

植物学报 ›› 2024, Vol. 59 ›› Issue (1): 34-53.DOI: 10.11983/CBB23004 cstr: 32102.14.CBB23004
顾家琦1,2,†, 朱福慧1,2,†, 谢沛豪1,2, 孟庆营1,2, 郑颖2, 张献龙1,2, 袁道军1,2,3,*( )
)
收稿日期:2023-01-02
接受日期:2023-07-08
出版日期:2024-01-10
发布日期:2024-01-10
通讯作者:
*E-mail: 作者简介:†共同第一作者
基金资助:
Jiaqi Gu1,2,†, Fuhui Zhu1,2,†, Peihao Xie1,2, Qingying Meng1,2, Ying Zheng2, Xianlong Zhang1,2, Daojun Yuan1,2,3,*( )
)
Received:2023-01-02
Accepted:2023-07-08
Online:2024-01-10
Published:2024-01-10
Contact:
*E-mail: About author:†These authors contributed equally to this paper
摘要: 光敏色素(PHY)作为植物体内重要的红光(R)和远红光(FR)受体, 在调节植物花期、提高作物产量以及调控植物抗逆性方面均具有重要作用。鉴定棉花PHY基因, 探究其驯化和改良的遗传调控网络, 发掘棉花光敏色素关键基因, 为早熟棉花品种的从头驯化和育种提供新见解。