

植物学报 ›› 2016, Vol. 51 ›› Issue (4): 473-487.DOI: 10.11983/CBB15148 cstr: 32102.14.CBB15148
刘宝玲1, 张莉1, 孙岩2, 薛金爱1, 高昌勇1, 苑丽霞1, 王计平1, 贾小云2, 李润植1,*( )
)
收稿日期:2015-08-18
接受日期:2016-03-25
出版日期:2016-07-01
发布日期:2016-08-05
通讯作者:
李润植
作者简介:# 共同第一作者
基金资助:Baoling Liu1, Li Zhang1, Yan Sun2, Jinai Xue1, Changyong Gao1, Lixia Yuan1, Jiping Wang1, Xiaoyun Jia2, Runzhi Li1*
Received:2015-08-18
Accepted:2016-03-25
Online:2016-07-01
Published:2016-08-05
Contact:
Li Runzhi
About author:# Co-first authors
摘要: bZIP蛋白是植物转录因子中最大和最保守的一类转录因子, 参与调控植物生长发育等多种生命活动。谷子(Setaria italica)是一种重要的C4杂粮作物, 其bZIP基因家族与功能报道较少。利用生物信息学工具, 从谷子全基因组中鉴定出73个SibZIP转录因子, 划分为A、B、C、D、E、G、H、I和X等亚家族。与已测序的禾谷类作物相比, 谷子SibZIP基因家族在进化中发生缩减。在谷子SibZIP蛋白中检测到25种不同的保守氨基酸基序。RNA-seq和定量PCR检测结果表明, 在干旱和盐胁迫条件下, 多数SibZIPs基因不同程度地被诱导表达, 预示着部分SibZIP成员在谷子干旱和盐胁迫响应中起重要作用。共表达关联性分析进一步揭示19个谷子SibZIP转录因子可通过与蛋白激酶或NPR1相关调节蛋白等互作介导谷子胁迫响应。研究结果为全面解析谷子SibZIPs基因结构与生物学功能、抗旱分子机制以及分子育种提供了新信息。
刘宝玲, 张莉, 孙岩, 薛金爱, 高昌勇, 苑丽霞, 王计平, 贾小云, 李润植. 谷子bZIP转录因子的全基因组鉴定及其在干旱和盐胁迫下的表达分析. 植物学报, 2016, 51(4): 473-487.
Baoling Liu, Li Zhang, Yan Sun,Jinai Xue, Changyong Gao, Lixia Yuan, Jiping Wang, Xiaoyun Jia, Runzhi Li. Genome-wide Characterization of bZIP Transcription Factors in Foxtail Millet and Their Expression Profiles in Response to Drought and Salt Stresses. Chinese Bulletin of Botany, 2016, 51(4): 473-487.
| Gene name | Forward primer (5'-3') | Reverse primer (5'-3') |
|---|---|---|
| 18S rRNA | CCACAGATTGAAAACCGCATTA | ACCTCCCACCAGCAGAACC |
| SibZIP2 | ATGGGACAGATGGCGATGG | AAGAGCACGCAGCCGAGA |
| SibZIP3 | ATGAGCTGCATTGGCAACG | CTCCTTGAGATGGTTCAGTTCG |
| SibZIP7 | GCCGTCCGACCACAACAG | CCTCTTGACACGCTTGGGA |
| SibZIP8 | CCCCAACAATCTCAGGGAAGG | GAGCGTGTTGTTCTCGTTCTGG |
| SibZIP21 | CCAGTCCTCCTCCTGCTCC | CTCCATCACCAGGTCCTTCTCC |
| SibZIP24 | ACGAGAAGGCCGAGCTGG | TGGTGCTGGTGGTGTTGTTG |
| SibZIP37 | TACGCCCGATGGTTAGAAGA | ATCGATCAGCCGTCGTTG |
| SibZIP45 | GTAGCCCGAGCCCTGACA | AGCTCGCGGTCGAAGGTC |
| SibZIP51 | AATCCCTCGGCTAAAA | ACCCACAAAGCATCACAAGG |
| SibZIP57 | GGCATCAGCACAGCCAGT | CCTGTCCTGTCGCTACCTTCA |
| SibZIP60 | CTCCAAACCCAGCATTCCA | GCTCATCCTCTGCCATCACC |
| SibZIP73 | AGCGTGGGCAGGGTAAATG | CAGGAGCACCCCAGTAATCTAC |
表1 谷子SibZIP基因实时荧光定量PCR分析所用引物
Table 1 List of primers used in quantitative real-time PCR analysis of SibZIP genes of Setaria italica
| Gene name | Forward primer (5'-3') | Reverse primer (5'-3') |
|---|---|---|
| 18S rRNA | CCACAGATTGAAAACCGCATTA | ACCTCCCACCAGCAGAACC |
| SibZIP2 | ATGGGACAGATGGCGATGG | AAGAGCACGCAGCCGAGA |
| SibZIP3 | ATGAGCTGCATTGGCAACG | CTCCTTGAGATGGTTCAGTTCG |
| SibZIP7 | GCCGTCCGACCACAACAG | CCTCTTGACACGCTTGGGA |
| SibZIP8 | CCCCAACAATCTCAGGGAAGG | GAGCGTGTTGTTCTCGTTCTGG |
| SibZIP21 | CCAGTCCTCCTCCTGCTCC | CTCCATCACCAGGTCCTTCTCC |
| SibZIP24 | ACGAGAAGGCCGAGCTGG | TGGTGCTGGTGGTGTTGTTG |
| SibZIP37 | TACGCCCGATGGTTAGAAGA | ATCGATCAGCCGTCGTTG |
| SibZIP45 | GTAGCCCGAGCCCTGACA | AGCTCGCGGTCGAAGGTC |
| SibZIP51 | AATCCCTCGGCTAAAA | ACCCACAAAGCATCACAAGG |
| SibZIP57 | GGCATCAGCACAGCCAGT | CCTGTCCTGTCGCTACCTTCA |
| SibZIP60 | CTCCAAACCCAGCATTCCA | GCTCATCCTCTGCCATCACC |
| SibZIP73 | AGCGTGGGCAGGGTAAATG | CAGGAGCACCCCAGTAATCTAC |
| Gene name | Phytozome identifier | Chromosome location (bp) | Protein length (aa) | Theore- tical pI | Molecular weight (Da) | Phylogeny group | NCBI accession No. |
|---|---|---|---|---|---|---|---|
| SibZIP1 | Si000955m | Chr.5: 42034648-42041849 | 538 | 6 | 58030.2 | D | XP_004970651 |
| SibZIP2 | Si001292m | Chr.5: 39537137-39542647 | 473 | 6 | 51362.5 | D | XP_004970302 |
| SibZIP3 | Si001731m | Chr.5: 42027056-42028626 | 399 | 6 | 42138.5 | A | XP_004970655 |
