

植物学报 ›› 2023, Vol. 58 ›› Issue (3): 404-416.DOI: 10.11983/CBB22105 cstr: 32102.14.CBB22105
收稿日期:2022-05-21
接受日期:2022-08-24
出版日期:2023-05-01
发布日期:2023-05-17
通讯作者:
*E-mail: zls83@zju.edu.cn
基金资助:
Fuhui Sun, Huiyi Fang, Xiaohui Wen, Liangsheng Zhang( )
)
Received:2022-05-21
Accepted:2022-08-24
Online:2023-05-01
Published:2023-05-17
Contact:
*E-mail: zls83@zju.edu.cn
摘要: 杜鹃花属具有极高的物种多样性和观赏价值, 其中马银花(Rhododendron ovatum)花型独特, 具有低海拔适应性, 在我国南方地区有良好的应用前景。基于杜鹃花目代表性植物的基因组信息, 探究MADS-box基因家族在杜鹃花目中的进化特点并挖掘马银花的花器官发育关键基因。系统进化分析表明, 杜鹃花科3个代表性物种(马银花、映山红(R. simsii)和马缨杜鹃(R. delavayi))中的MADS-box基因家族成员数目较为一致, 包括AP1、AP3、PI、AG和SEP等19个亚家族, 其中SVP、ANR1、Mα、Mβ和Mγ亚家族成员数量较多。马银花的AG和SEP基因有多个拷贝, 而AP3/PI基因的拷贝数目较少, 显示出更加保守的进化模式。基因表达分析表明, 马银花MADS-box基因的表达模式存在组织特异性。AP1、AP3/PI、AG、SEP和MIKC*分支基因在花器官中特异性表达。综上, 该文探讨了马银花MADS-box基因家族进化历史, 以及部分MADS-box基因可能具有的功能, 可为深入研究马银花的发育和MADS-box基因功能提供参考。
