

植物学报 ›› 2025, Vol. 60 ›› Issue (3): 377-392.DOI: 10.11983/CBB24117 cstr: 32102.14.CBB24117
杨莉, 曲茜彤, 陈子航, 邹婷婷, 王全华, 王小丽*( )
)
收稿日期:2024-07-30
接受日期:2024-12-26
出版日期:2025-05-10
发布日期:2024-12-27
通讯作者:
*王小丽, 上海师范大学生命科学学院副教授, 硕士生导师。主要从事园艺植物营养品质调控、遗传鉴定及新品种选育等工作。目前研究团队以菠菜为主要研究对象, 收集了世界各地菠菜种质资源千余份, 完成菠菜资源的深度转录组测序和全基因组测序工作, 利用生理生化、遗传学和分子生物学等手段对其重要农艺性状进行研究。E-mail: wangxl@shnu.edu.cn
基金资助:
Yang Li, Qu Xitong, Chen Zihang, Zou Tingting, Wang Quanhua, Wang Xiaoli*( )
)
Received:2024-07-30
Accepted:2024-12-26
Online:2025-05-10
Published:2024-12-27
Contact:
*E-mail: wangxl@shnu.edu.cn
摘要: AHL (AT-hook)蛋白家族在植物生长发育、器官构建、逆境胁迫及植物激素信号应答中发挥关键作用。为揭示菠菜(Spinacia oleracea) AHL基因家族的生物学功能, 在基因组范围内对SoAHL家族成员进行鉴定和分析, 并探究外源水杨酸对其表达水平的影响。结果表明, SoAHL家族包含19个成员, 在6条染色体间不均匀分布, 系统发育树将其分为2个亚族, I、II亚族分别包含10、9个SoAHL成员; 不同亚族间PPC和AT-hook保守基序序列组成有差异; SoAHL主要定位在细胞核、细胞质和线粒体; SoAHL I亚族成员无内含子, II亚族成员含4-5个内含子; SoAHL启动子上游分布有不同数量的植物激素和非生物胁迫响应相关顺式作用元件。SoAHL成员在根、叶片及叶柄中均有表达, 大多数在根中表达水平较高。水杨酸处理条件下, SOV6g041850.1和SOV2g038950.1受水杨酸显著诱导; SOV2g031340.1和SOV4g018880.1主要在地上部表达, 与叶酸含量的组织分布特征相似。瞬时过表达SOV4g018880.1能提高菠菜叶酸含量1.75倍。研究结果可为进一步解析SoAHL的功能奠定基础。
杨莉, 曲茜彤, 陈子航, 邹婷婷, 王全华, 王小丽. 菠菜AT-hook基因家族鉴定与表达谱分析. 植物学报, 2025, 60(3): 377-392.
Yang Li, Qu Xitong, Chen Zihang, Zou Tingting, Wang Quanhua, Wang Xiaoli. Identification of the Spinach AT-hook Gene Family and Analysis of Expression Profiles. Chinese Bulletin of Botany, 2025, 60(3): 377-392.
| Gene ID | Forward primer (5′-3′) | Reverse primer (5′-3′) |
|---|---|---|
| SOV4g002370.1 | GTTCTAACCATCTCCGCAAT | TGAATGGGTGAAATTGGCAT |
| SOV5g038710.1 | GAGCGGCTATGATGGAG | GGATCTGATCTGTCCATGTC |
| SOV1g029900.1 | TAAGATGGAGCAACAACAGG | TCGGGTTCTGATTATCCTCT |
| SOV6g041850.1 | ATATCGGCATCACAACATGG | AGTGGTTTTGGAGTTGAAGT |
| SOV2g030150.1 | ATGGAAGGCCTCTACCACC | GTGGTGAAACTGATGATGCTG |
| SOV6g047350.1 | GACTCTCGTGGCCCATCTGC | GTAAGGTGAATTTGATTGGTGG |
| SOV2g031340.1 | GAGAGGATTAGCCGAGTTAC | CCCGACATTTGATTGGAAAG |
| SOV3g001110.1 | GTAGTTCAGGGGTAACTGTG | TTTTCACCCCTAGCTGTTAC |
| SOV2g038950.1 | GATAACAATGCTCAAAACAATG | GTTGGAATTTGGAAGGATGAG |
| SOV4g018880.1 | CCGAATGAACAAAGCGGAC | GACATTCCATTGGTGGCGG |
| SOV5g038610.1 | TCTGAAAATGACGTCGGTAG | CGGCTTATAAACTGCATTCC |
| SOV6g041780.1 | GTAGTAAGGTCAGATGCTCC | CATACTTCCTTGGTCTACCC |
| SOV2g029980.1 | GGTGACAGTAATAGGAGCAG | TTACTGGAGTAACATCCCCA |
| SOV5g021850.1 | TCAGGACCTGGTAGCTTTTA | AAAGTCATCCCGGAATTAGG |
| SOV6g040610.1 | GGATGGGAGAGAAGGTATGGC | CCAACTGCCCCAGGAATTGT |
| SOV4g057830.1 | ATAACGCCCTGGTGCAACTG | TGTGGCCTCCATCCGAACACAT |
| SOV4g019010.1 | ACGAACAACCCGGACAACTT | ACCGCCATGATTCCATCCAT |
| SOV6g040700.1 | TTGCCTCCGGTCCTGTGGTTATTAT | TCAGGTCTTTGCCAGTGATCTACC |
| SOV6g047370.1 | TCCGGTCCTGTGGTTATTATGGC | TCAGGTCTTTGCCAGTGATCTACC |
| 18S | CCATAAACGATGCCGACCAG | AGCCTTGCGACCATACTCCC |
