | 13443 | 100 |
| Populus trichocarpa | 6550 | 5476(10952) | 2337(13368) | 14363 | 108 |
| Gossypium raimondii | 7582 | 3700(7400) | 2960(14806) | 14242 | 90 |
| Carica papaya | 10776 | 1505(3010) | 667(3948) | 12948 | 194 |
| Arabidopsis thaliana | 13278 | 2485(4970) | 1194(6144) | 16957 | 125 |
| A. lyrata | 12767 | 2605(5210) | 1327(6596) | 16699 | 67 |
| Cucumis sativus | 10152 | 1691(3382) | 795(4038) | 12638 | 38 |
| Prunus persica | 10822 | 1876(3752) | 1106(7192) | 13804 | 217 |
| Medicago truncatula | 9936 | 2812(5624) | 1948(13673) | 14696 | 308 |
| Glycine max | 4241 | 7735(15470) | 4206(23027) | 16182 | 153 |
| Phaseolus vulgaris | 11324 | 2873(5746) | 1430(7626) | 15569 | 132 |
表1 19种被子植物中直系同源基因家族(及基因)数目
Table 1 Number of homologous gene families (and genes) in 19 angiosperm species
| Species | Singletons | Two-gene families | Multigene families | Total gene families | Maximum gene family size |
|---|---|---|---|---|---|
| Amborella trichopoda | 9823 | 1061(2122) | 523(2935) | 11407 | 207 |
| Ananas comosus | 9059 | 2087(4174) | 1007(4916) | 12153 | 124 |
| Oryza sativa | 11966 | 2269(4538) | 1167(5805) | 15402 | 64 |
| Brachypodium distachyon | 11455 | 2264(4528) | 1209(6066) | 14928 | 50 |
| Sorghum bicolor | 12663 | 2529(5058) | 1399(8749) | 16591 | 416 |
| Zea mays | 10277 | 3568(7136) | 1964(10539) | 15809 | 297 |
| Solanum tuberosum | 11592 | 2390(4780) | 1399(10741) | 15381 | 1051 |
| S. lycopersicum | 12210 | 2448(4896) | 1371(7277) | 16029 | 72 |
| Vitis vinifera | 10408 | 1931(3862) | 1104(6483) | 13443 | 100 |
| Populus trichocarpa | 6550 | 5476(10952) | 2337(13368) | 14363 | 108 |
| Gossypium raimondii | 7582 | 3700(7400) | 2960(14806) | 14242 | 90 |