| SibZIP4 | Si001929m | Chr.5: 31392517-31397217 | 368 | 9 | 39273.7 | G | XP_004969366 |
| SibZIP5 | Si002173m | Chr.5: 39683613-39687077 | 333 | 9 | 36026.4 | A | XP_004970325 |
| SibZIP6 | Si002192m | Chr.5: 3576665-3581966 | 330 | 7 | 36678.2 | D | XP_004967673 |
| SibZIP7 | Si002311m | Chr.5: 13207621-13209504 | 314 | 6 | 34130.9 | E | XP_004968609 |
| SibZIP8 | Si002989m | Chr.5: 10278963-10280748 | 196 | 10 | 20861.9 | H | XP_004968380 |
| SibZIP9 | Si003247m | Chr.5: 42311957-42314982 | 147 | 10 | 16539.8 | A | XP_002458823 |
| SibZIP10 | Si003289m | Chr.5: 246101970-24611764 | 140 | 9 | 15506.5 | C | XP_004968894 |
| SibZIP11 | Si006417m | Chr.4: 33420130-33425120 | 449 | 9 | 48149.5 | D | XP_004965885 |
| SibZIP12 | Si006667m | Chr.4: 11743965-11745864 | 379 | 8 | 41521.2 | D | XP_004966886 |
| SibZIP13 | Si006889m | Chr.4: 6541497-4646365 | 325 | 9 | 35254.3 | A | XP_004964902 |
| SibZIP14 | Si006978m | Chr.4: 33015635-33017728 | 303 | 5 | 32514.6 | B | XP_004965858 |
| SibZIP15 | Si007043m | Chr.4: 30527846-30530681 | 285 | 6 | 30388.8 | C | XP_004965642 |
| SibZIP16 | Si007412m | Chr.4: 39706531-39709069 | 157 | 9 | 17268.5 | A | XP_004966480 |
| SibZIP17 | Si007892m | Chr.4: 12607851-12608351 | 167 | 9 | 18839.9 | C | XP_004965226 |
| SibZIP18 | Si010177m | Chr.7: 31096934-31105294 | 427 | 7 | 47577 | D | XP_004976974 |
| SibZIP19 | Si010440m | Chr.7: 32666098-32669917 | 359 | 9 | 39895 | E | XP_004977157 |
| SibZIP20 | Si010549m | Chr.7: 33666577-33670528 | 335 | 6 | 36005.8 | I | XP_004977320 |
| SibZIP21 | Si012481m | Chr.7: 34001430-34002276 | 185 | 9 | 19986.3 | X | XP_004978331 |
| SibZIP22 | Si013904m | Chr.6: 30497939-30500723 | 390 | 6 | 41467.1 | A | XP_004973666 |
| SibZIP23 | Si013962m | Chr.6: 3422261-3424756 | 367 | 7 | 40266.7 | D | XP_004972733 |
| SibZIP24 | Si014433m | Chr.6: 31421965-31422818 | 219 | 9 | 23535.9 | C | XP_004973757 |
| SibZIP25 | Si014523m | Chr.6: 21075712-21078072 | 175 | 9 | 19443.7 | C | XP_004973271 |
| SibZIP26 | Si014546m | Chr.6: 35057931-35059283 | 169 | 9 | 18500.6 | A | XP_004974151 |
| SibZIP27 | Si014861m | Chr.6: 34473052-34479032 | 553 | 8 | 60827.5 | D | XP_004978700 |
| SibZIP28 | Si017618m | Chr.1: 39059405-39061854 | 357 | 6 | 38234.6 | A | XP_004954028 |
| SibZIP29 | Si017637m | Chr.1: 8783667-8786862 | 354 | 5 | 37572.7 | G | XP_004952026 |
| SibZIP30 | Si017778m | Chr.1: 6341933-6345399 | 325 | 6 | 34446.2 | C | XP_004951739 |
| SibZIP31 | Si018161m | Chr.1: 1679857-1681771 | 252 | 6 | 26940.2 | E | XP_004951321 |
| SibZIP32 | Si018524m | Chr.1: 4311195-4313806 | 167 | 10 | 18441.4 | H | XP_004951525 |
| SibZIP33 | Si019099m | Chr.1: 5311182-5311658 | 158 | 6 | 18128.3 | C | XP_008645357 |
| SibZIP34 | Si019656m | Chr.1: 8856565-8857280 | 101 | 12 | 11586.4 | C | XP_006362088 |
| SibZIP35 | Si019897m | Chr.1: 36896832-36897341 | 169 | 8 | 19639.9 | C | XP_004953778 |
| SibZIP36 | Si021442m | Chr.3: 20717700-20721220 | 645 | 9 | 68096.2 | B | XP_004962093 |
| SibZIP37 | Si022106m | Chr.3: 18527822-18531435 | 431 | 6 | 48135.2 | D | XP_004961926 |
| SibZIP38 | Si022325m | Chr.3: 5860297-5865473 | 384 | 7 | 40757.5 | G | XP_004960729 |
| SibZIP39 | Si022331m | Chr.3: 48207088-48212169 | 383 | 6 | 41545.1 | C | XP_004963102 |
| SibZIP40 | Si022388m | Chr.3: 9871788-9876447 | 373 | 9 | 39693.2 | G | XP_004961173 |
| SibZIP41 | Si022626m | Chr.3: 14850120-14852158 | 331 | 8 | 36509.2 | D | XP_004961656 |
| SibZIP42 | Si022639m | Chr.3: 15026867-15029429 | 328 | 5 | 35423.8 | A | XP_004961686 |