表1 引物信息
Table 1 Sequences of the primers used in this study
| Gene ID | Forward primer (5′-3′) | Reverse primer (5′-3′) |
|---|---|---|
| SOV4g002370.1 | GTTCTAACCATCTCCGCAAT | TGAATGGGTGAAATTGGCAT |
| SOV5g038710.1 | GAGCGGCTATGATGGAG | GGATCTGATCTGTCCATGTC |
| SOV1g029900.1 | TAAGATGGAGCAACAACAGG | TCGGGTTCTGATTATCCTCT |
| SOV6g041850.1 | ATATCGGCATCACAACATGG | AGTGGTTTTGGAGTTGAAGT |
| SOV2g030150.1 | ATGGAAGGCCTCTACCACC | GTGGTGAAACTGATGATGCTG |
| SOV6g047350.1 | GACTCTCGTGGCCCATCTGC | GTAAGGTGAATTTGATTGGTGG |
| SOV2g031340.1 | GAGAGGATTAGCCGAGTTAC | CCCGACATTTGATTGGAAAG |
| SOV3g001110.1 | GTAGTTCAGGGGTAACTGTG | TTTTCACCCCTAGCTGTTAC |
| SOV2g038950.1 | GATAACAATGCTCAAAACAATG | GTTGGAATTTGGAAGGATGAG |
| SOV4g018880.1 | CCGAATGAACAAAGCGGAC | GACATTCCATTGGTGGCGG |
| SOV5g038610.1 | TCTGAAAATGACGTCGGTAG | CGGCTTATAAACTGCATTCC |
| SOV6g041780.1 | GTAGTAAGGTCAGATGCTCC | CATACTTCCTTGGTCTACCC |
| SOV2g029980.1 | GGTGACAGTAATAGGAGCAG | TTACTGGAGTAACATCCCCA |
| SOV5g021850.1 | TCAGGACCTGGTAGCTTTTA | AAAGTCATCCCGGAATTAGG |
| SOV6g040610.1 | GGATGGGAGAGAAGGTATGGC | CCAACTGCCCCAGGAATTGT |
| SOV4g057830.1 | ATAACGCCCTGGTGCAACTG | TGTGGCCTCCATCCGAACACAT |
| SOV4g019010.1 | ACGAACAACCCGGACAACTT | ACCGCCATGATTCCATCCAT |
| SOV6g040700.1 | TTGCCTCCGGTCCTGTGGTTATTAT | TCAGGTCTTTGCCAGTGATCTACC |
| SOV6g047370.1 | TCCGGTCCTGTGGTTATTATGGC | TCAGGTCTTTGCCAGTGATCTACC |
| 18S | CCATAAACGATGCCGACCAG | AGCCTTGCGACCATACTCCC |
| Gene ID | Genome location | Subcellular localization | Length (bp) | Amino acid (aa) | Molecular weight (kDa) | PI |
|---|---|---|---|---|---|---|
| SOV1g029900.1 | Chr. 1: 113135928-113136926 | NLS, Chl | 999 | 333 | 33.84 | 5.37 |
| SOV4g019010.1 | Chr. 4: 52394687-52395604 | NLS | 918 | 306 | 31.47 | 6.16 |
| SOV4g057830.1 | Chr. 4: 186409628-186410449 | NLS, Chl | 822 | 274 | 27.68 | 6.42 |
| SOV5g038710.1 | Chr. 5: 123243932-123247784 | NLS | 819 | 273 | 26.72 | 6.43 |
| SOV6g040700.1 | Chr. 6: 140433598-140434404 | NLS, Cyt | 807 | 269 | 28.36 | 9.07 |
| SOV6g047370.1 | Chr. 6: 148547185-148548063 | Chl, NLS | 879 | 293 | 30.18 | 6.04 |
| SOV2g030150.1 | Chr. 2: 101786656-101787666 | NLS, Chl | 1011 | 337 | 34.80 | 7.22 |
| SOV6g041850.1 | Chr. 6: 141988749-141989711 | NLS, Chl | 963 | 321 | 32.97 | 5.97 |
| SOV2g038950.1 | Chr. 2: 112654862-112655896 | Cyt, NLS | 1035 | 345 | 36.80 | 6.85 |
| SOV4g018880.1 | Chr. 4: 52004210-52011645 | NLS | 1041 | 347 | 36.10 | 8.46 |
| SOV2g031340.1 | Chr. 2: 103380711-103385136 | NLS | 990 | 330 | 34.05 | 9.58 |
| SOV6g040610.1 | Chr. 6: 140337052-140344304 | NLS, Cyt | 1044 | 348 | 35.29 | 9.89 |
| SOV2g029980.1 | Chr. 2: 101504484-101511484 | NLS, Chl | 1032 | 344 | 35.92 | 9.44 |
| SOV6g041780.1 | Chr. 6: 141900514-141905585 | NLS | 1026 | 342 | 35.70 | 8.33 |