| Carica papaya | 10776 | 1505(3010) | 667(3948) | 12948 | 194 |
| Arabidopsis thaliana | 13278 | 2485(4970) | 1194(6144) | 16957 | 125 |
| A. lyrata | 12767 | 2605(5210) | 1327(6596) | 16699 | 67 |
| Cucumis sativus | 10152 | 1691(3382) | 795(4038) | 12638 | 38 |
| Prunus persica | 10822 | 1876(3752) | 1106(7192) | 13804 | 217 |
| Medicago truncatula | 9936 | 2812(5624) | 1948(13673) | 14696 | 308 |
| Glycine max | 4241 | 7735(15470) | 4206(23027) | 16182 | 153 |
| Phaseolus vulgaris | 11324 | 2873(5746) | 1430(7626) | 15569 | 132 |
| Species | Singletons | Two-gene families | Multigene families | Species-specific genes | Maximum gene family size |
|---|---|---|---|---|---|
| Amborella trichopoda | 7892 | 547(1094) | 502(3447) | 12433 | 105 |
| Ananas comosus | 5685 | 483(966) | 297(2224) | 8875 | 94 |
| Oryza sativa | 10774 | 686(1372) | 292(1224) | 13370 | 29 |
| Brachypodium distachyon | 3485 | 235(470) | 125(548) | 4503 | 15 |
| Sorghum bicolor | 5682 | 350(700) | 254(1644) | 8026 | 103 |
| Zea mays | 7253 | 813(1626) | 552(2643) | 11522 | 65 |
| Solanum tuberosum | 7278 | 471(942) | 376(3688) | 11908 | 163 |
| S. lycopersicum | 7836 | 308(616) | 177(950) | 9402 | 51 |
| Vitis vinifera | 7238 | 445(890) | 229(1006) | 9134 | 44 |
| Populus trichocarpa | 7923 | 593(1186) | 281(1398) | 10507 | 31 |
| Gossypium raimondii | 5495 | 408(816) | 293(1406) | 7717 | 26 |
| Carica papaya | 7680 | 307(614) | 224(1653) | 9947 | 88 |
| Arabidopsis thaliana | 2751 | 105(210) | 57(261) | 3222 | 21 |
| A. lyrata | 5413 | 461(922) | 366(1759) | 8094 | 83 |
| Cucumis sativus | 3458 | 125(250) | 54(223) | 3931 | 13 |
| Prunus persica | 3347 | 242(484) | 195(2483) | 6314 | 838 |