| SibZIP43 | Si023448m | Chr.3: 19115835-19117133 | 170 | 9 | 18373.5 | A | XP_004961973 |
| SibZIP44 | Si023541m | Chr.3: 42857978-42859308 | 149 | 7 | 16621.5 | C | XP_004962768 |
| SibZIP45 | Si023562m | Chr.3: 3146171-3147458 | 145 | 9 | 15989 | C | XP_004960384 |
| SibZIP46 | Si024325m | Chr.3: 50281135-50281679 | 175 | 11 | 19023.7 | C | XP_004964234 |
| Gene name | Phytozome identifier | Chromosome location (bp) | Protein length (aa) | Theore- tical pI | Molecular weight (Da) | Phylogeny group | NCBI accession No. |
| SibZIP47 | Si026421m | Chr.8: 2241219-2245356 | 418 | 6 | 45444.7 | D | XP_004978700 |
| SibZIP48 | Si026452m | Chr.8: 2350696-2353211 | 403 | 7 | 42267.1 | X | XP_004978712 |
| SibZIP49 | Si026561m | Chr.8: 12073275-12076909 | 357 | 9 | 39171.5 | E | XP_004979095 |
| SibZIP50 | Si026605m | Chr.8: 3317921-3321817 | 336 | 7 | 35850.7 | I | XP_004978752 |
| SibZIP51 | Si029351m | Chr.2: 46950969-46953724 | 571 | 6 | 60447.1 | B | XP_004958502 |
| SibZIP52 | Si029606m | Chr.2: 35632657-35640776 | 497 | 7 | 55157.1 | D | XP_004957190 |
| SibZIP53 | Si029966m | Chr.2: 24028261-24036295 | 417 | 8 | 46056.5 | D | XP_004956460 |
| SibZIP54 | Si029970m | Chr.2: 4745736-4748783 | 416 | 5 | 45187.2 | C | XP_004955660 |
| SibZIP55 | Si030114m | Chr.2: 33169494-33171523 | 390 | 9 | 41890.8 | A | XP_004957010 |
| SibZIP56 | Si030123m | Chr.2: 7037626-7041762 | 389 | 7 | 39918.8 | G | XP_004955801 |
| SibZIP57 | Si030182m | Chr.2: 37345091-37349203 | 378 | 6 | 40638.3 | I | XP_004957354 |
| SibZIP58 | Si030224m | Chr.2: 48343483-48348262 | 371 | 6 | 39761.6 | I | XP_004958698 |
| SibZIP59 | Si031077m | Chr.2: 38684243-38685591 | 216 | 9 | 22457.3 | A | XP_004957504 |
| SibZIP60 | Si031238m | Chr.2: 34573960-34574626 | 181 | 7 | 20285.7 | C | XP_004957101 |
| SibZIP61 | Si031353m | Chr.2: 25296856-25298971 | 159 | 9 | 17806.8 | C | XP_004956521 |
| SibZIP62 | Si032429m | Chr.2: 1879467-1880494 | 315 | 10 | 33579.7 | C | XP_004958923 |
| SibZIP63 | Si032507m | Chr.2: 37624846-37627738 | 482 | 10 | 50744.7 | I | XP_002460540 |
| SibZIP64 | Si035062m | Chr.9: 3209713-3215709 | 541 | 5 | 56605 | C | XP_004981429 |
| SibZIP65 | Si035841m | Chr.9: 57321111-57323641 | 423 | 6 | 45305.5 | I | XP_004985885 |
| SibZIP66 | Si036380m | Chr.9: 47061608-47065486 | 353 | 6 | 38129.3 | I | XP_004984459 |
| SibZIP67 | Si036527m | Chr.9: 47985071-47991293 | 335 | 9 | 37389.2 | D | XP_004984573 |
| SibZIP68 | Si038381m | Chr.9: 47741259-47744465 | 70 | 10 | 8263.2 | A | XP_004987020 |
| SibZIP69 | Si039367m | Chr.9: 2420394-2422861 | 344 | 6 | 36158.7 | G | XP_004981331 |
| SibZIP70 | Si039558m | Chr.9: 9034726-9035118 | 131 | 12 | 14050 | C | XP_004986317 |
| SibZIP71 | Si039566m | Chr.9: 40545110-40547482 | 375 | 8 | 40649.5 | I | XP_004983840 |
| SibZIP72 | Si039895m | Chr.9: 48442410-48442913 | 167 | 10 | 18179 | C | XP_004987029 |
| SibZIP73 | Si040286m | Chr.9: 51600068-51604903 | 381 | 9 | 40899.1 | G | XP_004985055 |
表2 谷子SibZIPs基因家族成员
Table 2 Members of the SibZIPs gene family in Setaria italica
| Gene name | Phytozome identifier | Chromosome location (bp) | Protein length (aa) | Theore- tical pI | Molecular weight (Da) | Phylogeny group | NCBI accession No. |
|---|---|---|---|---|---|---|---|
| SibZIP1 | Si000955m | Chr.5: 42034648-42041849 | 538 | 6 | 58030.2 | D | XP_004970651 |
| SibZIP2 | Si001292m | Chr.5: 39537137-39542647 | 473 | 6 | 51362.5 | D | XP_004970302 |
| SibZIP3 | Si001731m | Chr.5: 42027056-42028626 | 399 | 6 | 42138.5 | A | XP_004970655 |