| SOV3g001110.1 | Chr. 3: 1145951-1151558 | Chl, NLS | 1089 | 363 | 37.96 | 5.73 |
| SOV5g021850.1 | Chr. 5: 39407625-39411618 | NLS, Chl | 1089 | 363 | 37.31 | 7.87 |
| SOV5g038610.1 | Chr. 5: 122545368-122554893 | NLS | 1092 | 364 | 37.09 | 9.87 |
| SOV6g047350.1 | Chr. 6: 148521377-148529101 | NLS, Chl | 1413 | 471 | 50.13 | 8.79 |
| SOV4g002370.1 | Chr. 4: 2764521-2765501 | NLS, Cyt | 981 | 327 | 36.06 | 6.04 |
表2 菠菜SoAHL基因家族成员基本信息
Table 2 Information on the SoAHL gene family members in spinach
| Gene ID | Genome location | Subcellular localization | Length (bp) | Amino acid (aa) | Molecular weight (kDa) | PI |
|---|---|---|---|---|---|---|
| SOV1g029900.1 | Chr. 1: 113135928-113136926 | NLS, Chl | 999 | 333 | 33.84 | 5.37 |
| SOV4g019010.1 | Chr. 4: 52394687-52395604 | NLS | 918 | 306 | 31.47 | 6.16 |
| SOV4g057830.1 | Chr. 4: 186409628-186410449 | NLS, Chl | 822 | 274 | 27.68 | 6.42 |
| SOV5g038710.1 | Chr. 5: 123243932-123247784 | NLS | 819 | 273 | 26.72 | 6.43 |
| SOV6g040700.1 | Chr. 6: 140433598-140434404 | NLS, Cyt | 807 | 269 | 28.36 | 9.07 |
| SOV6g047370.1 | Chr. 6: 148547185-148548063 | Chl, NLS | 879 | 293 | 30.18 | 6.04 |
| SOV2g030150.1 | Chr. 2: 101786656-101787666 | NLS, Chl | 1011 | 337 | 34.80 | 7.22 |
| SOV6g041850.1 | Chr. 6: 141988749-141989711 | NLS, Chl | 963 | 321 | 32.97 | 5.97 |
| SOV2g038950.1 | Chr. 2: 112654862-112655896 | Cyt, NLS | 1035 | 345 | 36.80 | 6.85 |
| SOV4g018880.1 | Chr. 4: 52004210-52011645 | NLS | 1041 | 347 | 36.10 | 8.46 |
| SOV2g031340.1 | Chr. 2: 103380711-103385136 | NLS | 990 | 330 | 34.05 | 9.58 |
| SOV6g040610.1 | Chr. 6: 140337052-140344304 | NLS, Cyt | 1044 | 348 | 35.29 | 9.89 |
| SOV2g029980.1 | Chr. 2: 101504484-101511484 | NLS, Chl | 1032 | 344 | 35.92 | 9.44 |
| SOV6g041780.1 | Chr. 6: 141900514-141905585 | NLS | 1026 | 342 | 35.70 | 8.33 |
| SOV3g001110.1 | Chr. 3: 1145951-1151558 | Chl, NLS | 1089 | 363 | 37.96 | 5.73 |
| SOV5g021850.1 | Chr. 5: 39407625-39411618 | NLS, Chl | 1089 | 363 | 37.31 | 7.87 |
| SOV5g038610.1 | Chr. 5: 122545368-122554893 | NLS | 1092 | 364 | 37.09 | 9.87 |
| SOV6g047350.1 | Chr. 6: 148521377-148529101 | NLS, Chl | 1413 | 471 | 50.13 | 8.79 |
| SOV4g002370.1 | Chr. 4: 2764521-2765501 | NLS, Cyt | 981 | 327 | 36.06 | 6.04 |
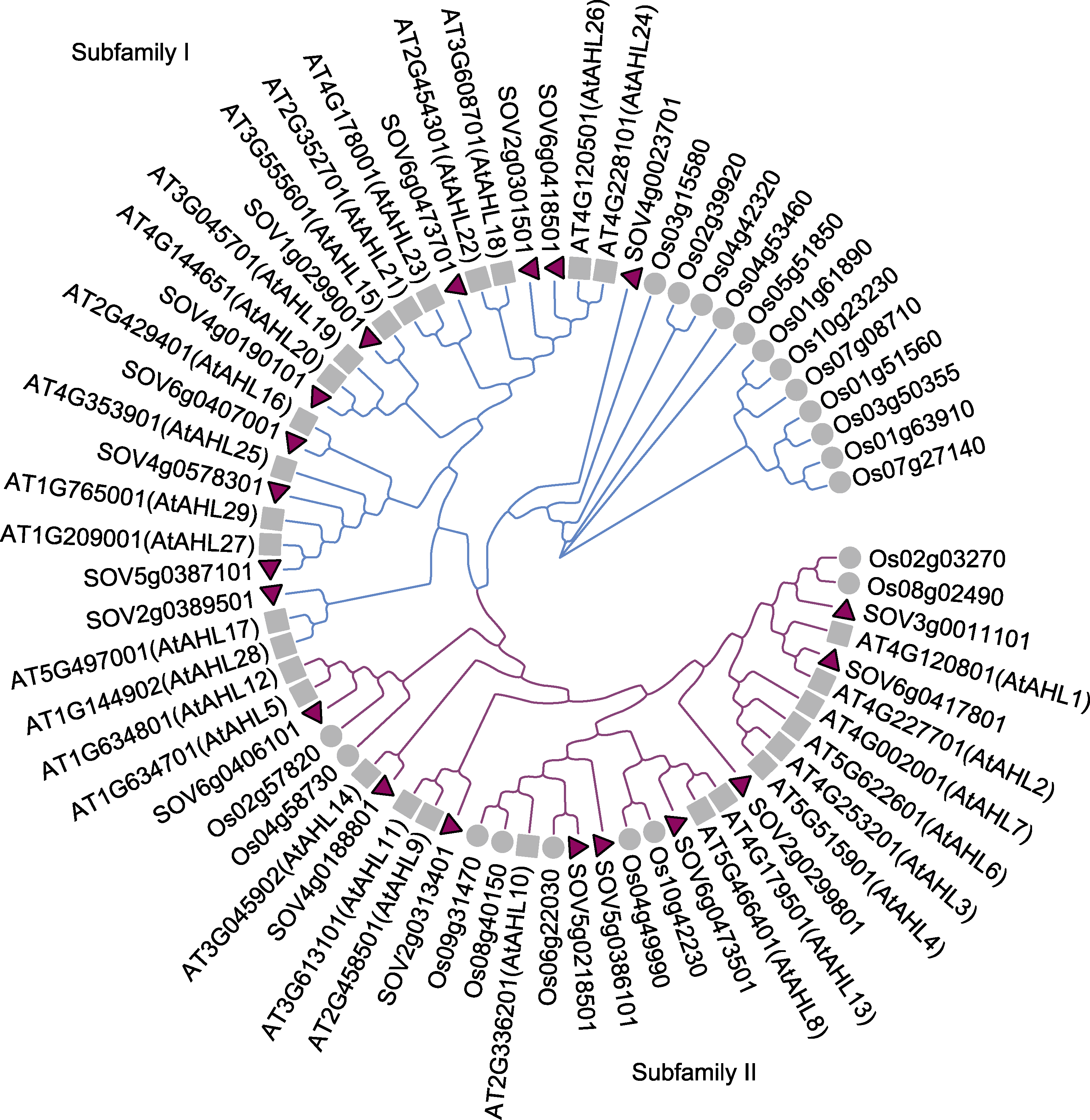
图1 菠菜、拟南芥和水稻AHL蛋白家族进化树分析 紫色表示I亚族, 蓝色表示II亚族。
Figure 1 Phylogenetic tree of AHL family members of spinach, Arabidopsis and rice Purple indicates subfamily I, and blue indicates subfamily II.
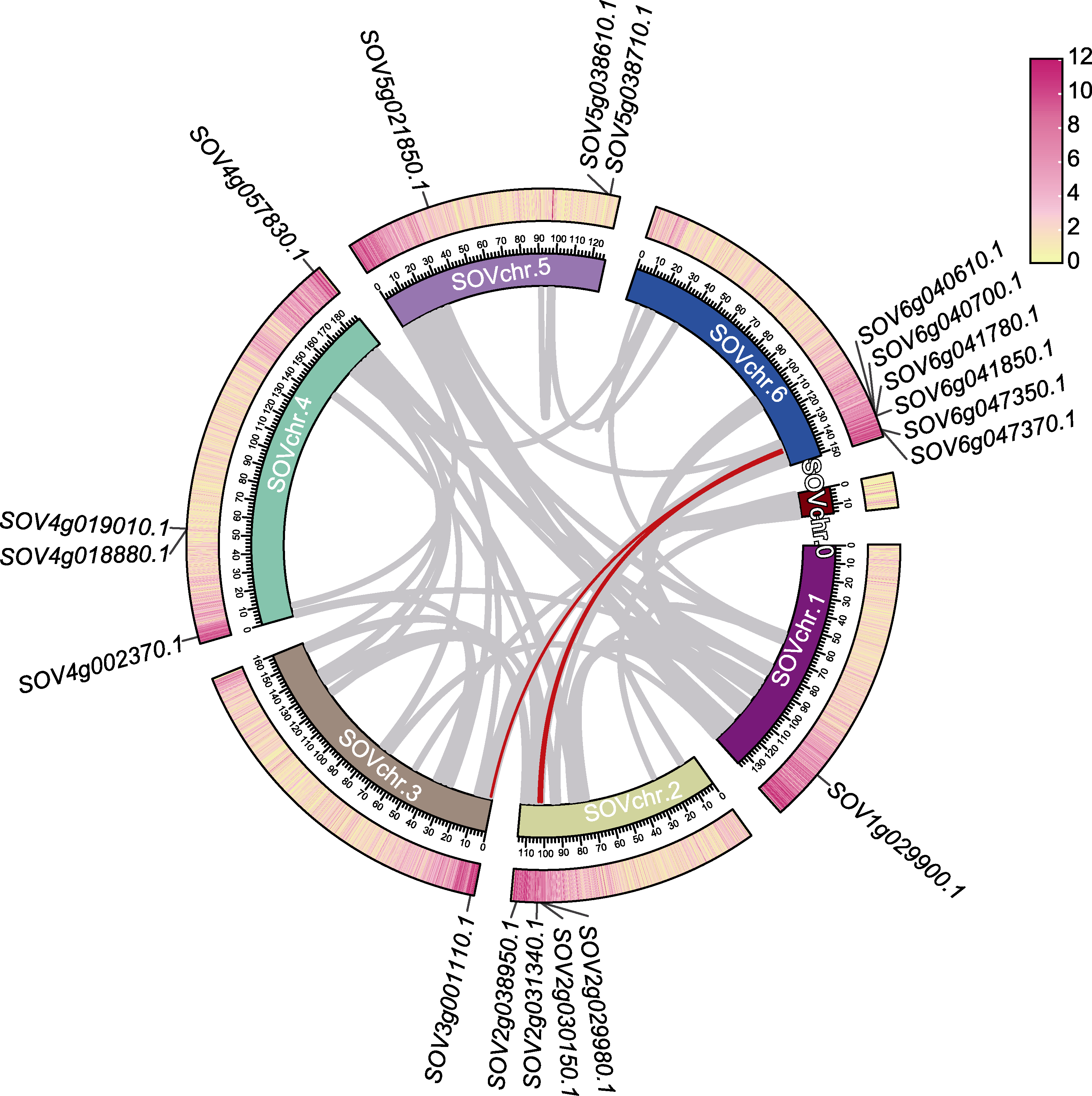
图4 菠菜SoAHL基因的区域共线性关系 具有同源性的染色体区域用红线连接SoAHL基因, 外环为基因密度热图。
Figure 4 Regional collinearity of the SoAHL gene in spinach The homologous chromosome regions were connected with the SoAHL genes by red lines, and the outer ring represents the gene density heatmap.