以5个拟南芥(Arabidopsis thaliana)光敏色素基因作为种子序列, 利用生物信息学方法, 对不同棉种中的PHY基因进行全基因组系统鉴定。系统发育分析表明, 锦葵科植物中的PHY基因包括PHYA、PHYB、PHYC与PHYE四个亚家族。PHY基因在陆地棉(Gossypium hirsutum)不同群体间的驯化选择分析结果表明, PHY基因的驯化过程可分为早期驯化和后期遗传改良2个阶段。对陆地棉野生种和栽培品种在长、短日照下的转录组分析显示, GhPHYA1Dt和GhPHYB1Dt在长、短日照下的表达差异显著; 在长日照处理14小时后, 栽培品种GhPHYC1At和GHPHYE1At的表达量显著低于野生种。研究结果为进一步揭示棉花PHY基因的驯化选择和功能机制奠定了基础, 为棉花早熟新品种选育和从头驯化提供了理论依据。
顾家琦, 朱福慧, 谢沛豪, 孟庆营, 郑颖, 张献龙, 袁道军. 棉属光敏色素PHY基因家族的全基因组鉴定与驯化选择分析. 植物学报, 2024, 59(1): 34-53.
Jiaqi Gu, Fuhui Zhu, Peihao Xie, Qingying Meng, Ying Zheng, Xianlong Zhang, Daojun Yuan. Genome-wide Identification and Domestication Analysis of the Phytochrome PHY Gene Family in Gossypium. Chinese Bulletin of Botany, 2024, 59(1): 34-53.
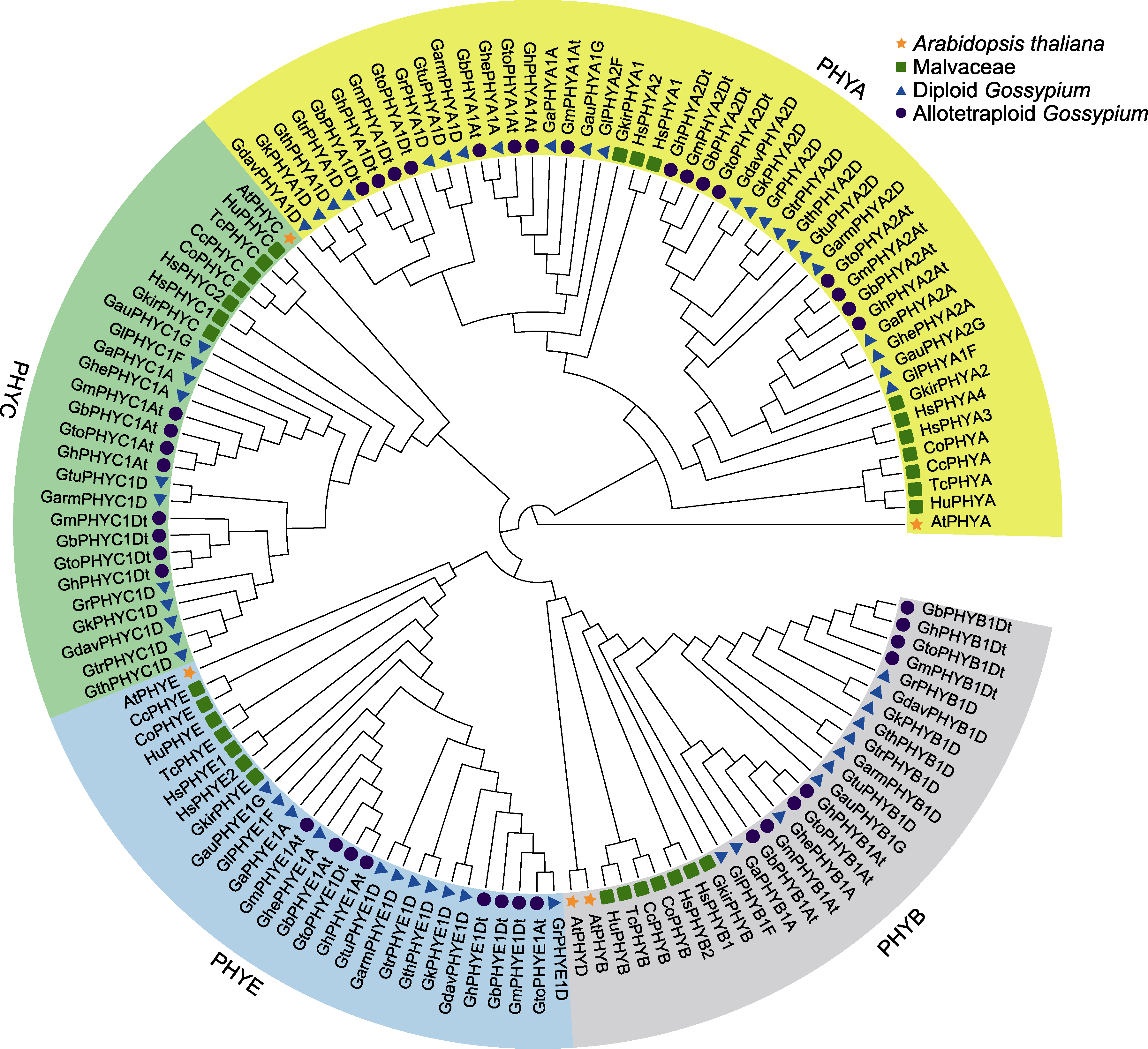