孙福辉, 方慧仪, 温小蕙, 张亮生. 马银花MADS-box基因家族系统进化与表达分析. 植物学报, 2023, 58(3): 404-416.
Fuhui Sun, Huiyi Fang, Xiaohui Wen, Liangsheng Zhang. Phylogenetic and Expression Analysis of MADS-box Gene Family in Rhododendron ovatum. Chinese Bulletin of Botany, 2023, 58(3): 404-416.
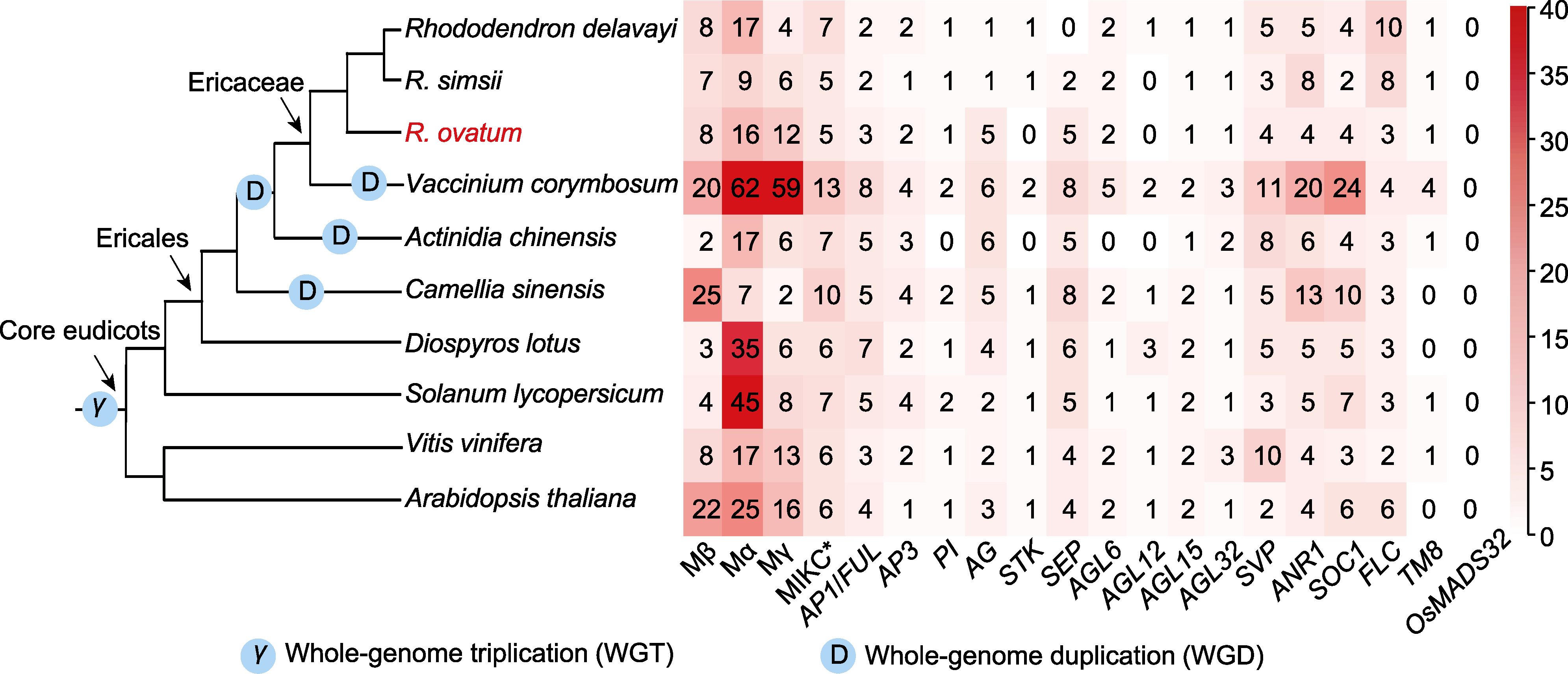
图1 马银花和其它9种被子植物的系统发育树和MADS-box基因数
Figure 1 The phylogenetic tree and MADS-box gene numbers of Rhododendron ovatum and nine other species in the context of angiosperms
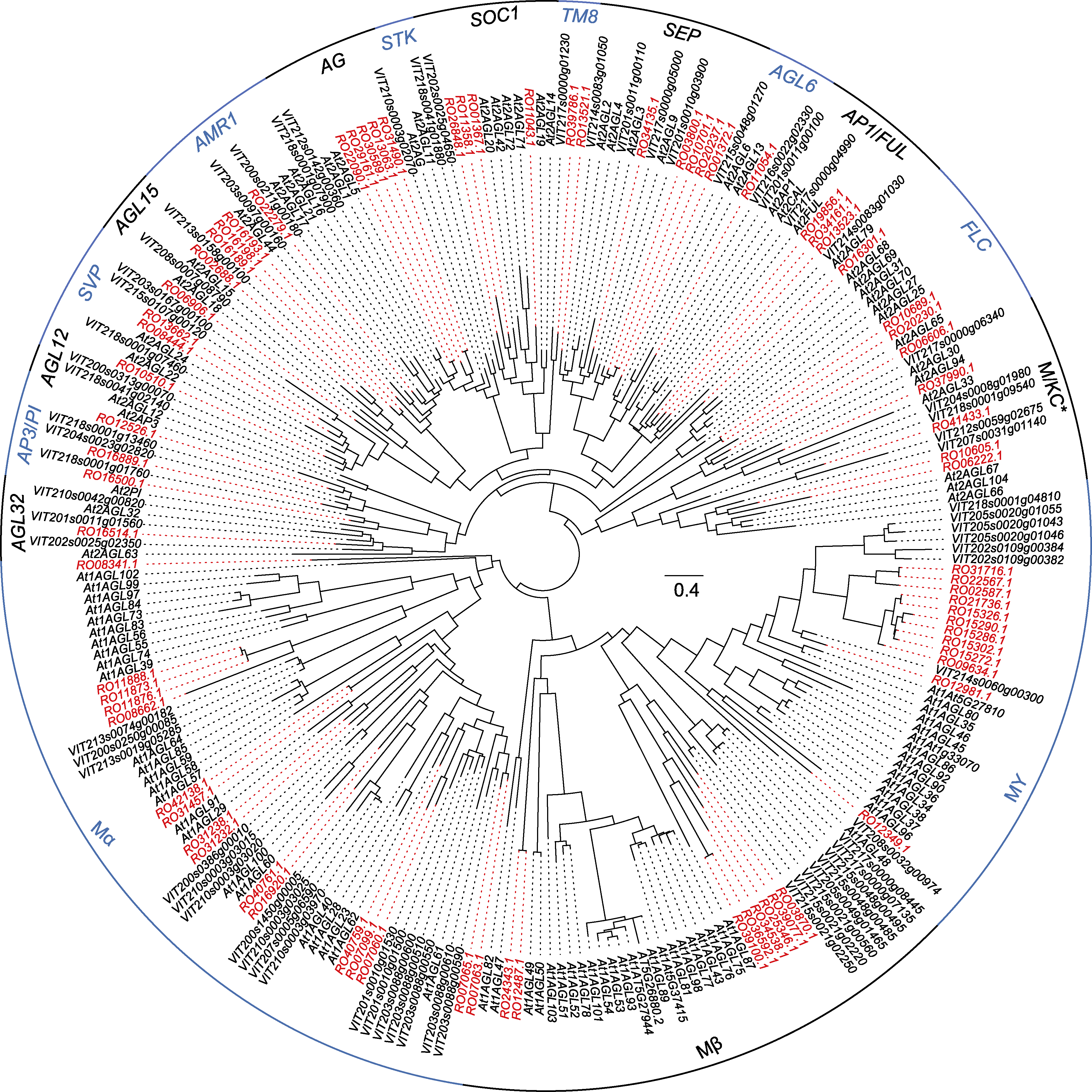
图2 马银花、拟南芥和葡萄MADS-box基因最大似然法系统发育树 红色字体表示马银花中的同源基因。
Figure 2 Maximum likelihood phylogenetic tree constructed by Rhododendron ovatum, Arabidopsis thaliana and Vitis vinifera MADS-box genes R. ovatum homologous genes are in red font.