| Medicago truncatula | 12763 | 962(1924) | 820(6524) | 21211 | 145 |
| Glycine max | 9961 | 476(952) | 118(523) | 11436 | 23 |
| Phaseolus vulgaris | 2013 | 85(170) | 58(318) | 2501 | 19 |
表2 19种被子植物中的orphan基因家族(及基因)数目
Table 2 Number of orphan gene families (and genes) in 19 angiosperm species
| Species | Singletons | Two-gene families | Multigene families | Species-specific genes | Maximum gene family size |
|---|---|---|---|---|---|
| Amborella trichopoda | 7892 | 547(1094) | 502(3447) | 12433 | 105 |
| Ananas comosus | 5685 | 483(966) | 297(2224) | 8875 | 94 |
| Oryza sativa | 10774 | 686(1372) | 292(1224) | 13370 | 29 |
| Brachypodium distachyon | 3485 | 235(470) | 125(548) | 4503 | 15 |
| Sorghum bicolor | 5682 | 350(700) | 254(1644) | 8026 | 103 |
| Zea mays | 7253 | 813(1626) | 552(2643) | 11522 | 65 |
| Solanum tuberosum | 7278 | 471(942) | 376(3688) | 11908 | 163 |
| S. lycopersicum | 7836 | 308(616) | 177(950) | 9402 | 51 |
| Vitis vinifera | 7238 | 445(890) | 229(1006) | 9134 | 44 |
| Populus trichocarpa | 7923 | 593(1186) | 281(1398) | 10507 | 31 |
| Gossypium raimondii | 5495 | 408(816) | 293(1406) | 7717 | 26 |
| Carica papaya | 7680 | 307(614) | 224(1653) | 9947 | 88 |
| Arabidopsis thaliana | 2751 | 105(210) | 57(261) | 3222 | 21 |
| A. lyrata | 5413 | 461(922) | 366(1759) | 8094 | 83 |
| Cucumis sativus | 3458 | 125(250) | 54(223) | 3931 | 13 |
| Prunus persica | 3347 | 242(484) | 195(2483) | 6314 | 838 |
| Medicago truncatula | 12763 | 962(1924) | 820(6524) | 21211 | 145 |
| Glycine max | 9961 | 476(952) | 118(523) | 11436 | 23 |
| Phaseolus vulgaris | 2013 | 85(170) | 58(318) | 2501 | 19 |
| Phylostratum internode | Genes (%) | Singletons | Two-genes | Multigenes |