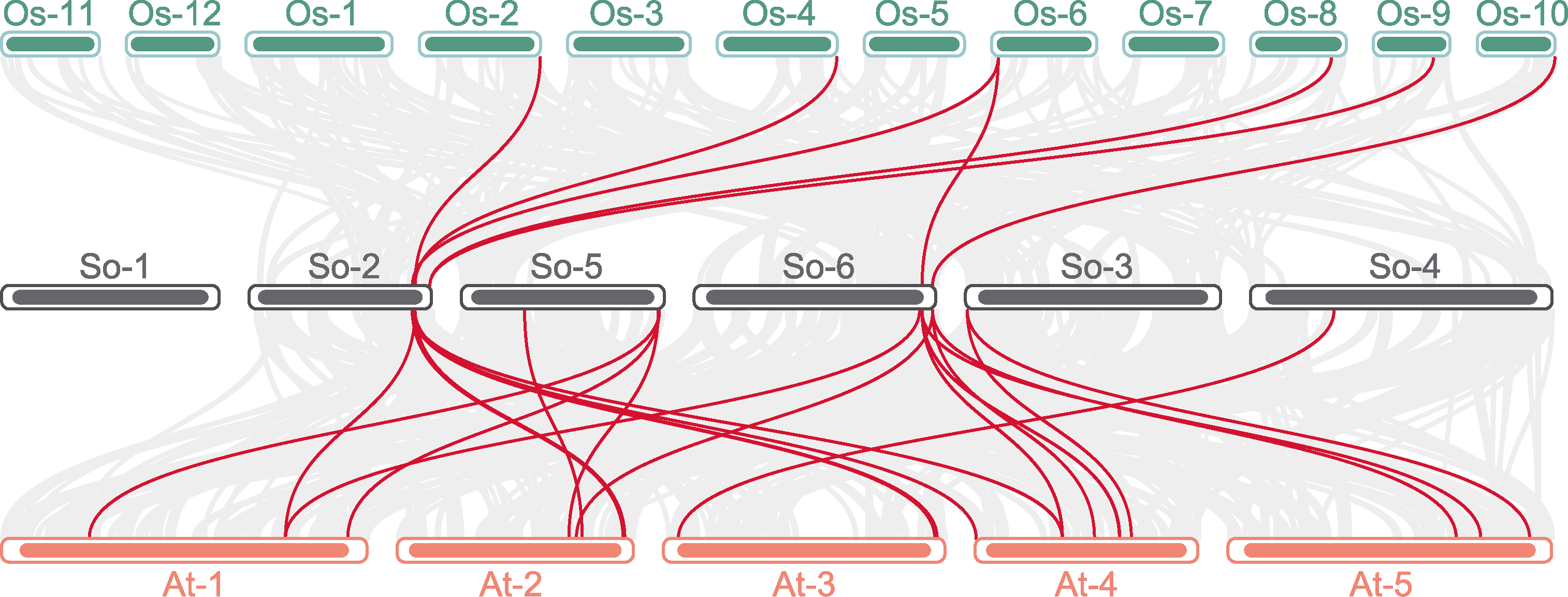
图5 AHL基因家族物种间共线性分析 红色线表示物种间AHL基因家族成员的共线性基因, 浅灰线表示各物种间包含的所有共线性基因。At: 拟南芥; Os: 水稻; So: 菠菜
Figure 5 Collinearity analysis of the AHL gene family in different species The red lines between the bars indicate collinear genes among members of the AHL gene family across species; the fine gray line shows all the collinear genes contained between species. At: Arabidopsis thaliana; Os: Oryza sativa; So: Spinacia oleracea
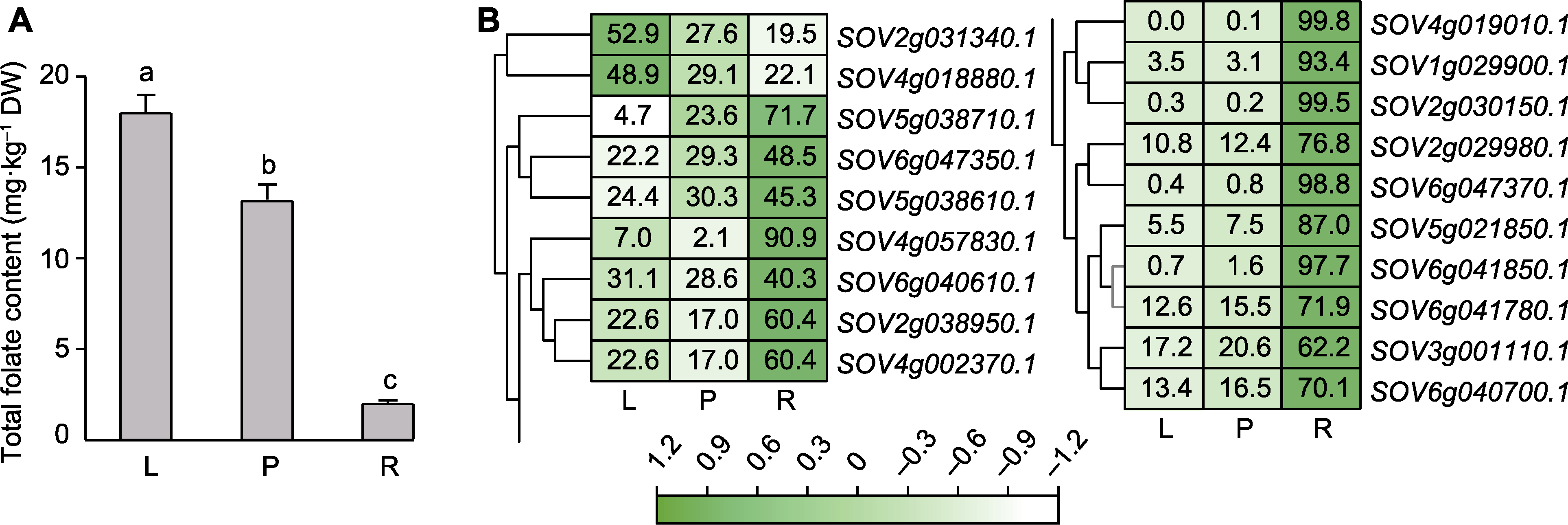
图7 菠菜3种组织叶酸含量(A)与SoAHL基因表达谱(B) L: 叶片; P: 叶柄; R: 根。不同小写字母表示差异显著(P<0.05)。
Figure 7 Folate content (A) and expression pattern of SoAHL gene (B) in three tissues of spinach L: Leaf; P: Petiole; R: Root. Different lowercase letters indicate significant differences(P<0.05).