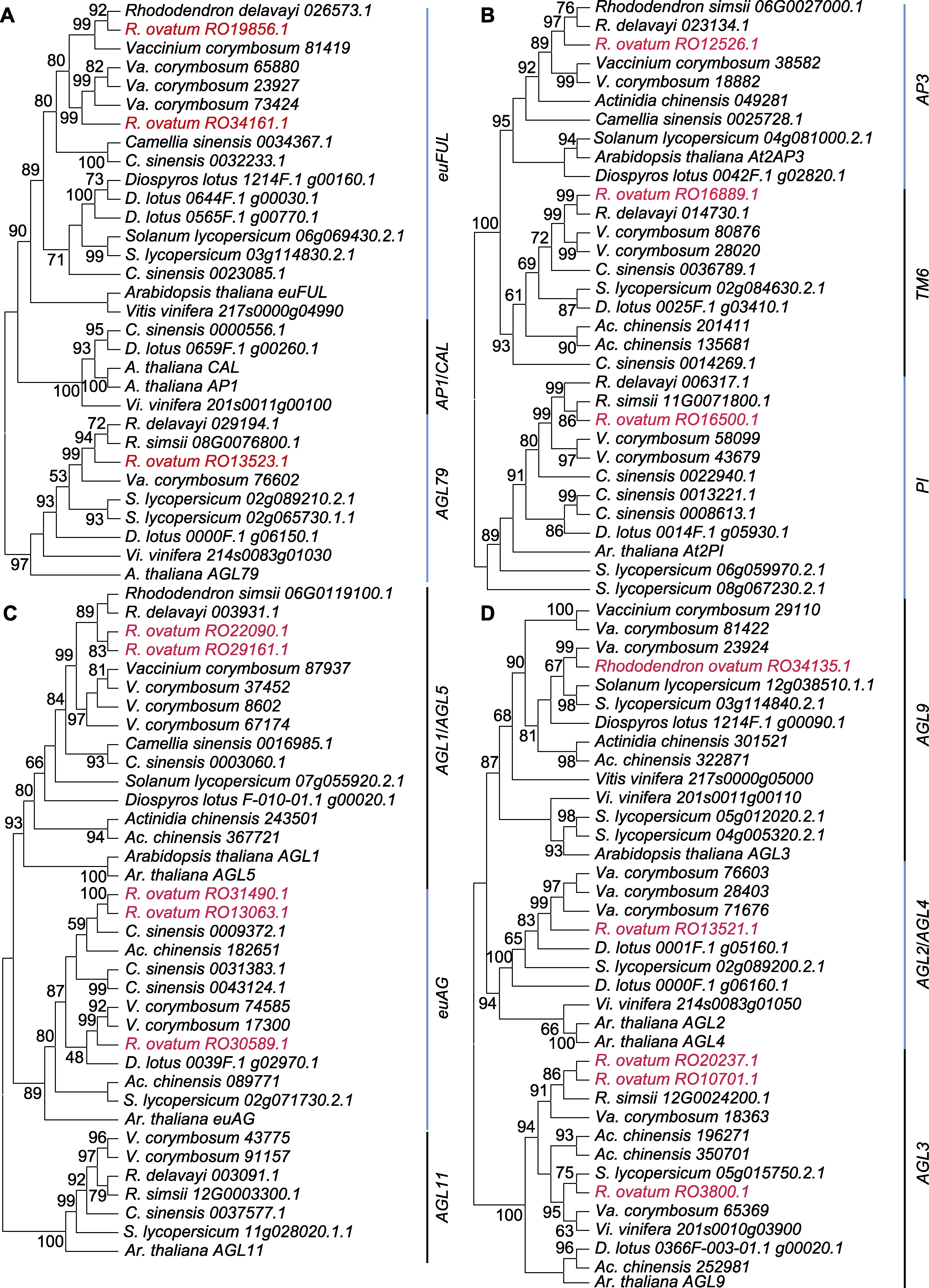
图3 花发育关键基因最大似然法系统发育树 (A) AP1/FUL系统发育树; (B) AP3/PI系统发育树; (C) AG系统发育树; (D) SEP系统发育树。红色字体表示马银花中的同源基因。
Figure 3 Maximum likelihood phylogenetic trees of floral identity genes (A) Phylogenetic tree of AP1/FUL genes; (B) Phylogenetic tree of AP3/PI genes; (C) Phylogenetic tree of AG genes; (D) Phylogenetic tree of SEP genes. Rhododendron ovatum homologous genes are in red font.
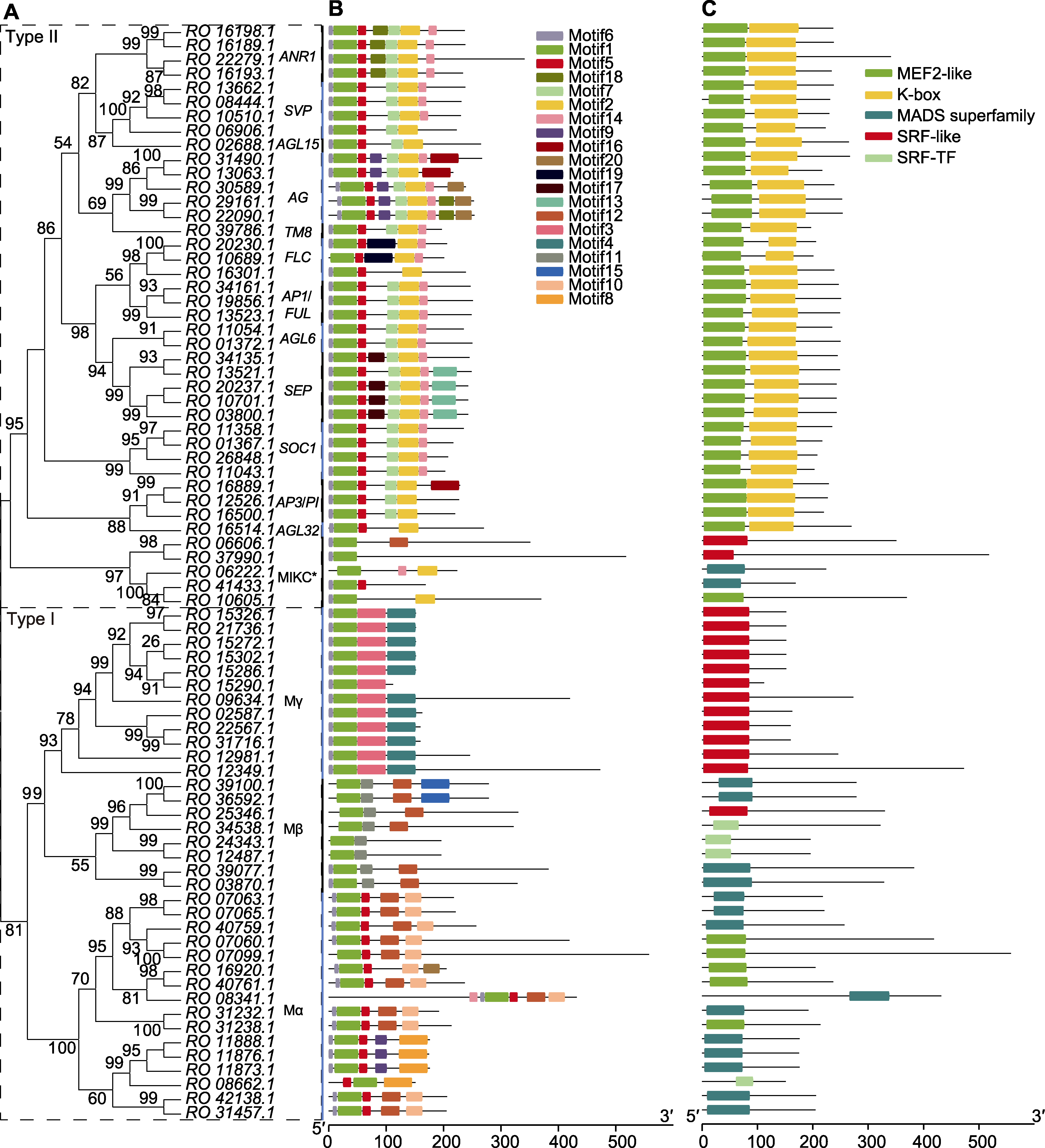