|---|---|---|---|---|
| Angiosperm (PS1) | 30932(58.7%) | 1982 | 5150(10300) | 3400(18650) |
| Mesangiosperm (PS2) | 4057(7.7%) | 508 | 708(1416) | 359(2133) |
| Eudicot (PS3) | 2356(4.5%) | 303 | 521(1042) | 206(1011) |
| Rosid (PS4) | 582(1.1%) | 109 | 181(362) | 31(111) |
| Legume (PS5) | 1780(3.4%) | 460 | 452(904) | 87(416) |
| Phaseoleae (PS6) | 1590(3.0%) | 568 | 400(800) | 49(222) |
| Soybean (PS7) | 11436(21.7%) | 9961 | 476(952) | 118(523) |
表3 定位到每个系统发育层级的大豆基因家族(和基因)数目
Table 3 Number of soybean gene families (and genes) assigned to each phylostratum
| Phylostratum internode | Genes (%) | Singletons | Two-genes | Multigenes |
|---|---|---|---|---|
| Angiosperm (PS1) | 30932(58.7%) | 1982 | 5150(10300) | 3400(18650) |
| Mesangiosperm (PS2) | 4057(7.7%) | 508 | 708(1416) | 359(2133) |
| Eudicot (PS3) | 2356(4.5%) | 303 | 521(1042) | 206(1011) |
| Rosid (PS4) | 582(1.1%) | 109 | 181(362) | 31(111) |
| Legume (PS5) | 1780(3.4%) | 460 | 452(904) | 87(416) |
| Phaseoleae (PS6) | 1590(3.0%) | 568 | 400(800) | 49(222) |
| Soybean (PS7) | 11436(21.7%) | 9961 | 476(952) | 118(523) |
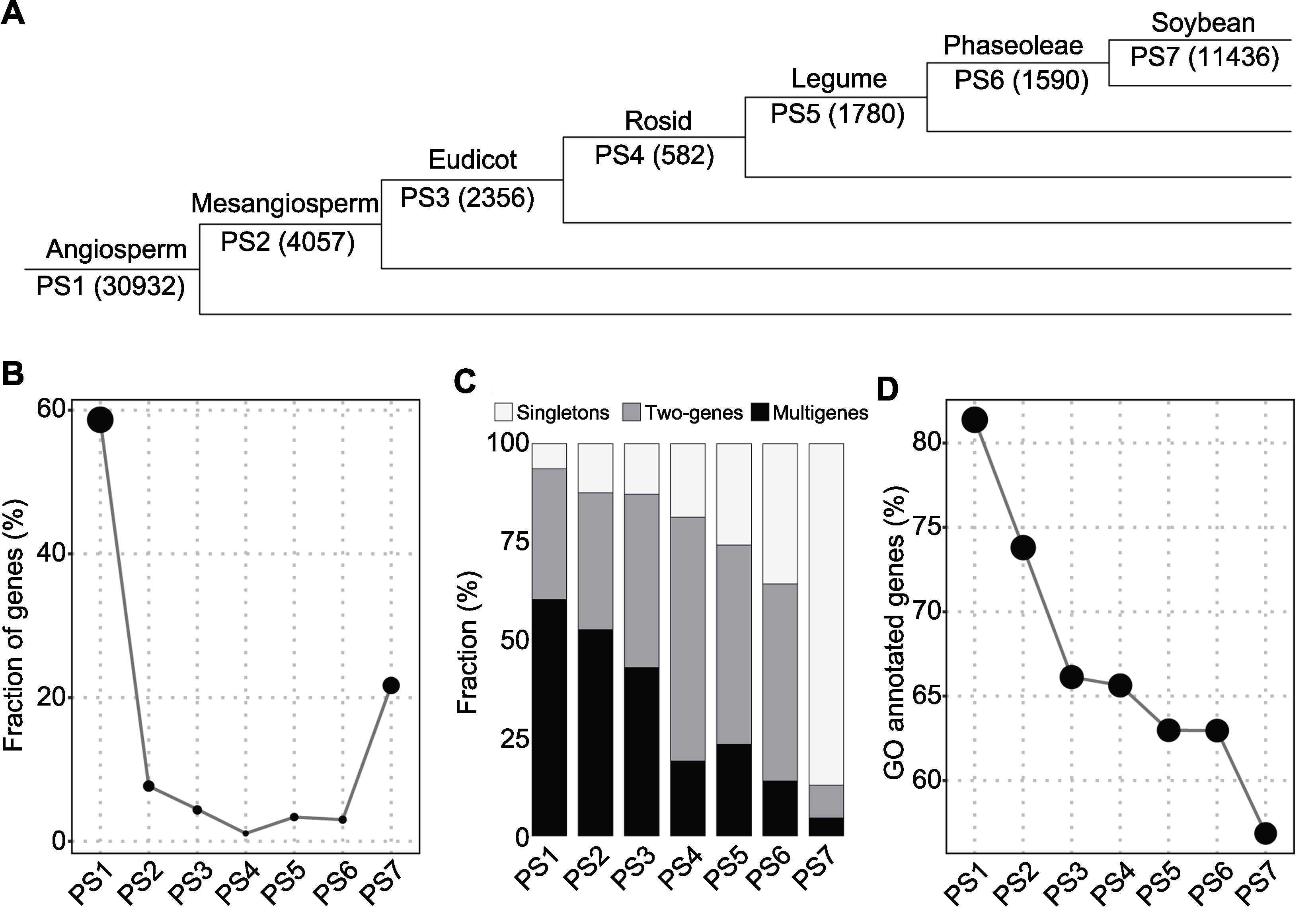
图2 大豆基因起源(A) 不同起源节点(PS1-PS7)基因数目; (B) 基因比例; (C) 基因拷贝数状态; (D) 基因GO注释
Figure 2 Origination of soybean genes(A) Numbers in parenthesis denote the number of genes per phylostratum (PS1-PS7); (B) Gene fraction; (C) Gene copy status; (D) Gene Ontology annotation
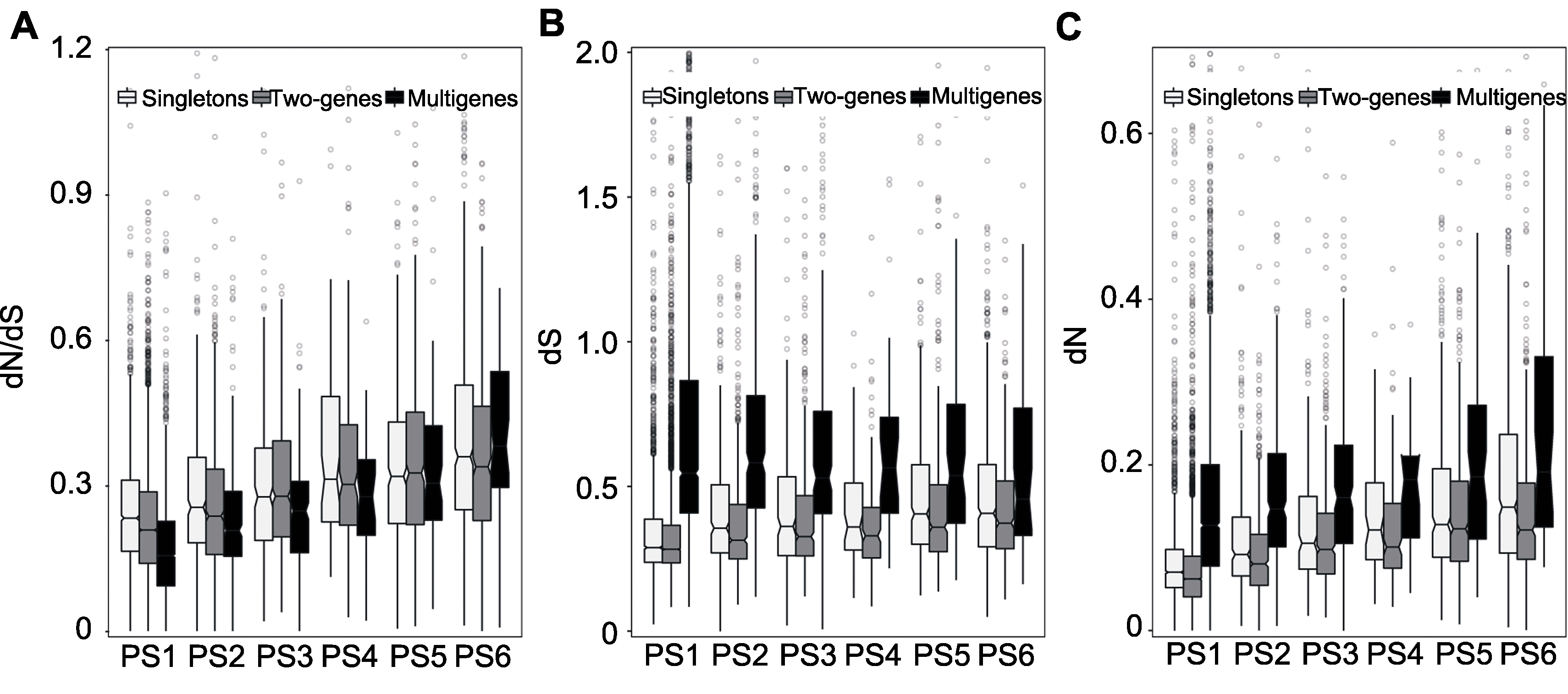
图3 大豆基因分歧程度通过大豆与菜豆同源基因对来评估选择压(dN/dS)(A)、同义替换率(dS) (B)和非同义替换率(dN) (C)。
Figure 3 Divergence degrees of soybean genes Estimated between soybean and common bean selection pressure (dN/dS)(A), synonymous substitution rate (dS) (B) and nonsynonymous substitution rate (dN) (C).