| Salicylic acid | Days of treatments (d) | Shoot fresh biomass (g) | Root fresh biomass (g) | Shoot dry biomass (g) | Root dry biomass (g) |
|---|---|---|---|---|---|
| Mock | 0 | 0.737±0.171 d | 0.053±0.004 b | 0.083±0.013 b | 0.004±0.001 a |
| 1 | 1.042±0.049 c | 0.076±0.016 b | 0.083±0.013 b | 0.030±0.036 a | |
| 3 | 1.097±0.125 c | 0.066±0.008 b | 0.095±0.014 b | 0.008±0.002 a | |
| 5 | 1.304±0.017 b | 0.079±0.016 b | 0.132±0.028 a | 0.012±0.008 a | |
| 7 | 1.511±0.054 a | 0.104±0.016 a | 0.111±0.020 ab | 0.010±0.003 a | |
| SA50 | 0 | 0.800±0.190 c | 0.061±0.003 a | 0.087±0.015 a | 0.007±0.003 a |
| 1 | 1.123±0.116 b | 0.077±0.015 a | 0.123±0.032 a | 0.012±0.005 a | |
| 3 | 1.083±0.115 b | 0.088±0.056 a | 0.339±0.441 a | 0.010±0.003 a | |
| 5 | 1.303±0.080 ab | 0.069±0.010 a | 0.096±0.018 a | 0.010±0.006 a | |
| 7 | 1.501±0.085 a | 0.077±0.015 a | 0.094±0.038 a | 0.049±0.070 a |
表3 水杨酸(SA)处理对菠菜生物量的影响
Table 3 Effects of salicylic acid (SA) treatment on the biomass of spinach
| Salicylic acid | Days of treatments (d) | Shoot fresh biomass (g) | Root fresh biomass (g) | Shoot dry biomass (g) | Root dry biomass (g) |
|---|---|---|---|---|---|
| Mock | 0 | 0.737±0.171 d | 0.053±0.004 b | 0.083±0.013 b | 0.004±0.001 a |
| 1 | 1.042±0.049 c | 0.076±0.016 b | 0.083±0.013 b | 0.030±0.036 a | |
| 3 | 1.097±0.125 c | 0.066±0.008 b | 0.095±0.014 b | 0.008±0.002 a | |
| 5 | 1.304±0.017 b | 0.079±0.016 b | 0.132±0.028 a | 0.012±0.008 a | |
| 7 | 1.511±0.054 a | 0.104±0.016 a | 0.111±0.020 ab | 0.010±0.003 a | |
| SA50 | 0 | 0.800±0.190 c | 0.061±0.003 a | 0.087±0.015 a | 0.007±0.003 a |
| 1 | 1.123±0.116 b | 0.077±0.015 a | 0.123±0.032 a | 0.012±0.005 a | |
| 3 | 1.083±0.115 b | 0.088±0.056 a | 0.339±0.441 a | 0.010±0.003 a | |
| 5 | 1.303±0.080 ab | 0.069±0.010 a | 0.096±0.018 a | 0.010±0.006 a | |
| 7 | 1.501±0.085 a | 0.077±0.015 a | 0.094±0.038 a | 0.049±0.070 a |
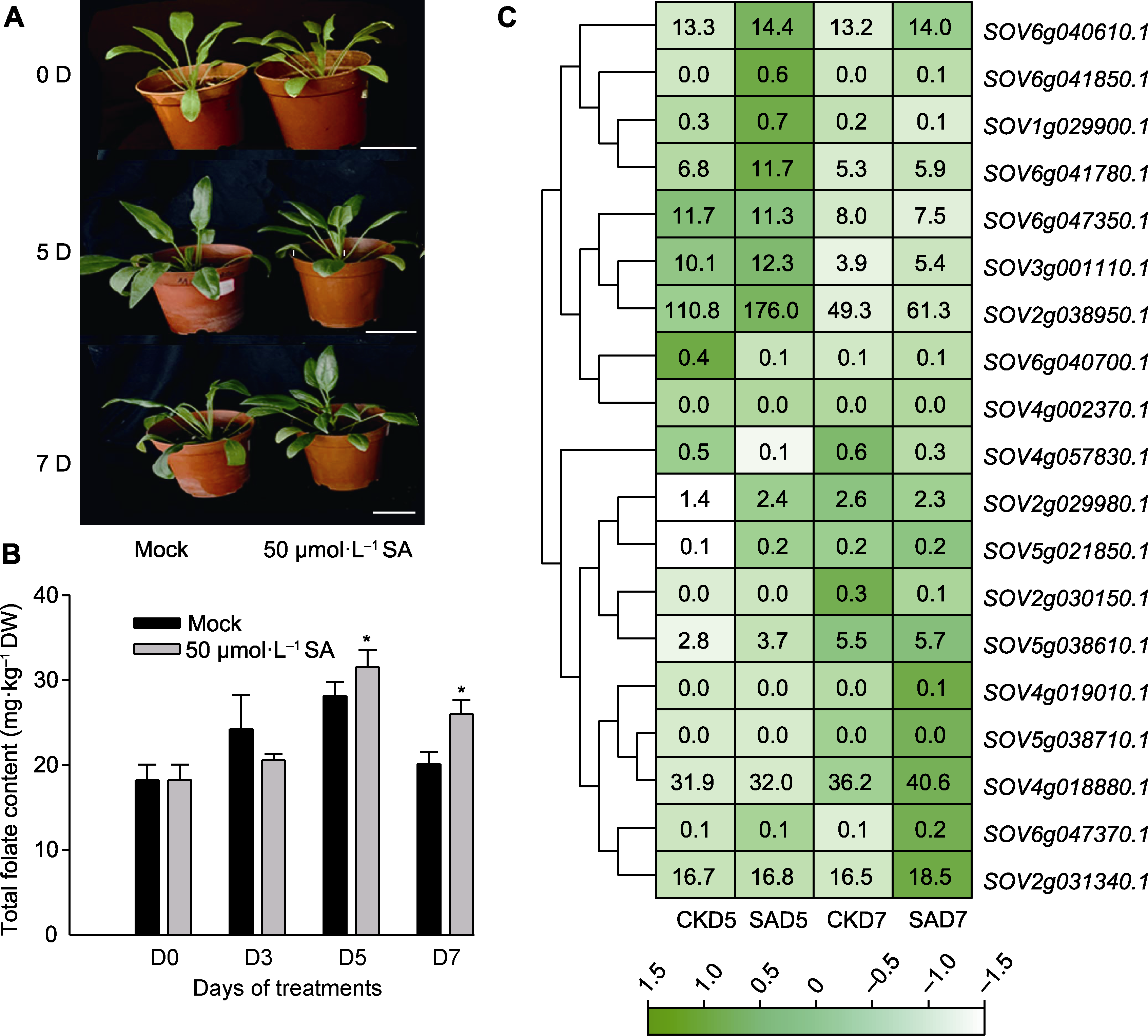
图8 水杨酸(SA)处理下菠菜表型(A)、叶酸含量(B)与SoAHL表达谱(C) CKD5: 对照组处理5天; SAD5: SA处理5天; CKD7: 对照组处理7天; SAD7: SA处理7天。星号表示同一时间下处理间差异显著(P<0.05, Tukey检验); 热图方框中数字为基因相对表达量。Bars=5 cm