图4 马银花MADS-box蛋白序列的结构特征分析 (A) 马银花MADS-box基因系统发育树; (B) 保守基序分析; (C) 保守结构域分析
Figure 4 Analysis of structure of Rhododendron ovatum MADS-box protein (A) Phylogenetic tree of R. ovatum MADS-box genes; (B) Conserved motif analysis; (C) Conserved domain analysis
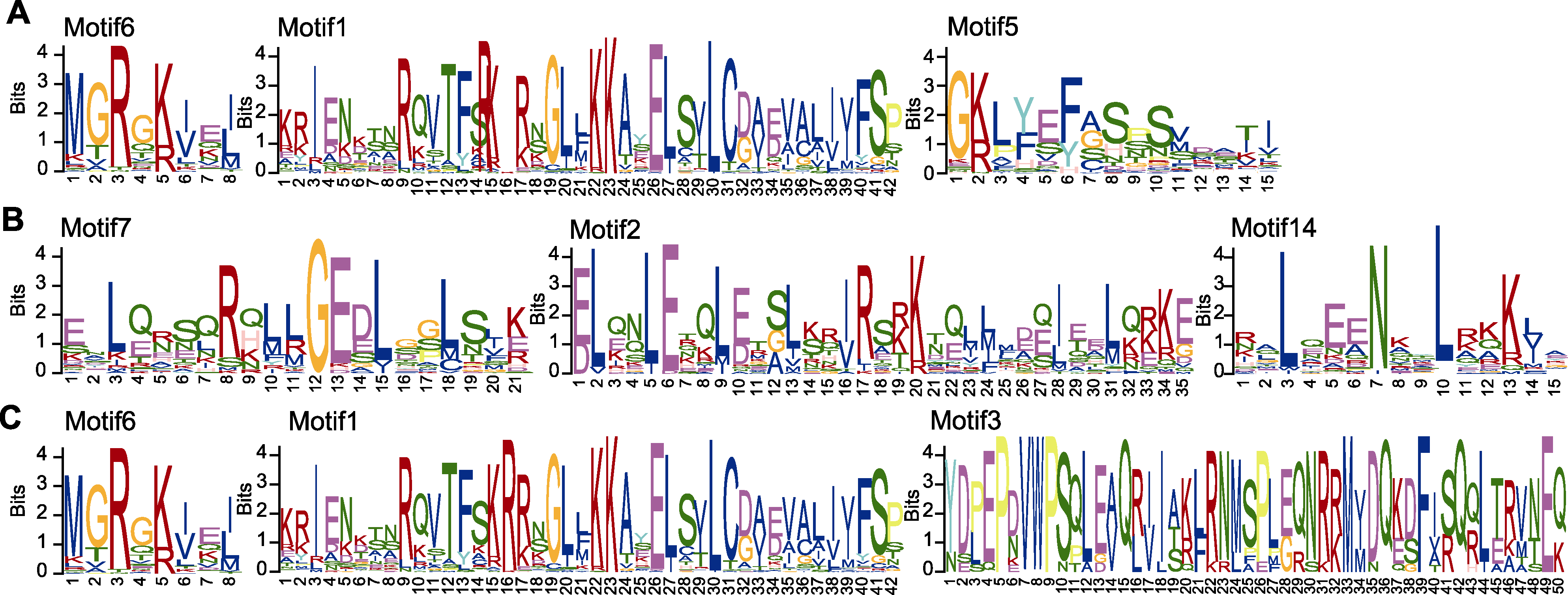
图5 马银花MADS-box蛋白的保守结构域 (A) MADS MEF2-like结构域; (B) K-box结构域; (C) MADS SRF-like结构域
Figure 5 Structures of Rhododendron ovaturn MADS-box proteins’ conserved