图1 棉属及其它植物中PHY家族成员的系统发育分析 At: 拟南芥; Cc: 圆果种黄麻; Co: 长果种黄麻; Ga: 亚洲棉; Garm: 辣根棉; Gau: 澳洲棉; Gb: 海岛棉; Gdav: 戴维逊棉; Gh: 陆地棉; Ghe: 草棉; Gk: 克劳茨基棉; Gl: 长萼棉; Gm: 黄褐棉; Gr: 雷蒙德氏棉; Gth: 瑟伯氏棉; Gto: 毛棉; Gtr: 三裂棉; Gtu: 特纳氏棉; Gkir: 叉柱棉; Hs: 木槿; Hu: 哥伦比亚锦葵; Tc: 可可
Figure 1 Phylogenetic analysis of PHY family members of Gossypium and other plants At: Arabidopsis thaliana; Cc: Corchorus capsularis; Co: C. olitorius; Ga: G. arboreum; Garm: G. armourianum; Gau: G. australe; Gb: G. barbadense; Gdav: G. davidsonii; Gh: G. hirsutum; Ghe: G. herbaceum; Gk: G. klotzschianum; Gl: G. longicalyx; Gm: G. mustelinum; Gr: G. raimondii; Gth: G. thurberi; Gto: G. tomentosum; Gtr: G. trilobum; Gtu: G. turneri; Gkir: Gossypioides kirkii; Hs: Hibiscus syriacus; Hu: Herrania umbratica; Tc: Theobroma cacao