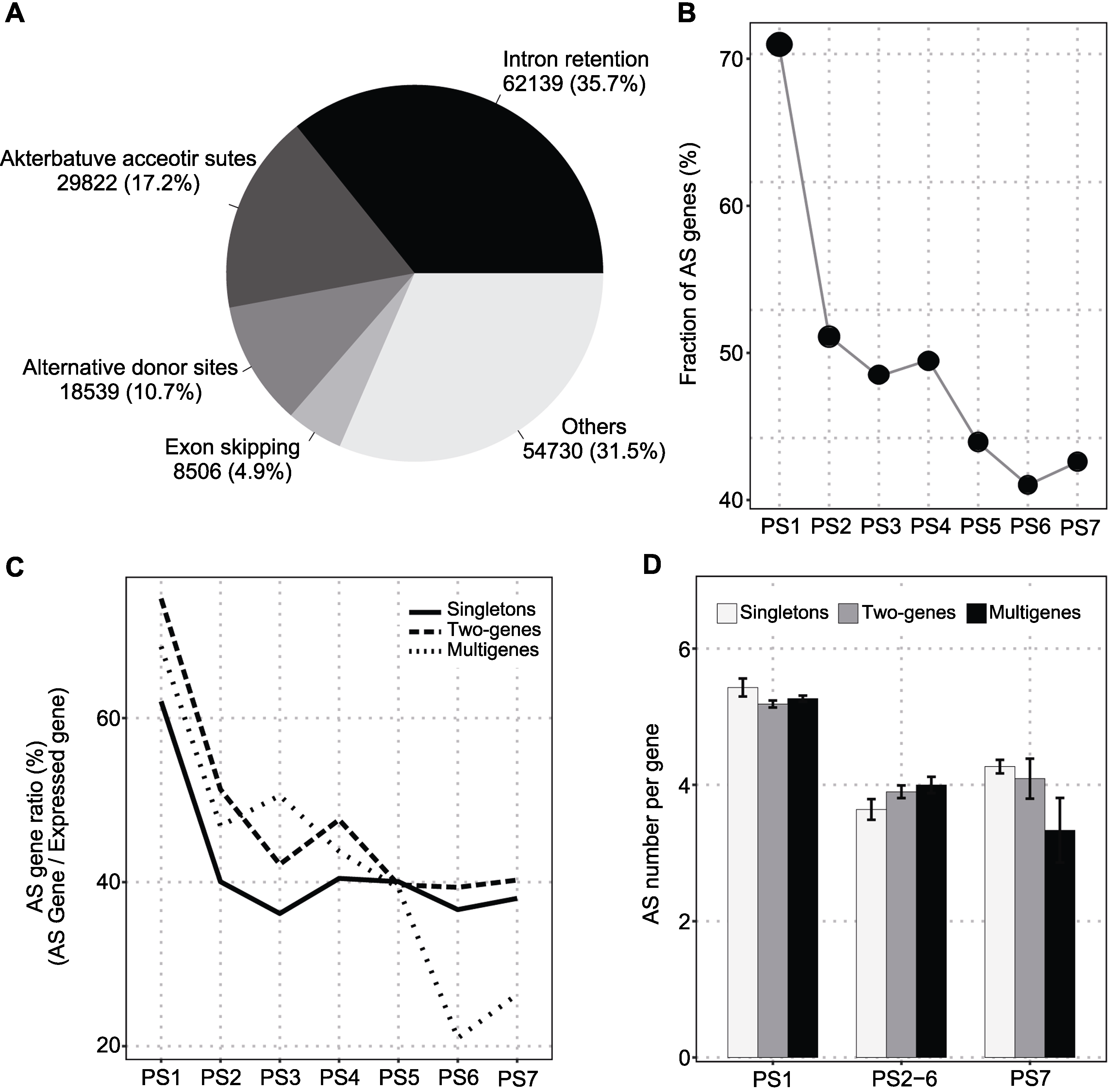
图5 大豆基因的选择性剪切(AS)(A) 选择性剪切事件; (B) 发生选择性剪切的基因比例; (C) 不同拷贝数状态下发生选择性剪切的基因; (D) 每个基因发生选择性剪切事件的数目
Figure 5 Alternative splicing (AS) of soybean genes(A) AS event; (B) AS genes ratio; (C) AS genes for different copy status; (D) AS number per gene
| [1] |
孙红正, 葛颂 ( 2010). 重复基因的进化——回顾与进展. 植物学报 45, 13-22.