domain (A) MADS MEF2-like domain; (B) K-box domain; (C) MADS SRF-like domain
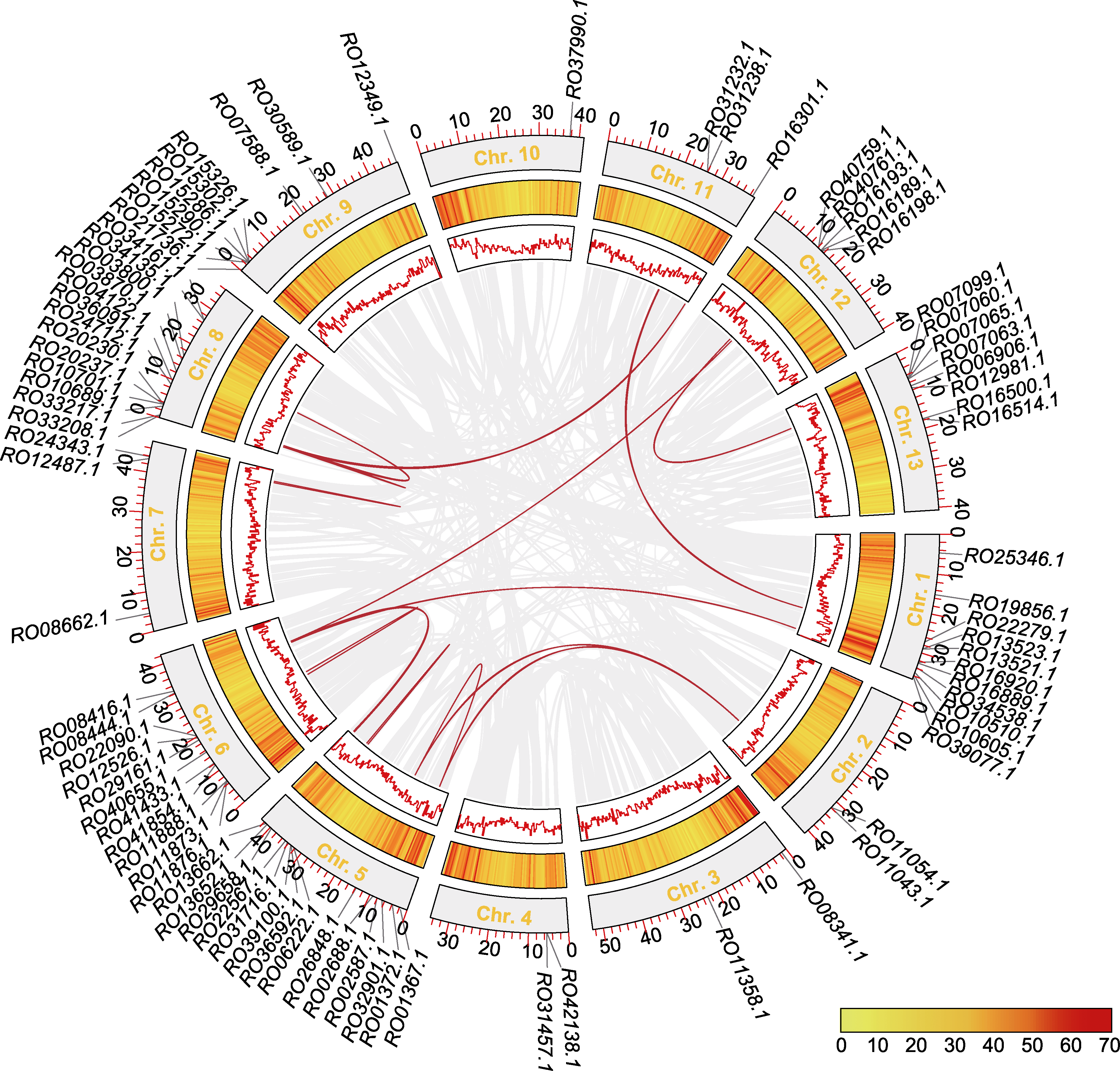
图6 马银花MADS-box基因的染色体定位和共线性分析 圆环从外到内依次代表基因在染色体上的定位、基因密度和共线性区域(红色连线为MADS-box基因)。
Figure 6 Chromosomal localization and collinearity analysis of Rhododendron ovatum MADS-box genes The tracks from outer to inner circles represent the gene location on chromosomes, gene density, and collinear regions (red lines indicate MADS-box genes).