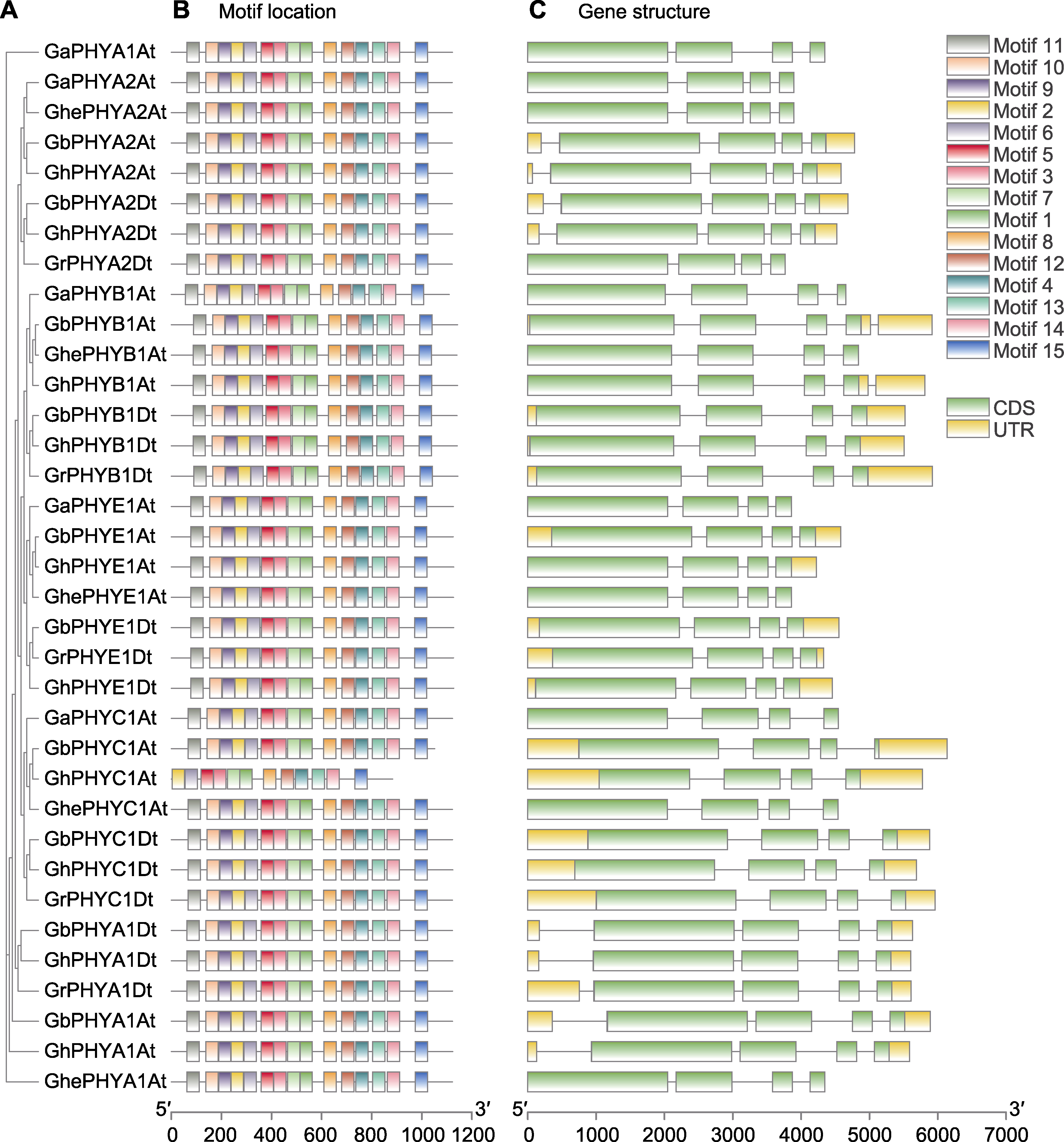
图2 棉花PHY基因结构和蛋白保守基序分析 (A) 系统进化树; (B) 蛋白保守基序; (C) 基因结构
Figure 2 Analysis of the PHY gene structure and protein conserved motifs of Gossypium (A) Phylogenetic tree; (B) Protein conserved motif; (C) Gene structure
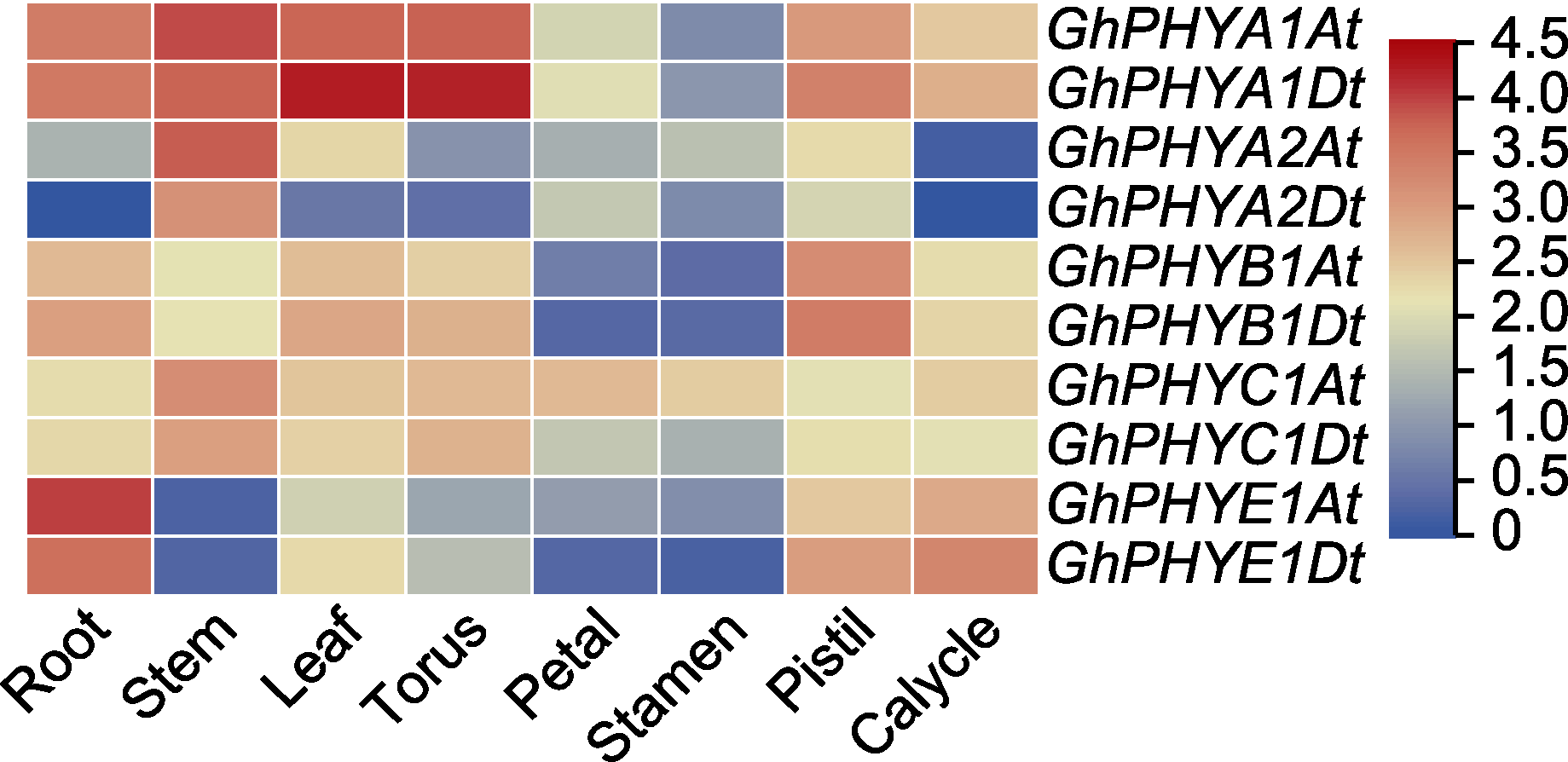
图4 陆地棉PHY基因在不同器官和组织中的表达聚类分析 不同颜色的矩形代表陆地棉不同PHY基因的表达水平, 即log2 (1+FPKM)。
Figure 4 Clustering analysis of the expression of Gossypium hirsutum PHY genes in different organs and tissues The rectangles with different colors represent the expression levels of different PHY genes in upland cotton, namely log2(1+ FPKM).