DOI URL |
| [2] | Albalat R, Cañestro C ( 2016). Evolution by gene loss. Nat Rev Genet 17, 379-391. |
| [3] |
Amborella Genome Project ( 2013). The Amborella genome and the evolution of flowering plants. Science 342, 124-1089.
DOI URL PMID |
| [4] |
Bolger AM, Lohse M, Usadel B ( 2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114-2120.
DOI URL PMID |
| [5] |
Cai JJ, Borenstein E, Chen R, Petrov DA ( 2009). Similarly strong purifying selection acts on human disease genes of all evolutionary ages. Genome Biol Evol 1, 131-144.
DOI URL PMID |
| [6] |
Chen SD, Krinsky BH, Long MY ( 2013). New genes as drivers of phenotypic evolution. Nat Rev Genet 14, 645-660.
DOI URL PMID |
| [7] |
Chen TW, Wu TH, Ng WV, Lin WC ( 2011). Interrogation of alternative splicing events in duplicated genes during evolution. BMC Genomics 12(Suppl3), S16.
DOI URL PMID |
| [8] |
Domazet-Lošo T, Brajković J, Tautz D ( 2007). A phylostratigraphy approach to uncover the genomic history of major adaptations in metazoan lineages. Trends Genet 23, 533-539.
DOI URL PMID |
| [9] |
Doyle JJ, Luckow MA ( 2003). The rest of the iceberg. Legume diversity and evolution in a phylogenetic context. Plant Physiol 131, 900-910.
DOI URL |
| [10] |
Enright AJ, Van Dongen S, Ouzounis CA ( 2002). An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res 30, 1575-1584.
URL PMID |
| [11] |
Foissac S, Sammeth M ( 2007). ASTALAVISTA: dynamic and flexible analysis of alternative splicing events in custom gene datasets. Nucleic Acids Res 35, W297-W299.
DOI URL PMID |
| [12] |
Freeling M ( 2009). Bias in plant gene content following different sorts of duplication: tandem, whole-genome, segmental, or by transposition. Annu Rev Plant Biol 60, 433-453.
DOI URL |
| [13] |
Guo YL ( 2013). Gene family evolution in green plants with emphasis on the origination and evolution of Arabidopsis thaliana genes. Plant J 73, 941-951.
DOI URL PMID |
| [14] |
Jiao YN, Paterson AH ( 2014). Polyploidy-associated genome modifications during land plant evolution. Philos Trans R Soc Lond B Biol Sci 369, 20130355.
DOI URL PMID |
| [15] |
Kaessmann H ( 2010). Origins, evolution, and phenotypic impact of new genes. Genome Res 20, 1313-1326.
DOI URL |
| [16] |
Keren H, Lev-Maor G, Ast G ( 2010). Alternative splicing and evolution: diversification, exon definition and function. Nat Rev Genet 11, 345-355.
DOI URL PMID |
| [17] |
Kim D, Langmead B, Salzberg SL ( 2015). HISAT: a fast spliced aligner with low memory requirements. Nat Me- thods 12, 357-360.