Figure 8 Phenotype (A), total folate content (B) of spinach and expression analysis of SoAHL (C) under salicylic acid (SA) treatment CKD5: Control group for 5 days; SAD5: SA treatment for 5 days; CKD7: Control group for 7 days; SAD7: SA treatment for 7 days. The stars indicate significant differences between treatments at the same time (P<0.05, Tukey test); the numbers in the heat map boxes respresent gene relative expression. Bars=5 cm
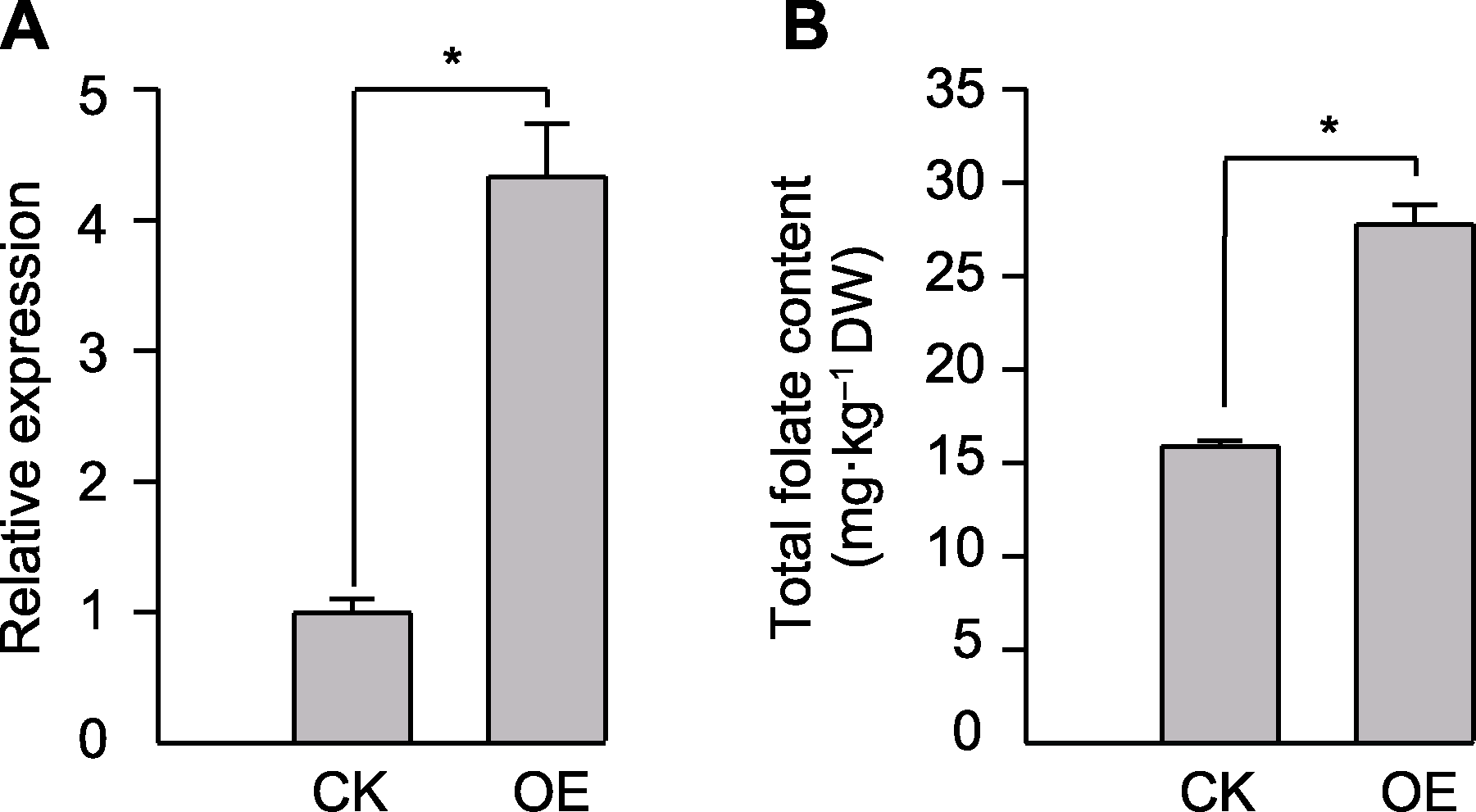
图9 SoAHLa瞬时过表达后基因的相对表达量(A)及菠菜叶片叶酸含量变化(B) CK: 注射空载体pEG100-SW的菠菜叶片; OE: 注射含SOV4g018880.1重组过表达载体的菠菜叶片。* 表示同一时间下处理间差异显著(P<0.05, Tukey检验)。
Figure 9 Changes in the relative expression level of the SoAHLa (A) and the folate content of leaf (B) after transient overexpression of SoAHLa CK: Spinach leaves injected with empty pEG100-SW; OE: Spinach leaves injected with vector containing SOV4g018880.1. * indicate significant differences between treatments at the same time (P<0.05, Tukey test).
| [1] |
Bishop EH, Kumar R, Luo F, Saski C, Sekhon RS (2020). Genome-wide identification, expression profiling, and network analysis of AT-hook gene family in maize. Genomics 112, 1233-1244.
DOI PMID |
| [2] |
Cai GQ, Kim SC, Li JW, Zhou YM, Wang XM (2020). Transcriptional regulation of lipid catabolism during seedling establishment. Mol Plant 13, 984-1000.
DOI PMID |
| [3] |
Chen CJ, Wu Y, Li JW, Wang X, Zeng ZH, Xu J, Liu YL, Feng JT, Chen H, He YH, Xia R (2023). TBtools-II: a "one for all, all for one" bioinformatics platform for biological big-data mining. Mol Plant 16, 1733-1742.
DOI PMID |
| [4] | Delaney SK, Orford SJ, Martin-Harris M, Timmis JN (2007). The fiber specificity of the cotton FSltp4 gene promoter is regulated by an AT-rich promoter region and the AT-hook transcription factor GhAT1. Plant Cell Physiol 48, 1426-1437. |
| [5] |
Ding LX, Li T, Li ZL, Xu XW, Li Y, Wang HM, Wang YF, Ma SM, Li ZX (2016). Genome-wide identification and expression analysis in oxidative stress of AT-hook gene family in tomato. J Plant Genet Resour 17, 303-315. (in Chinese)
DOI |
|
丁丽雪, 李涛, 李植良, 徐小万, 李颖, 王恒明, 王永飞, 马三梅, 黎振兴 (2016). 番茄AT-hook基因家族的鉴定及胁迫条件下的表达分析. 植物遗传资源学报 17, 303-315.