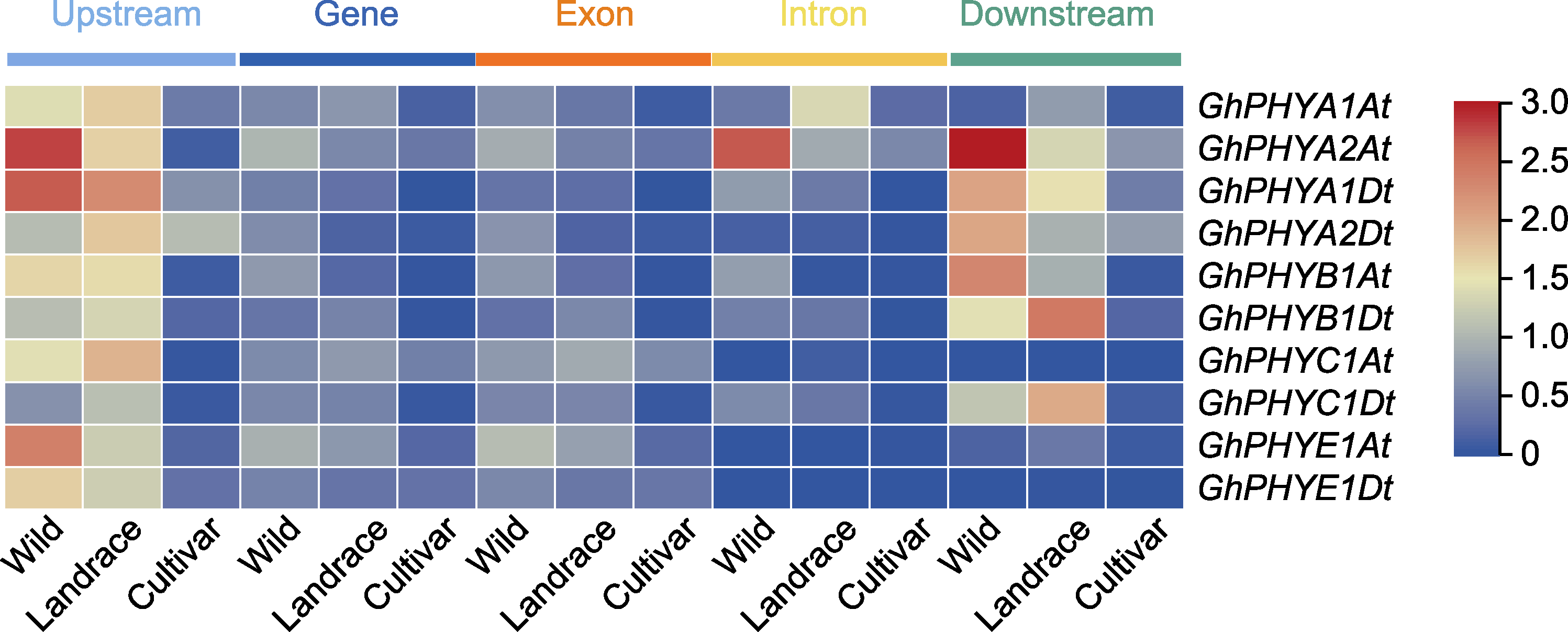
图5 陆地棉PHY基因在不同群体间的核苷酸多样性分析 不同颜色的矩形代表陆地棉不同材料中每个基因及其上、下游、外显子和内含子的核苷酸多样性(Pi×10-4)。
Figure 5 Analysis of nucleotide diversity of Gossypium hirsutum PHY genes among different populations Different color rectangles represent the nucleotide diversity (Pi×10-4 value) of each gene and its upstream, downstream, exon and intron in different materials of upland cotton.