| SibZIP4 | Si001929m | Chr.5: 31392517-31397217 | 368 | 9 | 39273.7 | G | XP_004969366 |
| SibZIP5 | Si002173m | Chr.5: 39683613-39687077 | 333 | 9 | 36026.4 | A | XP_004970325 |
| SibZIP6 | Si002192m | Chr.5: 3576665-3581966 | 330 | 7 | 36678.2 | D | XP_004967673 |
| SibZIP7 | Si002311m | Chr.5: 13207621-13209504 | 314 | 6 | 34130.9 | E | XP_004968609 |
| SibZIP8 | Si002989m | Chr.5: 10278963-10280748 | 196 | 10 | 20861.9 | H | XP_004968380 |
| SibZIP9 | Si003247m | Chr.5: 42311957-42314982 | 147 | 10 | 16539.8 | A | XP_002458823 |
| SibZIP10 | Si003289m | Chr.5: 246101970-24611764 | 140 | 9 | 15506.5 | C | XP_004968894 |
| SibZIP11 | Si006417m | Chr.4: 33420130-33425120 | 449 | 9 | 48149.5 | D | XP_004965885 |
| SibZIP12 | Si006667m | Chr.4: 11743965-11745864 | 379 | 8 | 41521.2 | D | XP_004966886 |
| SibZIP13 | Si006889m | Chr.4: 6541497-4646365 | 325 | 9 | 35254.3 | A | XP_004964902 |
| SibZIP14 | Si006978m | Chr.4: 33015635-33017728 | 303 | 5 | 32514.6 | B | XP_004965858 |
| SibZIP15 | Si007043m | Chr.4: 30527846-30530681 | 285 | 6 | 30388.8 | C | XP_004965642 |
| SibZIP16 | Si007412m | Chr.4: 39706531-39709069 | 157 | 9 | 17268.5 | A | XP_004966480 |
| SibZIP17 | Si007892m | Chr.4: 12607851-12608351 | 167 | 9 | 18839.9 | C | XP_004965226 |
| SibZIP18 | Si010177m | Chr.7: 31096934-31105294 | 427 | 7 | 47577 | D | XP_004976974 |
| SibZIP19 | Si010440m | Chr.7: 32666098-32669917 | 359 | 9 | 39895 | E | XP_004977157 |
| SibZIP20 | Si010549m | Chr.7: 33666577-33670528 | 335 | 6 | 36005.8 | I | XP_004977320 |
| SibZIP21 | Si012481m | Chr.7: 34001430-34002276 | 185 | 9 | 19986.3 | X | XP_004978331 |
| SibZIP22 | Si013904m | Chr.6: 30497939-30500723 | 390 | 6 | 41467.1 | A | XP_004973666 |
| SibZIP23 | Si013962m | Chr.6: 3422261-3424756 | 367 | 7 | 40266.7 | D | XP_004972733 |
| SibZIP24 | Si014433m | Chr.6: 31421965-31422818 | 219 | 9 | 23535.9 | C | XP_004973757 |
| SibZIP25 | Si014523m | Chr.6: 21075712-21078072 | 175 | 9 | 19443.7 | C | XP_004973271 |
| SibZIP26 | Si014546m | Chr.6: 35057931-35059283 | 169 | 9 | 18500.6 | A | XP_004974151 |
| SibZIP27 | Si014861m | Chr.6: 34473052-34479032 | 553 | 8 | 60827.5 | D | XP_004978700 |
| SibZIP28 | Si017618m | Chr.1: 39059405-39061854 | 357 | 6 | 38234.6 | A | XP_004954028 |
| SibZIP29 | Si017637m | Chr.1: 8783667-8786862 | 354 | 5 | 37572.7 | G | XP_004952026 |
| SibZIP30 | Si017778m | Chr.1: 6341933-6345399 | 325 | 6 | 34446.2 | C | XP_004951739 |
| SibZIP31 | Si018161m | Chr.1: 1679857-1681771 | 252 | 6 | 26940.2 | E | XP_004951321 |
| SibZIP32 | Si018524m | Chr.1: 4311195-4313806 | 167 | 10 | 18441.4 | H | XP_004951525 |
| SibZIP33 | Si019099m | Chr.1: 5311182-5311658 | 158 | 6 | 18128.3 | C | XP_008645357 |
| SibZIP34 | Si019656m | Chr.1: 8856565-8857280 | 101 | 12 | 11586.4 | C | XP_006362088 |
| SibZIP35 | Si019897m | Chr.1: 36896832-36897341 | 169 | 8 | 19639.9 | C | XP_004953778 |
| SibZIP36 | Si021442m | Chr.3: 20717700-20721220 | 645 | 9 | 68096.2 | B | XP_004962093 |
| SibZIP37 | Si022106m | Chr.3: 18527822-18531435 | 431 | 6 | 48135.2 | D | XP_004961926 |
| SibZIP38 | Si022325m | Chr.3: 5860297-5865473 | 384 | 7 | 40757.5 | G | XP_004960729 |
| SibZIP39 | Si022331m | Chr.3: 48207088-48212169 | 383 | 6 | 41545.1 | C | XP_004963102 |
| SibZIP40 | Si022388m | Chr.3: 9871788-9876447 | 373 | 9 | 39693.2 | G | XP_004961173 |
| SibZIP41 | Si022626m | Chr.3: 14850120-14852158 | 331 | 8 | 36509.2 | D | XP_004961656 |
| SibZIP42 | Si022639m | Chr.3: 15026867-15029429 | 328 | 5 | 35423.8 | A | XP_004961686 |
| SibZIP43 | Si023448m | Chr.3: 19115835-19117133 | 170 | 9 | 18373.5 | A | XP_004961973 |