DOI |
|
| [6] | Eckner R, Birnstiel ML (1989). Cloning of cDNAs coding for human HMG I and HMG Y proteins: both are capable of binding to the octamer sequence motif. Nucl Acids Res 17, 5947-5959. |
| [7] |
Favero DS, Kawamura A, Shibata M, Takebayashi A, Jung JH, Suzuki T, Jaeger KE, Ishida T, Iwase A, Wigge PA, Neff MM, Sugimoto K (2020). AT-hook transcription factors restrict petiole growth by antagonizing PIFs. Curr Biol 30, 1454-1466.
DOI PMID |
| [8] | Feng CY, Li T, Li ZX, Li ZL, Xu XW, Ding LX, Wang YF (2015). Expression analysis of root-specific genes in tomato. Plant Physiol J 51, 921-934. (in Chinese) |
| 冯婵莹, 李涛, 黎振兴, 李植良, 徐小万, 丁丽雪, 王永飞 (2015). 番茄根特异基因的表达分析. 植物生理学报 51, 921-934. | |
| [9] | Hu DX, Liu H, Liang XQ, Wu ZM, Fang JH (2021). Bioinformatics analysis of AT-hook genes family in peanut (Arachis hypogaea L.). Chin J Trop Crops 42, 649-659. (in Chinese) |
|
胡冬秀, 刘浩, 梁炫强, 吴自明, 方加海 (2021). 花生AT-hook家族基因的生物信息学分析. 热带作物学报 42, 649-659.
DOI |
|
| [10] | Huang PX, Dong Z, Guo PR, Zhang X, Qiu YP, Li BS, Wang YC, Guo HW (2020). Salicylic acid suppresses apical hook formation via NPR1-mediated repression of EIN3 and EIL1in Arabidopsis. Plant Cell 32, 612-629. |
| [11] | Jacques CN, Favero DS, Kawamura A, Suzuki T, Sugimoto K, Neff MM (2022). SUPPRESSOR OF PHYTOCHROME B-4 #3 reduces the expression of PIF-activated genes and increases expression of growth repressors to regulate hypocotyl elongation in short days. BMC Plant Biol 22, 399. |
| [12] |
Jeffares DC, Penkett CJ, Bähler J (2008). Rapidly regulated genes are intron poor. Trends Genet 24, 375-378.
DOI PMID |
| [13] | Jeong HN, Sun HJ, Zuo ZF, Lee DH, Song PS, Kang HG, Lee HY (2020). Overexpression of ATHG1/AHL23 and ATPG3/AHL20,Arabidopsis AT-hook motif nuclear-localized genes, confers salt tolerance in transgenic Zoysia japonica. Plant Biotechnol Rep 14, 351-361. |
| [14] | Kim SY, Kim YC, Seong ES, Lee YH, Park JM, Choi D (2007). The chili pepper CaATL1: an AT-hook motif-containing transcription factor implicated in defence responses against pathogens. Mol Plant Pathol 8, 761-771. |
| [15] | Li XL, He HH, Wang H, Wu XQ, Wang H, Mao J (2021). Identification and expression analysis of the AHL gene family in grape (Vitis vinifera). Plant Gene 26, 100285. |
| [16] | Liu HH, Chong PF (2023). Bioinformatics analysis of the AHL gene family in Populus euphratica and its expression characteristics under stress. Acta Agrestia Sin 31, 741-750. (in Chinese) |
|
刘行行, 种培芳 (2023). 胡杨AHL基因家族生物信息学分析及逆境胁迫下的表达特征. 草地学报 31, 741-750.
DOI |
|
| [17] |
Livak KJ, Schmittgen TD (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCTmethod. Methods 25, 402-408.
DOI PMID |
| [18] | Lu HB, Zou Y, Feng N (2010). Overexpression of AHL20 negatively regulates defenses in Arabidopsis. J Integr Plant Biol 52, 801-808. |
| [19] | Pan X (2023). Screening and Identification of Upstream Regulatory Factors of SoGCH1 and SoPTAR Genes in Spinach. Master's thesis. Shanghai: Shanghai Normal University. pp. 52-60. (in Chinese) |
| 潘曦 (2023). 菠菜SoGCH1与SoPTAR基因上游调控因子筛选及鉴定. 硕士论文. 上海: 上海师范大学. pp. 52-60. | |
| [20] | Pan X, Sun F, Zhang JY, Yang F, Wang QH, Wang XL (2023). Research progress on folate accumulation in horticultural plants. China Veg (4), 29-38. (in Chinese) |
| 潘曦, 孙飞, 张玖漪, 杨帆, 王全华, 王小丽 (2023). 园艺植物叶酸积累规律研究进展. 中国蔬菜 (4), 29-38. | |
| [21] |
Puthusseri B, Divya P, Lokesh V, Neelwarne B (2013). Salicylic acid-induced elicitation of folates in coriander (Coriandrum sativum L.) improves bioaccessibility and reduces pro-oxidant status. Food Chem 136, 569-575.
DOI PMID |
| [22] | Scott J, Rébeillé F, Fletcher J (2000). Folic acid and folates: the feasibility for nutritional enhancement in plant foods. J Sci Food Agric 80, 795-824. |
| [23] | Širl M, Šnajdrová T, Gutiérrez-Alanís D, Dubrovsky JG, Vielle-Calzada JP, Kulich I, Soukup A (2020). AT-hook motif nuclear localised protein 18 as a novel modulator of root system architecture. Int J Mol Sci 21, 1886. |
| [24] | Sun JL (2023). Function Analysis of PuAHL17 Gene for Drought Tolerance in Populus ussuriensis. Master's thesis. Harbin:Northeast Forestry University. pp. 32-50. (in Chinese) |
| 孙佳丽 (2023). 大青杨PuAHL17基因抗旱功能研究. 硕士论文. 哈尔滨: 东北林业大学. pp. 32-50. | |
| [25] |
Tayengwa R, Sharma Koirala P, Pierce CF, Werner BE, Neff MM (2020). Overexpression of AtAHL20 causes delayed flowering in Arabidopsis via repression of FT expression. BMC Plant Biol 20, 559.