| Gene name | Gene ID | PiWild/ PiLandrace | PiLandrace/ PiCultivar |
|---|---|---|---|
| GhPHYA1At | Gohir.A11G261900 | 0.7891 | 5.1234 |
| GhPHYA2At | Gohir.A13G172900 | 1.8274 | 1.4509 |
| GhPHYA1Dt | Gohir.D11G271900 | 1.4826 | 99.2593 |
| GhPHYA2Dt | Gohir.D13G178700 | 3.5917 | 2.3415 |
| GhPHYB1At | Gohir.A10G172500 | 3.4313 | 16.8376 |
| GhPHYB1Dt | Gohir.D10G179000 | 0.6808 | 38.3027 |
| GhPHYC1At | Gohir.A11G149100 | 0.7955 | 1.5879 |
| GhPHYC1Dt | Gohir.D11G156200 | 1.0780 | 15.8177 |
| GhPHYE1At | Gohir.A10G037800 | 1.3580 | 3.4484 |
| GhPHYE1Dt | Gohir.D10G038800 | 1.4530 | 1.0696 |
表1 陆地棉不同群体间PHY基因的核苷酸多样性(Pi)比值
Table 1 Nucleotide diversity (Pi) ratio of PHY genes in Gossypium hirsutum among different populations
| Gene name | Gene ID | PiWild/ PiLandrace | PiLandrace/ PiCultivar |
|---|---|---|---|
| GhPHYA1At | Gohir.A11G261900 | 0.7891 | 5.1234 |
| GhPHYA2At | Gohir.A13G172900 | 1.8274 | 1.4509 |
| GhPHYA1Dt | Gohir.D11G271900 | 1.4826 | 99.2593 |
| GhPHYA2Dt | Gohir.D13G178700 | 3.5917 | 2.3415 |
| GhPHYB1At | Gohir.A10G172500 | 3.4313 | 16.8376 |
| GhPHYB1Dt | Gohir.D10G179000 | 0.6808 | 38.3027 |
| GhPHYC1At | Gohir.A11G149100 | 0.7955 | 1.5879 |
| GhPHYC1Dt | Gohir.D11G156200 | 1.0780 | 15.8177 |
| GhPHYE1At | Gohir.A10G037800 | 1.3580 | 3.4484 |
| GhPHYE1Dt | Gohir.D10G038800 | 1.4530 | 1.0696 |
| Gene name | Gene ID | FstWild/ FstLandrace | FstLandrace/ FstCultivar |