| SibZIP44 | Si023541m | Chr.3: 42857978-42859308 | 149 | 7 | 16621.5 | C | XP_004962768 |
| SibZIP45 | Si023562m | Chr.3: 3146171-3147458 | 145 | 9 | 15989 | C | XP_004960384 |
| SibZIP46 | Si024325m | Chr.3: 50281135-50281679 | 175 | 11 | 19023.7 | C | XP_004964234 |
| Gene name | Phytozome identifier | Chromosome location (bp) | Protein length (aa) | Theore- tical pI | Molecular weight (Da) | Phylogeny group | NCBI accession No. |
| SibZIP47 | Si026421m | Chr.8: 2241219-2245356 | 418 | 6 | 45444.7 | D | XP_004978700 |
| SibZIP48 | Si026452m | Chr.8: 2350696-2353211 | 403 | 7 | 42267.1 | X | XP_004978712 |
| SibZIP49 | Si026561m | Chr.8: 12073275-12076909 | 357 | 9 | 39171.5 | E | XP_004979095 |
| SibZIP50 | Si026605m | Chr.8: 3317921-3321817 | 336 | 7 | 35850.7 | I | XP_004978752 |
| SibZIP51 | Si029351m | Chr.2: 46950969-46953724 | 571 | 6 | 60447.1 | B | XP_004958502 |
| SibZIP52 | Si029606m | Chr.2: 35632657-35640776 | 497 | 7 | 55157.1 | D | XP_004957190 |
| SibZIP53 | Si029966m | Chr.2: 24028261-24036295 | 417 | 8 | 46056.5 | D | XP_004956460 |
| SibZIP54 | Si029970m | Chr.2: 4745736-4748783 | 416 | 5 | 45187.2 | C | XP_004955660 |
| SibZIP55 | Si030114m | Chr.2: 33169494-33171523 | 390 | 9 | 41890.8 | A | XP_004957010 |
| SibZIP56 | Si030123m | Chr.2: 7037626-7041762 | 389 | 7 | 39918.8 | G | XP_004955801 |
| SibZIP57 | Si030182m | Chr.2: 37345091-37349203 | 378 | 6 | 40638.3 | I | XP_004957354 |
| SibZIP58 | Si030224m | Chr.2: 48343483-48348262 | 371 | 6 | 39761.6 | I | XP_004958698 |
| SibZIP59 | Si031077m | Chr.2: 38684243-38685591 | 216 | 9 | 22457.3 | A | XP_004957504 |
| SibZIP60 | Si031238m | Chr.2: 34573960-34574626 | 181 | 7 | 20285.7 | C | XP_004957101 |
| SibZIP61 | Si031353m | Chr.2: 25296856-25298971 | 159 | 9 | 17806.8 | C | XP_004956521 |
| SibZIP62 | Si032429m | Chr.2: 1879467-1880494 | 315 | 10 | 33579.7 | C | XP_004958923 |
| SibZIP63 | Si032507m | Chr.2: 37624846-37627738 | 482 | 10 | 50744.7 | I | XP_002460540 |
| SibZIP64 | Si035062m | Chr.9: 3209713-3215709 | 541 | 5 | 56605 | C | XP_004981429 |
| SibZIP65 | Si035841m | Chr.9: 57321111-57323641 | 423 | 6 | 45305.5 | I | XP_004985885 |
| SibZIP66 | Si036380m | Chr.9: 47061608-47065486 | 353 | 6 | 38129.3 | I | XP_004984459 |
| SibZIP67 | Si036527m | Chr.9: 47985071-47991293 | 335 | 9 | 37389.2 | D | XP_004984573 |
| SibZIP68 | Si038381m | Chr.9: 47741259-47744465 | 70 | 10 | 8263.2 | A | XP_004987020 |
| SibZIP69 | Si039367m | Chr.9: 2420394-2422861 | 344 | 6 | 36158.7 | G | XP_004981331 |
| SibZIP70 | Si039558m | Chr.9: 9034726-9035118 | 131 | 12 | 14050 | C | XP_004986317 |
| SibZIP71 | Si039566m | Chr.9: 40545110-40547482 | 375 | 8 | 40649.5 | I | XP_004983840 |
| SibZIP72 | Si039895m | Chr.9: 48442410-48442913 | 167 | 10 | 18179 | C | XP_004987029 |
| SibZIP73 | Si040286m | Chr.9: 51600068-51604903 | 381 | 9 | 40899.1 | G | XP_004985055 |
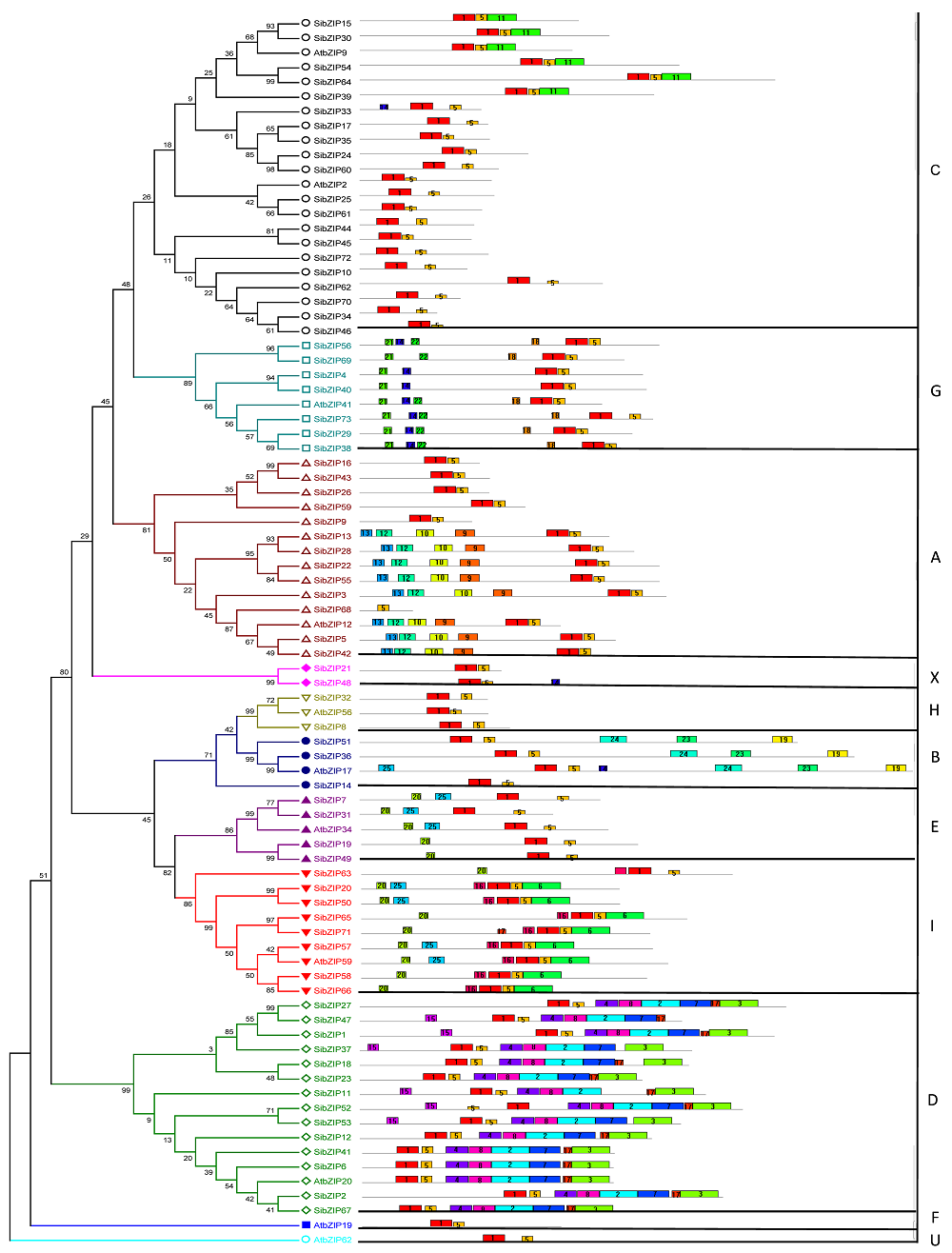
图1 谷子73条SibZIPs蛋白序列的MEME保守基序分析及系统发育树
Figure 1 The MEME conserved motifs analysis and phylogenetic tree of 73 SibZIPs protein sequences of Setaria italica
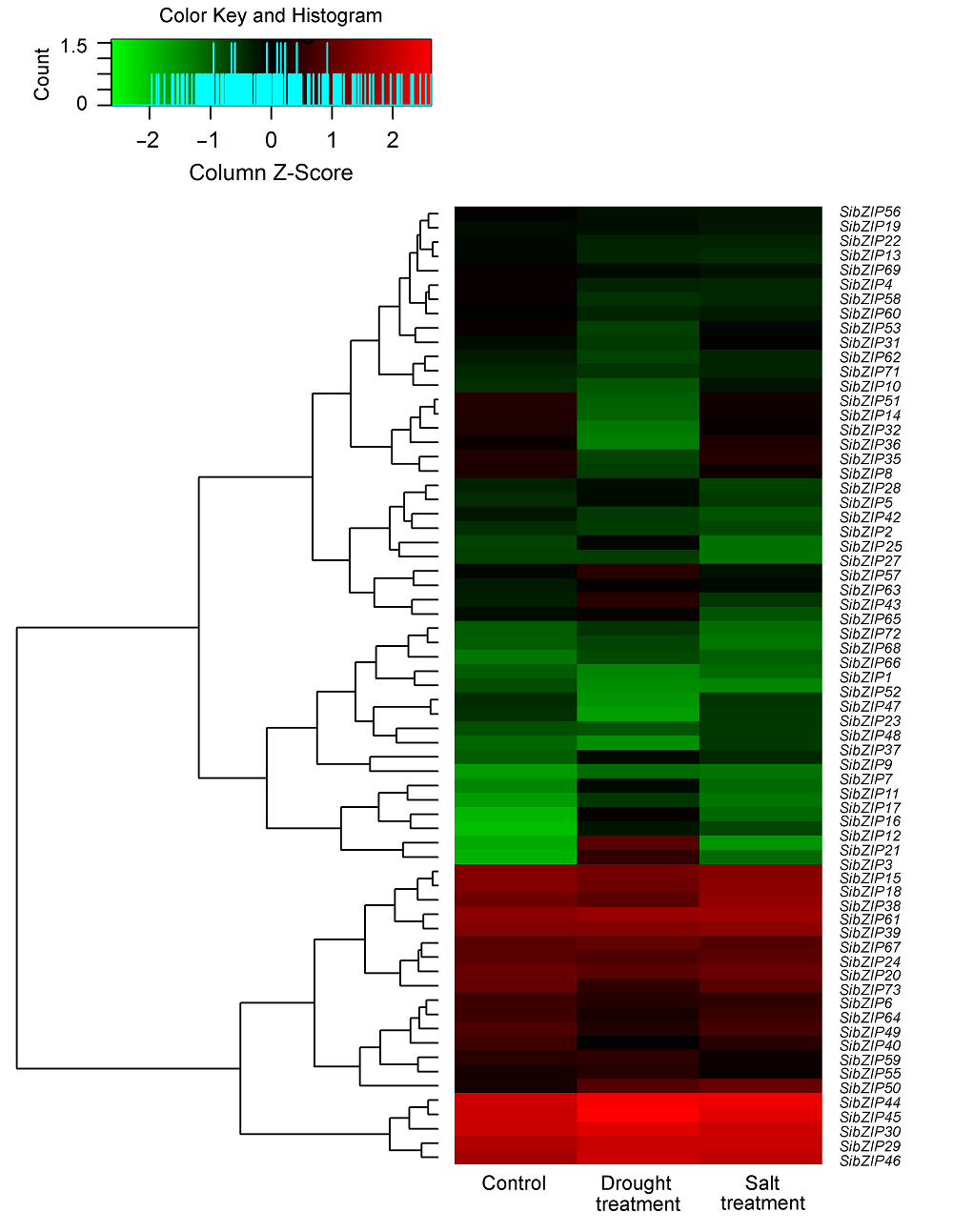
图2 干旱和盐胁迫下谷子SibZIP基因的表达谱 红色和绿色长方块分别代表高和低水平的SibZIP转录因子基因表达情况。表达差异大于1.5倍视为显著差异(P<0.05)。
Figure 2 Expression profiles of 73 SibZIP genes in Setaria italica leaves under drought and salt stresses Blocks with colors indicate upregulated (red) and downregulated (green) transcript accumulation of SibZIPs relative to the control. The expression level changed more than 1.5 folds is considered as significant difference (P<0.05).