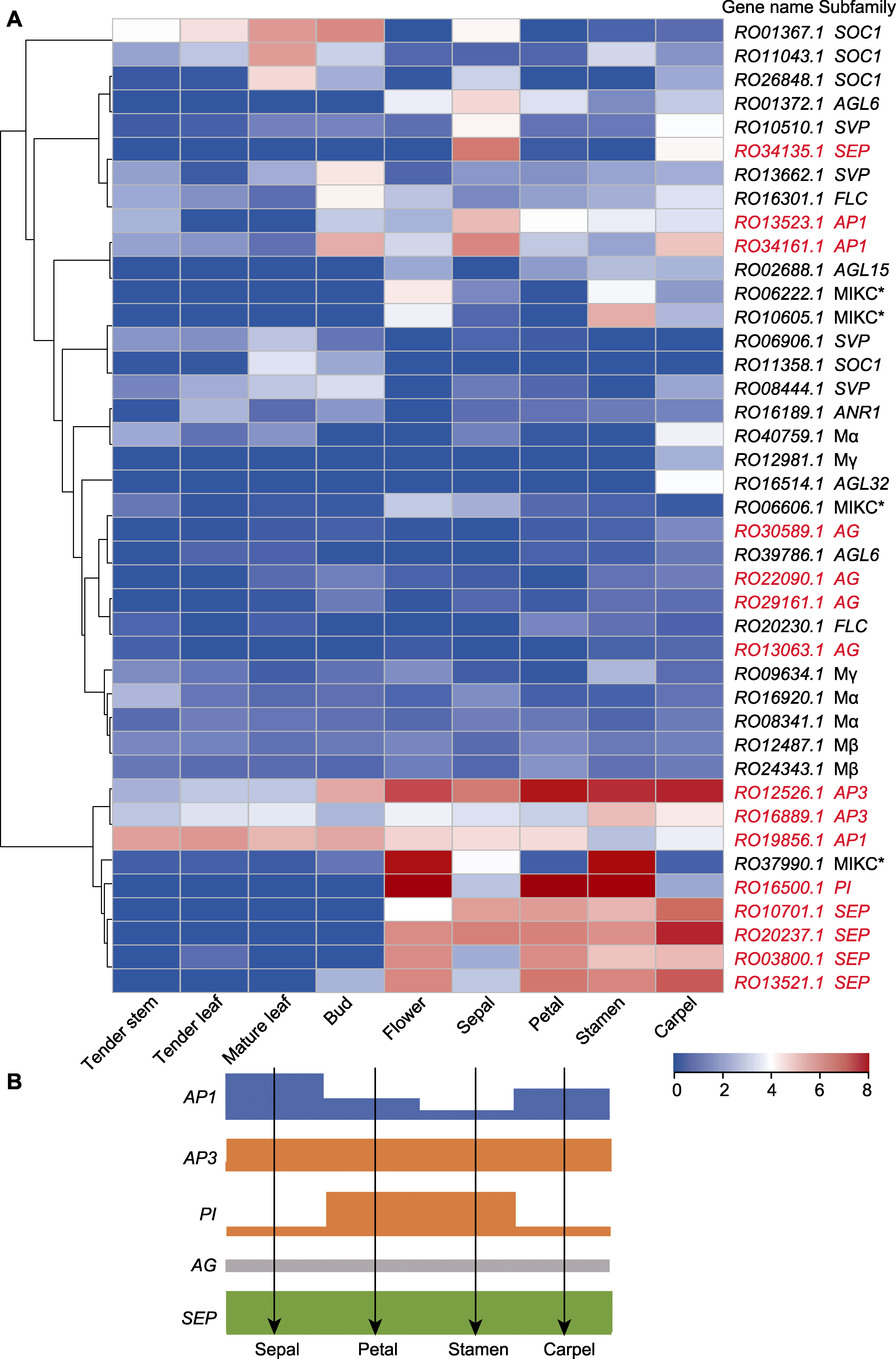
图7 马银花MADS-box基因表达分析 (A) 马银花MADS-box基因在不同组织中的表达(红色基因名为花发育相关基因); (B) 马银花ABCE花发育模型(方块高度代表基因在花器官中的表达量)
Figure 7 Expression analysis of Rhododendron ovatum MADS-box genes (A) Expression analysis of R. ovatum MADS-box genes in different tissues (the floral identity genes are in red); (B) Proposed ABCE flower development model of R. ovatum (the bar height represents the expression level)
| [1] | 丁献华, 陈伟, 张直峰 (2020). 荷花B类MADS-box基因家族NnDEF基因的克隆及表达分析. 上海农业学报 36, 1-7. |
| [2] |
高虎虎, 张云霄, 胡胜武, 郭媛 (2017). 甘蓝型油菜MADS- box基因家族的鉴定与系统进化分析. 植物学报 52, 699-712.
DOI |
| [3] |
高玮林, 张力曼, 薛超玲, 张垚, 刘孟军, 赵锦 (2022). 枣E类MADS基因在花和果中的表达及其蛋白互作研究. 园艺学报 49, 739-748.
DOI |
| [4] | 耿兴敏, 宦智群, 苏家乐, 刘晓青 (2021). 杜鹃花属植物种质创新研究进展. 分子植物育种 19, 604-613. |
| [5] | 景丹龙, 郭启高, 陈薇薇, 夏燕, 吴頔, 党江波, 何桥, 梁国鲁 (2018). 被子植物花器官发育的模型演变和分子调控. 植物生理学报 54, 355-362. |
| [6] |
景丹龙, 夏燕, 张守攻, 王军辉 (2016). 黄金树B类MADS- box基因表达特征分析. 植物学报 51, 210-217.
DOI |
| [7] |
王莹, 穆艳霞, 王锦 (2021). MADS-box基因家族调控植物花器官发育研究进展. 浙江农业学报 33, 1149-1158.