DOI PMID |
| [26] | Wang LY, Li TT, Liu N, Liu XC (2023). Identification of tomato AHL gene families and functional analysis their roles in fruit development and abiotic stress response. Plant Physiol Biochem 202, 107931. |
| [27] | Wang M, Chen BW, Zhou W, Xie LN, Wang LS, Zhang YL, Zhang QZ (2021a). Genome-wide identification and expression analysis of the AT-hook Motif Nuclear Localized gene family in soybean. BMC Genomics 22, 361. |
| [28] | Wang XL, Cai XF, Xu CX, Wang QH (2021b). Identification and characterization of the NPF, NRT2 and NRT3 in spinach. Plant Physiol Biochem 158, 297-307. |
| [29] | Xiao CW, Chen FL, Yu XH, Lin CT, Fu YF (2009). Over-expression of an AT-hook gene, AHL22, delays flowering and inhibits the elongation of the hypocotyl in Arabidopsis thaliana. Plant Mol Biol 71, 39-50. |
| [30] | Yun J, Kim YS, Jung JH, Seo PJ, Park CM (2012). The AT-hook motif-containing protein AHL22 regulates flowering initiation by modifying FLOWERING LOCUS chromatin in Arabidopsis. J Biol Chem 287, 15307-15316. |
| [31] | Zeng QK, Song L, Xia MZ, Zheng Z, Chen ZA, Che XM, Liu D (2023). Overexpression of AHL proteins enhances root hair production by altering the transcription of RHD6- downstream genes. Plant Direct 7, e517. |
| [32] | Zhang DY, Qi WC, Wan Q, Liu J, Xu ZL, Huang YH, Shao HB (2017). Cloning and localization analysis on five AT- hook genes GmAHLs from Glycine max. J Plant Resour Environ 26(4), 1-7. (in Chinese) |
| 张大勇, 戚维聪, 万群, 刘佳, 徐照龙, 黄益洪, 邵宏波 (2017). 5个大豆AT-hook基因GmAHLs的克隆与定位分析. 植物资源与环境学报 26(4), 1-7. | |
| [33] | Zhang GW, Zeng Y, Guo W, Luo Q (2014). Bioinformatics analysis of the AT-hook gene family in rice. Chin Bull Bot 49, 49-62. (in Chinese) |
|
张贵慰, 曾珏, 郭维, 罗琼 (2014). 水稻AT-hook基因家族生物信息学分析. 植物学报 49, 49-62.
DOI |
|
| [34] | Zhang JY, Cai XF, Xu CX, Wang QH, Wang XL (2020). Gene identification and expression profiling analysis of spinach folate anabolic pathway. J Shanghai Norm Univ (Nat Sci) 49, 637-649. (in Chinese) |
| 张玖漪, 蔡晓锋, 徐晨曦, 王全华, 王小丽 (2020). 菠菜叶酸合成代谢途径基因鉴定及表达谱分析. 上海师范大学学报(自然科学版) 49, 637-649. | |
| [35] | Zhang SL, Wang T, Lima RM, Pettkó-Szandtner A, Ker- eszt A, Downie JA, Kondorosi E (2023). Widely conserved AHL transcription factors are essential for NCR gene expression and nodule development in Medicago. Nat Plants 9, 280-288. |
| [36] | Zhang WM, Cheng XZ, Fang D, Cao J (2022). AT-hook motif nuclear localized (AHL) proteins of ancient origin radiate new functions. Int J Biol Macromol 214, 290-300. |
| [37] | Zhang YW, Wang XT, Zhang XL, Tang XN, Liu J, Guo JC (2024). Cloning and expression analysis of MeAHL17 gene in cassava. Chi J Trop Crops 45, 1303-1313. (in Chinese) |
|
张亚文, 王晓彤, 张兴龙, 唐湘宁, 刘姣, 郭建春 (2024). 木薯MeAHL17基因的克隆及表达分析. 热带作物学报 45, 1303-1313.
DOI |
|
| [38] | Zhao JF, Favero DS, Peng H, Neff MM (2013). Arabidopsis thaliana AHL family modulates hypocotyl growth redundantly by interacting with each other via the PPC/DUF296 domain. Proc Natl Acad Sci USA 110, E4688-E4697. |
| [39] | Zhao LJ, Lü YJ, Chen W, Yao JB, Li Y, Li QL, Pan JW, Fang ST, Sun J, Zhang YS (2020). Genome-wide identification and analyses of the AHL gene family in cotton (Gossypium). BMC Genomics 21, 69. |
| [40] | Zhou YS, Zhang XM, Chen J, Guo XP, Wang HY, Zhen WB, Zhang JL, Hu ZB, Zhang XB, Botella JR, Ito T, Guo SY (2022). Overexpression of AHL9 accelerates leaf senescence in Arabidopsis thaliana. BMC Plant Biol 22, 248. |
| [41] |
Zhu M, Yan BW, Hu YJ, Cui ZB, Wang XX (2020). Genome-wide identification and phylogenetic analysis of rice FTIP gene family. Genomics 112, 3803-3814.
DOI PMID |
| [1] | 史世肸, 严顺平. 高效液相色谱法检测水杨酸的优化[J]. 植物学报, 2025, 60(5): 1-0. |
| [2] | 粟思琳 唐先宇 陈祎 王婷 夏石头. 系统获得性抗性的转录调控[J]. 植物学报, 2025, 60(5): 1-0. |
| [3] | 叶灿, 姚林波, 金莹, 高蓉, 谭琪, 李旭映, 张艳军, 陈析丰, 马伯军, 章薇, 张可伟. 水稻水杨酸代谢突变体高通量筛选方法的建立与应用[J]. 植物学报, 2025, 60(4): 1-0. |
| [4] | 袁民航, 辛秀芳. 烽火狼烟: 水杨酸甲酯介导的植物间通讯和气传性免疫的机制解析[J]. 植物学报, 2023, 58(5): 682-686. |
| [5] | 园园, 恩和巴雅尔, 齐艳华. 植物GH3基因家族生物学功能研究进展[J]. 植物学报, 2023, 58(5): 770-782. |
| [6] | 赵宇慧, 李秀秀, 陈倬, 鲁宏伟, 刘羽诚, 张志方, 梁承志. 生物信息学分析方法I: 全基因组关联分析概述[J]. 植物学报, 2020, 55(6): 715-732. |
| [7] | 李格,孟小庆,李宗芸,朱明库. 甘薯盐胁迫响应基因IbMYB3的表达特征及生物信息学分析[J]. 植物学报, 2020, 55(1): 38-48. |
| [8] | 曹栋栋,陈珊宇,秦叶波,吴华平,阮关海,黄玉韬. 水杨酸调控盐胁迫下羽衣甘蓝种子萌发的机理[J]. 植物学报, 2020, 55(1): 49-61. |
| [9] | 徐悦,曹英萍,王玉,付春祥,戴绍军. 发根农杆菌介导的菠菜毛状根遗传转化体系的建立[J]. 植物学报, 2019, 54(4): 515-521. |
| [10] | 程广前,贾克利,李娜,邓传良,李书粉,高武军. 石刁柏核质体DNA的生物信息学分析及染色体定位[J]. 植物学报, 2019, 54(3): 328-334. |
| [11] | 代宇佳,罗晓峰,周文冠,陈锋,帅海威,杨文钰,舒凯. 生物和非生物逆境胁迫下的植物系统信号[J]. 植物学报, 2019, 54(2): 255-264. |
| [12] | 刘魏, 童永鳌, 白洁. 水稻雄配子体发育过程中tRNA片段的生物信息学分析[J]. 植物学报, 2018, 53(5): 625-633. |
| [13] | 贾乐东, 李施蒙, 许代香, 曲存民, 李加纳, 王瑞. 甘蓝型油菜BnMYB80基因的生物信息学分析[J]. 植物学报, 2016, 51(5): 620-630. |
| [14] | 程甜, 魏强, 李广林. 中粒咖啡萜类合成酶基因家族的生物信息学分析[J]. 植物学报, 2016, 51(2): 235-250. |
| [15] | 李冬梅, 王路雅, 张澜玥, 帖子阳, 毛惠平. 拟南芥短肽激素PROPEP基因家族在根生长中的作用机理[J]. 植物学报, 2016, 51(2): 202-209. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||