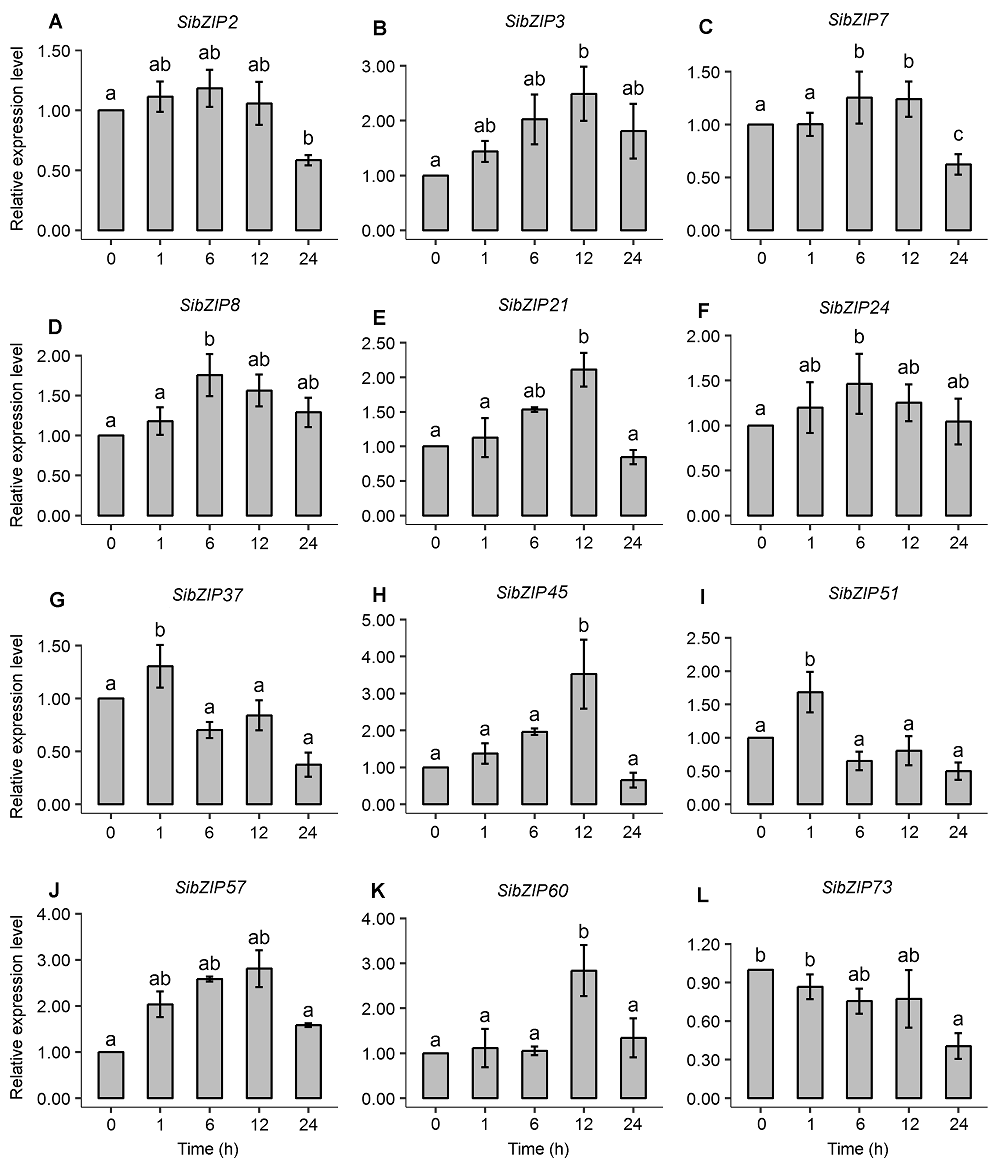
图3 12个SibZIP基因在谷子幼苗叶片中的实时荧光定量PCR表达 不同字母代表Duncan检验法样品间差异显著(P<0.05)。
Figure 3 Expression analysis of 12 SibZIP genes in leaves of Setaria italic seedlings using real-time quantitative RT-PCR The different letters indicate significant difference (P<0.05) among samples by Duncan’s test.