DOI |
| [8] | 徐启江, 关录飞, 吴笑女, 孙丽, 谭文勃, 聂玉哲, 李玉花 (2008). 草原龙胆MADS-box基因的克隆及表达分析. 植物学通报 25, 415-429. |
| [9] | 杨华, 宋绪忠 (2016). 高温胁迫对马银花的生理指标影响. 林业科技通讯 01, 3-6. |
| [10] | 杨丽, 严露露, 李慧娥, 杜致辉, 郑淇元 (2021). 映山红杜鹃MADS基因家族的鉴定与分析. 分子植物育种 19, 6290-6301. |
| [11] |
Airoldi CA, Davies B (2012). Gene duplication and the evolution of plant MADS-box transcription factors. J Genet Genomics 39, 157-165.
DOI PMID |
| [12] |
Alvarez-Buylla ER, Pelaz S, Liljegren SJ, Gold SE, Burgeff C, Ditta GS, De Pouplana LR, Martínez-Castilla L, Yanofsky MF (2000). An ancestral MADS-box gene duplication occurred before the divergence of plants and animals. Proc Natl Acad Sci USA 97, 5328-5333.
PMID |
| [13] |
Chen CJ, Chen H, Zhang Y, Thomas HR, Frank MH, He YH, Xia R (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol Plant 13, 1194-1202.
DOI PMID |
| [14] |
Chen F, Zhang XT, Liu X, Zhang LS (2017). Evolutionary analysis of MIKCC-type MADS-box genes in gymnosperms and angiosperms. Front Plant Sci 8, 895.
DOI PMID |
| [15] |
Duarte JM, Cui LY, Wall PK, Zhang Q, Zhang XH, Leebens-Mack J, Ma H, Altman N, dePamphilis CW (2006). Expression pattern shifts following duplication indicative of subfunctionalization and neofunctionalization in regulatory genes of Arabidopsis. Mol Biol Evol 23, 469-478.
PMID |
| [16] |
Edgar RC (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32, 1792-1797.
DOI PMID |
| [17] |
Lee JH, Ryu HS, Chung KS, Posé D, Kim S, Schmid M, Ahn JH (2013). Regulation of temperature-responsive flowering by MADS-box transcription factor repressors. Science 342, 628-632.
DOI PMID |
| [18] |
Litt A, Kramer EM (2010). The ABC model and the diversification of floral organ identity. Semin Cell Dev Biol 21, 129-137.
DOI PMID |
| [19] |
Liu Y, Cui SJ, Wu F, Yan S, Lin XL, Du XQ, Chong K, Schilling S, Theißen G, Meng Z (2013). Functional conservation of MIKC*-type MADS box genes in Arabidopsis and rice pollen maturation. Plant Cell 25, 1288-1303.
DOI URL |
| [20] |
Pařenicová L, De Folter S, Kieffer M, Horner DS, Favalli C, Busscher J, Cook HE, Ingram RM, Kater MM, Davies B, Angenent GC, Colombo L (2003). Molecular and phylogenetic analyses of the complete MADS-box transcription factor family in Arabidopsis: new openings to the MADS world. Plant Cell 15, 1538-1551.
DOI URL |
| [21] |
Price MN, Dehal PS, Arkin AP (2009). FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol 26, 1641-1650.
DOI PMID |
| [22] | Qiu YC, Köhler C (2022). Endosperm evolution by duplicated and neofunctionalized Type I MADS-box transcription factors. Mol Biol Evol 39, msab355. |
| [23] |
Ruelens P, Zhang ZC, Van Mourik H, Maere S, Kaufmann K, Geuten K (2017). The origin of floral organ identity quartets. Plant Cell 29, 229-242.
DOI URL |
| [24] |
Shan HY, Cheng J, Zhang R, Yao X, Kong HZ (2019). Developmental mechanisms involved in the diversification of flowers. Nat Plants 5, 917-923.
DOI PMID |
| [25] |
Smaczniak C, Immink RGH, Angenent GC, Kaufmann K (2012). Developmental and evolutionary diversity of plant MADS-domain factors: insights from recent studies. Development 139, 3081-3098.
DOI PMID |
| [26] |
Theißen G, Kim JT, Saedler H (1996). Classification and phylogeny of the MADS-box multigene family suggest defined roles of MADS-box gene subfamilies in the morphological evolution of eukaryotes. J Mol Evol 43, 484-516.
PMID |
| [27] |
Theißen G, Melzer R, Rümpler F (2016). MADS-domain transcription factors and the floral quartet model of flower development: linking plant development and evolution. Development 143, 3259-3271.
DOI PMID |
| [28] |
Vekemans D, Proost S, Vanneste K, Coenen H, Viaene T, Ruelens P, Maere S, Van De Peer Y, Geuten K (2012). Gamma paleohexaploidy in the stem lineage of core eudicots: significance for MADS-box gene and species diversification. Mol Biol Evol 29, 3793-3806.
DOI PMID |
| [29] |
Wang XY, Gao Y, Wu XP, Wen XH, Li DQ, Zhou H, Li Z, Liu B, Wei JF, Chen F, Chen F, Zhang CJ, Zhang LS, Xia YP (2021). High-quality evergreen azalea genome reveals tandem duplication-facilitated low-altitude adaptability and floral scent evolution. Plant Biotechnol J 19, 2544-2560.