DOI URL PMID |
| [18] |
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG ( 2007). Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947-2948.
DOI URL |
| [19] |
Li L, Stoeckert CJ Jr, Roos DS ( 2003). OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res 13, 2178-2189.
DOI URL |
| [20] | Long M, Betrán E, Thornton K, Wang W ( 2003). The origin of new genes: glimpses from the young and old. Nat Rev Genet 4, 865-875. |
| [21] |
Lynch M, Conery JS ( 2000). The evolutionary fate and consequences of duplicate genes. Science 290, 1151-1155.
DOI URL PMID |
| [22] |
Merkin J, Russell C, Chen P, Burge CB ( 2012). Evolutionary dynamics of gene and isoform regulation in mam- malian tissues. Science 338, 1593-1599.
DOI URL PMID |
| [23] |
Michael TP, Jackson S ( 2013). The first 50 plant genomes. Plant Gen 6, 2.
DOI URL |
| [24] |
Michael TP, VanBuren R ( 2015). Progress, challenges and the future of crop genomes. Curr Opin Plant Biol 24, 71-81.
DOI URL PMID |
| [25] | Ohno S ( 1970). Evolution by Gene Duplication. Berlin, Heidelberg: Springer. pp. 1-160. |
| [26] |
Panchy N, Lehti-Shiu M, Shiu SH ( 2016). Evolution of gene duplication in plants. Plant Physiol 171, 2294-2316.
DOI URL PMID |
| [27] |
Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL ( 2015). StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol 33, 290-295.
DOI URL PMID |
| [28] |
Quint M, Drost HG, Gabel A, Ullrich KK, Bönn M, Grosse I ( 2012). A transcriptomic hourglass in plant embryogenesis. Nature 490, 98-101.
DOI URL PMID |
| [29] |
Reddy ASN, Marquez Y, Kalyna M, Barta A ( 2013). Complexity of the alternative splicing landscape in plants. Plant Cell 25, 3657-3683.
DOI URL |
| [30] |
Schmutz J, Cannon SB, Schlueter J, Ma JX, Mitros T, Nelson W, Hyten DL, Song QJ, Thelen JJ, Cheng JL, Xu D, Hellsten U, May GD, Yu Y, Sakurai T, Umezawa T, Bhattacharyya MK, Sandhu D, Valliyodan B, Lindquist E, Peto M, Grant D, Shu SQ, Goodstein D, Barry K, Futrell-Griggs M, Abernathy B, Du JC, Tian ZX, Zhu LC, Gill N, Joshi T, Libault M, Sethuraman A, Zhang XC, Shinozaki K, Nguyen HT, Wing RA, Cregan P, Specht J, Grimwood J, Rokhsar D, Stacey G, Shoemaker RC, Jackson SA ( 2010). Genome sequence of the palaeopolyploid soybean. Nature 463, 178-183.
DOI URL |
| [31] |
Shen YT, Zhou ZK, Wang Z, Li WY, Fang C, Wu M, Ma YM, Liu TF, Kong LA, Peng DL, Tian ZX ( 2014). Global dissection of alternative splicing in paleopolyploid soybean. Plant Cell 26, 996-1008.
DOI URL PMID |
| [32] |
Suyama M, Torrents D, Bork P ( 2006). PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res 34, W609-W612.
DOI URL PMID |
| [33] |
Tasdighian S, Van Bel M, Li Z, Van de Peer Y, Carretero-Paulet L, Maere S ( 2017). Reciprocally retained genes in the angiosperm lineage show the hallmarks of dosage balance sensitivity. Plant Cell 29, 2766-2785.
DOI URL PMID |
| [34] |
Tautz D, Domazet-Lošo T ( 2011). The evolutionary origin of orphan genes. Nat Rev Genet 12, 692-702.
DOI URL PMID |
| [35] |
Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L ( 2010). Transcript assembly and quantification by RNA-seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28, 511-515.