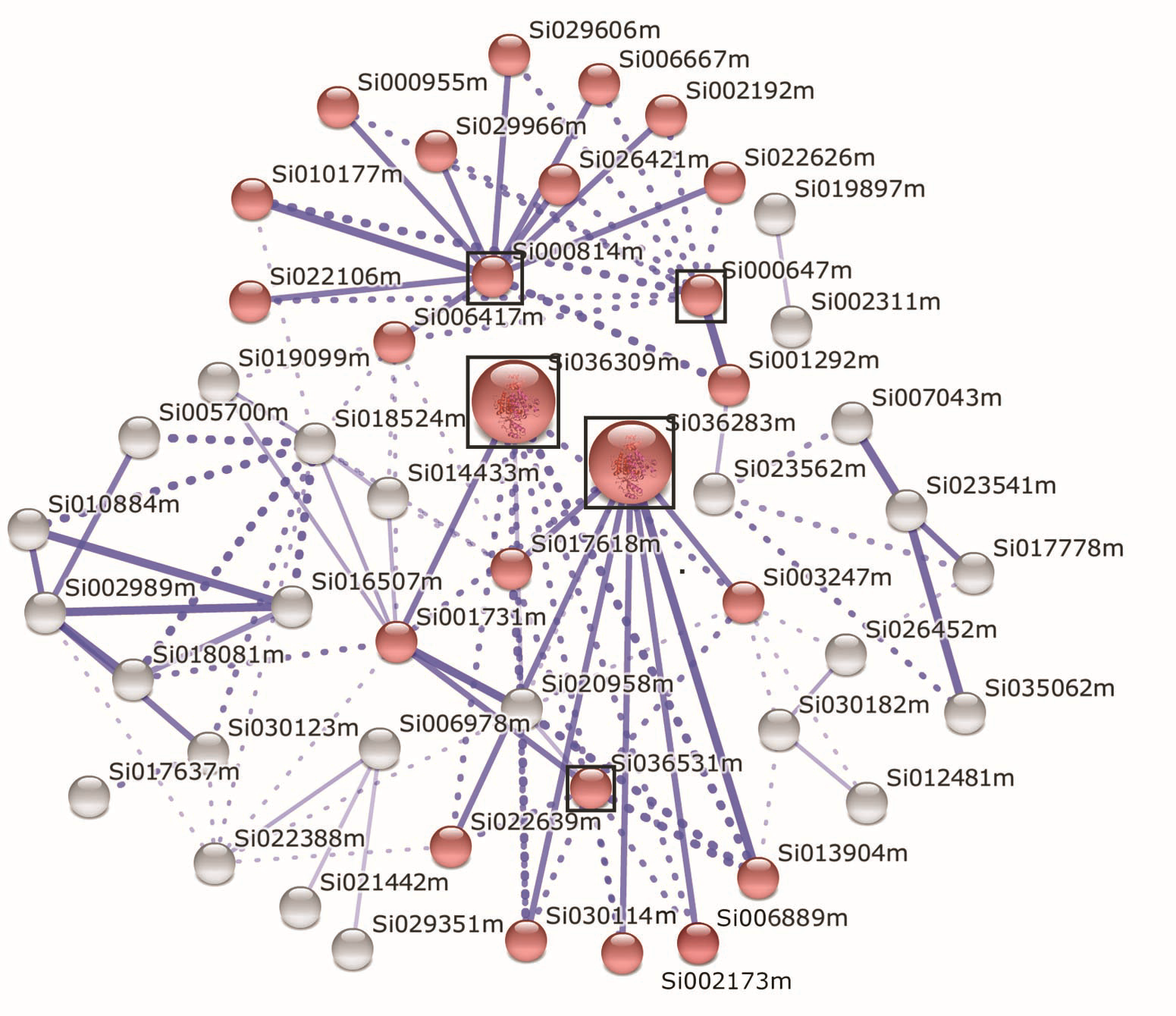
图4 胁迫条件下谷子SibZIP基因家族共表达关联性 逆境胁迫下谷子编码蛋白基因的表达数据来自Setaria italica v2.1数据库。利用String功能蛋白互作网站(http://string-db.org/)相关工具对谷子SibZIP转录因子基因共表达关联性进行分析。红色标记为参与逆境反应相关蛋白基因。圆圈大小表示该蛋白功能重要与否, 体积越大表示在胁迫相关代谢通路中越重要。蓝色实线表示蛋白之间显著关联, 线越粗表示蛋白之间关联度越紧密。蓝色虚线表示蛋白之间可能具有相互作用。对应于谷子蛋白(红色圆球代表)编号的各SibZIP成员名称见表2。Si036309m: SRK2D相关的丝氨酸/苏氨酸蛋白激酶; Si036283m: SF160-丝氨酸/苏氨酸蛋白激酶; Si036531m: 丝氨酸/苏氨酸蛋白激酶SAPK3; Si000814m: NPR1相关的SF66-调控蛋白; Si000647m: KOG0512-胚胎球蛋白诱导因子
Figure 4 Correlation analysis of co-expression of Setaria italica SibZIP genes under stressed conditions The expression data of the protein coding genes in foxtail millet under stressed conditions were derived from Setaria italica v2.1 database. Bioinformatics tools in String Functional Protein Interaction website (http://string-db.org/) were employed for correlation analysis of co-expression of SibZIP genes. Red circle represents SibZIP genes and the related genes involved in regulation network responsible for stress responses. The size of circle indicates the significance of the protein with bigger size having more importance in the related pathway. Blue solid line shows correlation between the related proteins with thinker line meaning stronger correlation. Blue dotted line shows possible interraction between the related proteins. The corresponding SibZIP protein name for each foxtail millet protein (red color circle representive) are list in Table 2. Si036309m: serine/threonine-protein kinase SRK2D-related; Si036283m: SF160-serine/threonine-protein kinase; Si036531m: serine/threonine-protein kinase SAPK3; Si000814m: SF66-regulatory protein NPR1-related; Si000647m: KOG0512-fetal globin-inducing factor
| 1 | 曹红利, 岳川, 王新超, 杨亚军 (2012). bZIP转录因子与植物抗逆性研究进展. 南方农业学报 43, 1094-1100. |
| 2 | 杨颖, 高世庆, 唐益苗, 冶晓芳, 王永波, 刘美英, 赵昌平 (2009). 植物bZIP转录因子的研究进展. 麦类作物学报 29, 730-737. |
| 3 | 张水军, 曾千春, 卢秀萍, 李文正 (2010). 植物富含甘氨酸蛋白的研究进展. 中国农学通报 26(4), 54-58. |
| 4 | 朱芸晔, 薛冰, 王安全, 王文杰, 周昂, 黄胜雄, 刘永胜 (2014). 番茄bZIP转录因子家族的生物信息学分析. 应用与环境生物学报 (5), 767-774. |
| 5 | Abe M, Kobayashi Y, Yamamoto S, Daimon Y, Yamaguchi A, Ikeda Y, Ichinoki H, Notaguchi M, Goto K, Araki T (2005). FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex.Science 309, 1052-1056. |
| 6 | Baloglu MC, Eldem V, Hajyzadeh M, Unver T (2014). Genome-wide analysis of the bZIP transcription factors in cucumber.PLoS One 9, e96014. |
| 7 | Bennetzen JL, Schmutz J, Wang H, Percifield R, Hawkins J, Pontaroli AC, Estep M, Feng L, Vaughn JN, Grimwood J, Jenkins J, Barry K, Lindquist E, Hellsten U, Deshpande S, Wang X, Wu X, Mitros T, Triplett J, Yang X, Ye CY, Mauro-Herrera M, Wang L, Li P, Sharma M, Sharma R, Ronald PC, Panaud O, Kellogg EA, Brutnell TP, Doust AN, Tuskan GA, Rokhsar D, Devos KM (2012). Reference genome sequence of the model plant Setaria.Nat Biotechnol 30, 555-561. |
| 8 | Chuang CF, Running MP, Williams RW, Meyerowitz EM (1999). The PERIANTHIA gene encodes a bZIP protein involved in the determination of floral organ number in Arabidopsis thaliana.Genes Devel 13, 334-344. |
| 9 | Corrêa LG, Riaño-Pachón DM, Schrago CG, Dos Santos RV, Mueller-Roeber B, Vincentz M (2008). The role of bZIP transcription factors in green plant evolution: adaptive features emerging from four founder genes.PLoS One 3, e2944. |
| 10 | Fujita Y, Fujita M, Satoh R, Maruyama K, Parvez MM, Seki M, Hiratsu K, Ohme-Takagi M, Shinozaki K, Yamaguchi-Shinozaki K (2005). AREB1 is a transcription activator of novel ABRE-dependent ABA signaling that enhances drought stress tolerance in Arabidopsis.Plant Cell 17, 3470-3488. |
| 11 | Gibalová A, Renák D, Matczuk K, Dupl'áková N, Cháb D, Twell D, Honys D (2009). AtbZIP34 is required for Arabidopsis pollen wall patterning and the control of several metabolic pathways in developing pollen.Plant Mol Biol 70, 581-601. |
| 12 | Guiltinan MJ, Marcotte WR Jr, Quatrano RS (1990). A plant leucine zipper protein that recognizes an abscisic acid response element.Science 250, 267-271. |
| 13 | Iven T, Strathmann A, Böttner S, Zwafink T, Heinekamp T, Guivarc'h A, Roitsch T, Dröge-Laser W (2010). Homo- and heterodimers of tobacco bZIP proteins counteract as positive or negative regulators of transcription during pollen development.Plant J Cell Mol Biol 63, 155-166. |
| 14 | Izawa T, Foster R, Nakajima M, Shimamoto K, Chua NH (1994). The rice bZIP transcriptional activator RITA-1 is highly expressed during seed development.Plant Cell 6, 1277-1287. |
| 15 | Jakoby M, Weisshaar B, Dröge-Laser W, Vicente-Carba- josa J, Tiedemann J, Kroj T, Parcy F, BZIPRG (2002). bZIP transcription factors in Arabidopsis.Trends Plant Sci 7, 106-111. |