DOI PMID |
| [30] | Wen XH, Li JZ, Wang LL, Lu CF, Gao Q, Xu P, Pu Y, Zhang QL, Hong Y, Hong L, Huang H, Xin HG, Wu XY, Kang DR, Gao K, Li YJ, Ma CF, Li XM, Zheng HK, Wang ZC, Jiao YN, Zhang LS, Dai SL (2022). The Chrysanthemum lavandulifolium genome and the molecular mechanism underlying diverse capitulum types. Hortic Res 9, uhab022. |
| [31] |
Wu WW, Huang XT, Cheng J, Li ZG, De Folter S, Huang ZR, Jiang XQ, Pang HX, Tao SH (2011). Conservation and evolution in and among SRF- and MEF2-type MADS domains and their binding sites. Mol Biol Evol 28, 501-511.
DOI PMID |
| [32] |
Yang FS, Nie S, Liu H, Shi TL, Tian XC, Zhou SS, Bao YT, Jia KH, Guo JF, Zhao W, An N, Zhang RG, Yun QZ, Wang XZ, Mannapperuma C, Porth I, El-Kassaby YA, Street NR, Wang XR, Van De Peer Y, Mao JF (2020). Chromosome-level genome assembly of a parent species of widely cultivated azaleas. Nat Commun 11, 5269.
DOI |
| [33] |
Zhang LS, Chen F, Zhang XT, Li Z, Zhao YY, Lohaus R, Chang XJ, Dong W, Ho SYW, Liu X, Song AX, Chen JH, Guo WL, Wang ZJ, Zhuang YY, Wang HF, Chen XQ, Hu J, Liu YH, Qin Y, Wang K, Dong SS, Liu Y, Zhang SZ, Yu XX, Wu Q, Wang LS, Yan XQ, Jiao YN, Kong HZ, Zhou XF, Yu CW, Chen YC, Li F, Wang JH, Chen W, Chen XL, Jia QD, Zhang C, Jiang YF, Zhang WB, Liu GH, Fu JY, Chen F, Ma H, Van De Peer Y, Tang HB (2020). The water lily genome and the early evolution of flowering plants. Nature 577, 79-84.
DOI |
| [1] | 曹婕, 卢秋连, 翟健平, 刘宝辉, 方超, 李世晨, 苏彤. 大豆TPS基因家族在盐胁迫下的表达变化及单倍型选择规律分析(长英文摘要)[J]. 植物学报, 2025, 60(2): 172-185. |
| [2] | 李巧峡, 李有龙, 李纪纲, 陈晨龙, 孙坤. 光周期调控维西堇菜与裂叶堇菜开放花和闭锁花的发育[J]. 生物多样性, 2024, 32(6): 23484-. |
| [3] | 段政勇, 丁敏, 王宇卓, 丁艺冰, 陈凌, 王瑞云, 乔治军. 糜子SBP基因家族全基因组鉴定及表达分析[J]. 植物学报, 2024, 59(2): 231-244. |
| [4] | 范业赓,丘立杭,黄杏,周慧文,甘崇琨,李杨瑞,杨荣仲,吴建明,陈荣发. 甘蔗节间伸长过程赤霉素生物合成关键基因的表达及相关植物激素动态变化[J]. 植物学报, 2019, 54(4): 486-496. |
| [5] | 马骊, 孙万仓, 袁金海, 刘自刚, 武军艳, 方彦, 许耀照, 蒲媛媛, 白静, 董小云, 何辉立. 白菜型冬油菜β-1,3-葡聚糖酶基因在低温胁迫下的表达[J]. 植物学报, 2017, 52(5): 568-578. |
| [6] | 刘宝玲, 张莉, 孙岩, 薛金爱, 高昌勇, 苑丽霞, 王计平, 贾小云, 李润植. 谷子bZIP转录因子的全基因组鉴定及其在干旱和盐胁迫下的表达分析[J]. 植物学报, 2016, 51(4): 473-487. |
| [7] | 景丹龙, 夏燕, 张守攻, 王军辉. 黄金树B类MADS-box基因表达特征分析[J]. 植物学报, 2016, 51(2): 210-217. |
| [8] | 蔡元保, 杨祥燕, 孙光明, 黄强, 刘业强, 李绍鹏, 张治礼. 菠萝花发育相关基因AcMADS1的克隆与组织表达特性分析[J]. 植物学报, 2014, 49(6): 692-703. |
| [9] | 翟莹, 杨晓杰, 孙天国, 赵艳, 余春粉, 王秀文. 大豆转录因子GmERF5的克隆、表达及功能分析[J]. 植物学报, 2013, 48(5): 498-506. |
| [10] | 景丹龙, 马江, 张博, 韩逸洋, 刘志雄, 陈发菊. 红花玉兰MwAG基因在花发育不同时期的表达[J]. 植物学报, 2013, 48(2): 145-150. |
| [11] | 张俊红, 张守攻, 吴涛, 韩素英, 杨文华, 齐力旺. 落叶松体胚发育中5个miRNA前体与成熟体的表达[J]. 植物学报, 2012, 47(5): 462-473. |
| [12] | 袁伟, 万红建, 杨悦俭. 植物实时荧光定量PCR内参基因的特点及选择[J]. 植物学报, 2012, 47(4): 427-436. |
| [13] | 于明明;李兴国;张宪省. APETALA1 启动子驱动AtIPT4 在转基因拟南芥中表达导致花和花器官发育异常[J]. 植物学报, 2009, 44(01): 59-68. |
| [14] | 张冰玉;苏晓华*;周祥明. 林木花发育的基因调控[J]. 植物学报, 2008, 25(04): 476-482. |
| [15] | 陆露 王红 李德铢. 杜鹃花科白珠树属分子系统学和生物地理学研究进展[J]. 植物学报, 2005, 22(06): 658-667. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||