DOI URL PMID |
| [36] |
Yanai I, Benjamin H, Shmoish M, Chalifa-Caspi V, Shklar M, Ophir R, Bar-Even A, Horn-Saban S, Safran M, Domany E, Lancet D, Shmueli O ( 2005). Genome-wide midrange transcription profiles reveal expression level re- lationships in human tissue specification. Bioinformatics 21, 650-659.
DOI URL PMID |
| [37] |
Yang ZH ( 2007). PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol 24, 1586-1591.
DOI URL PMID |
| [38] |
Zhang JZ ( 2003). Evolution by gene duplication: an update. Trends Ecol Evol 18, 292-298.
DOI URL |
| [1] | 郑立媛, 徐茜竹, 尹嘉淇, 孙小雯, 王艳. 沈阳城郊近河农田退耕地野大豆群落生态位和种间联结研究[J]. 植物生态学报, 2025, 49(濒危植物的保护与恢复): 1-. |
| [2] | 杨莉, 曲茜彤, 陈子航, 邹婷婷, 王全华, 王小丽. 菠菜AT-hook基因家族鉴定与表达谱分析[J]. 植物学报, 2025, 60(3): 377-392. |
| [3] | 曹婕, 卢秋连, 翟健平, 刘宝辉, 方超, 李世晨, 苏彤. 大豆TPS基因家族在盐胁迫下的表达变化及单倍型选择规律分析(长英文摘要)[J]. 植物学报, 2025, 60(2): 172-185. |
| [4] | 何花, 谭敦炎, 杨晓琛. 被子植物隐性雌雄异株性系统的多样性、系统演化及进化意义[J]. 生物多样性, 2024, 32(6): 24149-. |
| [5] | 顾家琦, 朱福慧, 谢沛豪, 孟庆营, 郑颖, 张献龙, 袁道军. 棉属光敏色素PHY基因家族的全基因组鉴定与驯化选择分析[J]. 植物学报, 2024, 59(1): 34-53. |
| [6] | 张飞飞, 杨天凤, 陈莉荣, 刘冬梅, 杨柳园, 杨杜宇, 鞠鹏, 陆露. 被子植物花粉颜色多样性及应用研究进展[J]. 生物多样性, 2024, 32(1): 23346-. |
| [7] | 陈佳欣, 梅浩, 黄彩翔, 梁宗原, 全依桐, 李东鹏, 布威麦尔耶姆·赛麦提, 李欣欣, 廖红. 利用转基因毛状根高效培育大豆嵌合植株的方法[J]. 植物学报, 2024, 59(1): 89-98. |
| [8] | 孙福辉, 方慧仪, 温小蕙, 张亮生. 马银花MADS-box基因家族系统进化与表达分析[J]. 植物学报, 2023, 58(3): 404-416. |
| [9] | 王菲菲, 周振祥, 洪益, 谷洋洋, 吕超, 郭宝健, 朱娟, 许如根. 大麦NF-YC基因鉴定及在盐胁迫下的表达分析[J]. 植物学报, 2023, 58(1): 140-149. |
| [10] | 张慧, 梁红凯, 智慧, 张林林, 刁现民, 贾冠清. 谷子β-胡萝卜素异构酶家族基因的表达与变异分析[J]. 植物学报, 2023, 58(1): 34-50. |
| [11] | 王芸芸, 郝占庆. 被子植物性系统的多样性、生态功能及分布规律[J]. 生物多样性, 2022, 30(7): 22065-. |
| [12] | 钱宏, 张健, 赵静超. 世界上已知维管植物有多少种? 基于多个全球植物数据库的整合[J]. 生物多样性, 2022, 30(7): 22254-. |
| [13] | 寄玲, 谢宜飞, 李中阳, 许廷晨, 杨波, 李波. 江西省野生维管植物名录[J]. 生物多样性, 2022, 30(6): 22057-. |
| [14] | 张淑梅, 李微, 李丁男. 辽宁省高等植物多样性编目[J]. 生物多样性, 2022, 30(6): 22038-. |
| [15] | 王韫慧, 王一帆, 蔺佳雨, 李金红, 姚士恩, 冯湘池, 曹振林, 王俊, 李美娜. 植物驱动蛋白: 从微管阵列到生理活动调控[J]. 植物学报, 2022, 57(3): 358-374. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