| 16 | Jin Z, Xu W, Liu A (2014). Genomic surveys and expression analysis of bZIP gene family in castor bean (Ricinus communis L.).Planta 239, 299-312. |
| 17 | Johnson C, Boden E, Arias J (2003). Salicylic acid and NPR1 induce the recruitment of trans-activating TGA factors to a defense gene promoter in Arabidopsis.Plant Cell 15, 1846-1858. |
| 18 | Lee SC, Choi HW, Hwang IS, Choi DS, Hwang BK (2006). Functional roles of the pepper pathogen-induced bZIP transcription factor, CAbZIP1, in enhanced resistance to pathogen infection and environmental stresses.Planta 224, 1209-1225. |
| 19 | Liao Y, Zou HF, Wei W, Hao YJ, Tian AG, Huang J, Liu YF, Zhang JS, Chen SY (2008). Soybean GmbZIP44, GmbZIP62 and GmbZIP78 genes function as negative regulator of ABA signaling and confer salt and freezing tolerance in transgenic Arabidopsis.Planta 228, 225-240. |
| 20 | Liu J, Chen N, Chen F, Cai B, Dal Santo S, Tornielli GB, Pezzotti M, Cheng ZM (2014). Genome-wide analysis and expression profile of the bZIP transcription factor gene family in grapevine (Vitis vinifera).BMC Genomics 15, 281. |
| 21 | Liu JX, Srivastava R, Howell SH (2008). Stress-induced expression of an activated form of AtbZIP17 provides protection from salt stress in Arabidopsis.Plant Cell Environ 31, 1735-1743. |
| 22 | Nijhawan A, Jain M, Tyagi AK, Khurana JP (2008). Genomic survey and gene expression analysis of the basic leucine zipper transcription factor family in rice.Plant Physiol 146, 333-350. |
| 23 | Pourabed E, Ghane Golmohamadi F, Soleymani Monfared P, Razavi SM, Shobbar ZS (2015). Basic leucine zipper family in barley: genome-wide characterization of members and expression analysis.Mol Biotechnol 57, 12-26. |
| 24 | Schultz J, Milpetz F, Bork P, Ponting CP (1998). SMART, a simple modular architecture research tool: identification of signaling domains.Proc Natl Acad Sci USA 95, 5857-5864. |
| 25 | Schwechheimer C, Zourelidou M, Bevan MW (1998). Plant transcription factor studies.Annu Rev Plant Physiol Plant Mol Biol 49, 127-150. |
| 26 | Singh K, Foley RC, Oñate-Sánchez L (2002). Transcription factors in plant defense and stress responses.Curr Opin Plant Biol 5, 430-436. |
| 27 | Stanković B, Vian A, Henry-Vian C, Davies E (2000). Molecular cloning and characterization of a tomato cDNA encoding a systemically wound-inducible bZIP DNA- binding protein.Planta 212, 60-66. |
| 28 | Toh S, McCourt P, Tsuchiya Y (2012). HY5 is involved in strigolactone-dependent seed germination in Arabidopsis.Plant Signal Behavior 7, 556-558. |
| 29 | Uno Y, Furihata T, Abe H, Yoshida R, Shinozaki K, Yamaguchi-Shinozaki K (2000). Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid- dependent signal transduction pathway under drought and high-salinity conditions.Proc Natl Acad Sci USA 97, 11632-11637. |
| 30 | Wang J, Zhou J, Zhang B, Vanitha J, Ramachandran S, Jiang SY (2011). Genome-wide expansion and expression divergence of the basic leucine zipper transcription factors in higher plants with an emphasis on sorghum.J Integr Plant Biol 53, 212-231. |
| 31 | Wei K, Chen J, Wang Y, Chen Y, Chen S, Lin Y, Pan S, Zhong X, Xie D (2012). Genome-wide analysis of bZIP- encoding genes in maize.DNA Res 19, 463-476. |
| 32 | Xiang Y, Tang N, Du H, Ye H, Xiong L (2008). Characterization of OsbZIP23 as a key player of the basic leucine zipper transcription factor family for conferring abscisic acid sensitivity and salinity and drought tolerance in rice.Plant Physiol 148, 1938-1952. |
| 33 | Xu J, Li Y, Ma X, Ding J, Wang K, Wang S, Tian Y, Zhang H, Zhu XG (2013). Whole transcriptome analysis using next-generation sequencing of model species Setaria viridis to support C4 photosynthesis research.Plant Mol Biol 83, 77-87. |
| 34 | Ying S, Zhang DF, Fu J, Shi YS, Song YC, Wang TY, Li Y (2012). Cloning and characterization of a maize bZIP transcription factor, ZmbZIP72, confers drought and salt tolerance in transgenic Arabidopsis.Planta 235, 253-266. |
| 35 | Yoshida T, Fujita Y, Sayama H, Kidokoro S, Maruyama K, Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K (2010). AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation.Plant J 61, 672-685. |
| 36 | Zhang G, Liu X, Quan Z, Cheng S, Xu X, Pan S, Xie M, Zeng P, Yue Z, Wang W, Tao Y, Bian C, Han C, Xia Q, Peng X, Cao R, Yang X, Zhan D, Hu J, Zhang Y, Li H, Li H, Li N, Wang J, Wang C, Wang R, Guo T, Cai Y, Liu C, Xiang H, Shi Q, Huang P, Chen Q, Li Y, Wang J, Zhao Z, Wang J (2012). Genome sequence of foxtail millet (Setaria italica) provides insights into grass evolution and biofuel potential.Nat Biotechnol 30, 549-554. |
| No related articles found! |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||