|---|---|---|---|
| GhPHYA1At | Gohir.A11G261900 | 0.5843 | 0.2320 |
| GhPHYA2At | Gohir.A13G172900 | 0.4386 | 0.1680 |
| GhPHYA1Dt | Gohir.D11G271900 | 0.0569 | 0.2137 |
| GhPHYA2Dt | Gohir.D13G178700 | 0.6049 | 0.0374 |
| GhPHYB1At | Gohir.A10G172500 | 0.7068 | 0.5372 |
| GhPHYB1Dt | Gohir.D10G179000 | 0.5158 | 0.7092 |
| GhPHYC1At | Gohir.A11G149100 | 0.2304 | 0.4461 |
| GhPHYC1Dt | Gohir.D11G156200 | 0.5470 | 0.8466 |
| GhPHYE1At | Gohir.A10G037800 | 0.4756 | 0.4709 |
| GhPHYE1Dt | Gohir.D10G038800 | 0.4096 | 0.1872 |
表2 陆地棉PHY基因在不同群体间的遗传分化系数(Fst)分析
Table 2 Analysis of genetic differentiation coefficients (Fst) of PHY genes of Gossypium hirsutum among different populations
| Gene name | Gene ID | FstWild/ FstLandrace | FstLandrace/ FstCultivar |
|---|---|---|---|
| GhPHYA1At | Gohir.A11G261900 | 0.5843 | 0.2320 |
| GhPHYA2At | Gohir.A13G172900 | 0.4386 | 0.1680 |
| GhPHYA1Dt | Gohir.D11G271900 | 0.0569 | 0.2137 |
| GhPHYA2Dt | Gohir.D13G178700 | 0.6049 | 0.0374 |
| GhPHYB1At | Gohir.A10G172500 | 0.7068 | 0.5372 |
| GhPHYB1Dt | Gohir.D10G179000 | 0.5158 | 0.7092 |
| GhPHYC1At | Gohir.A11G149100 | 0.2304 | 0.4461 |
| GhPHYC1Dt | Gohir.D11G156200 | 0.5470 | 0.8466 |
| GhPHYE1At | Gohir.A10G037800 | 0.4756 | 0.4709 |
| GhPHYE1Dt | Gohir.D10G038800 | 0.4096 | 0.1872 |
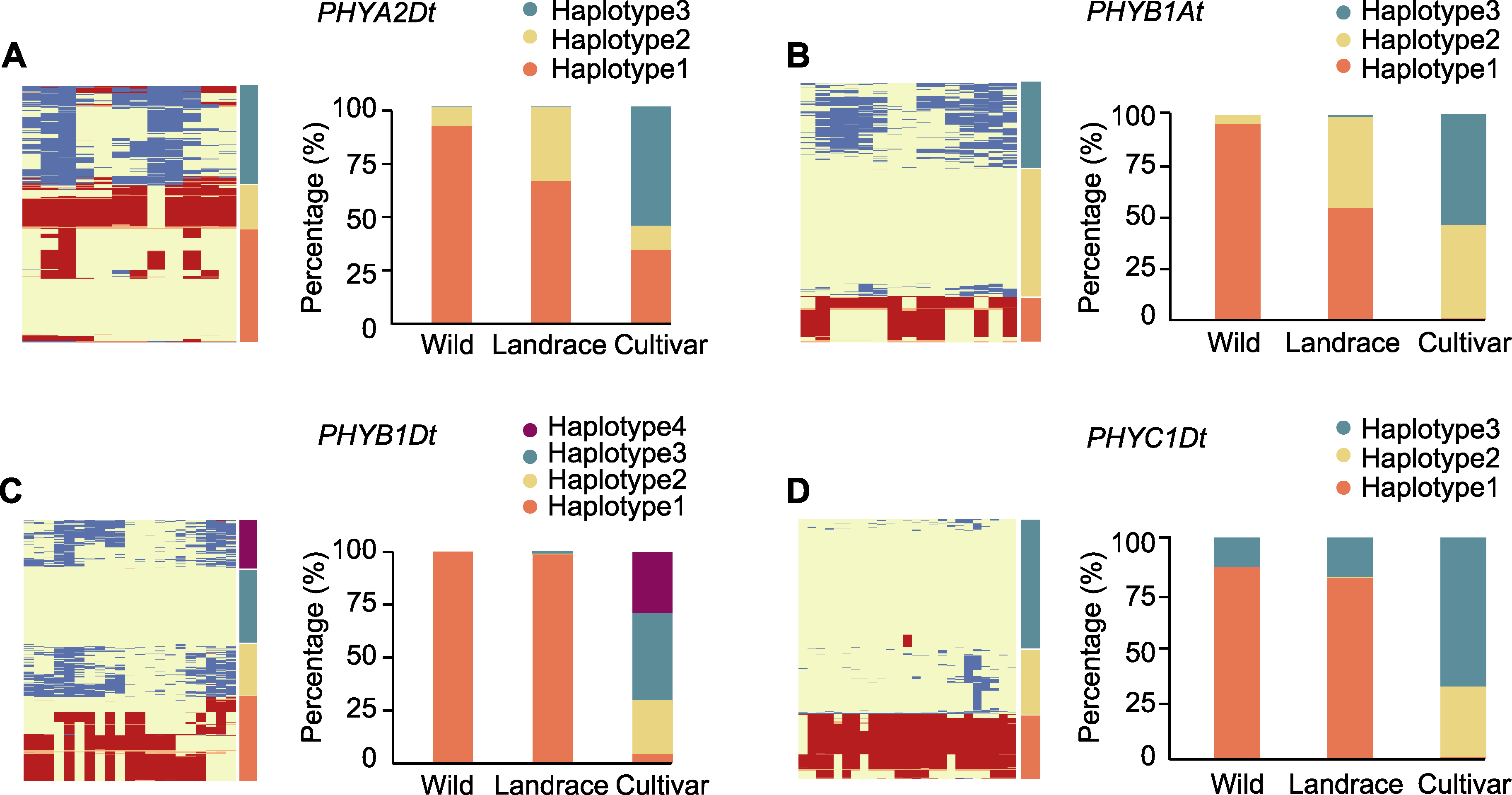
图6 PHY基因在野生种、半野生种及栽培品种中的单倍型分析 左图代表不同材料的聚类, 右图代表不同单倍型在野生种、半野生种及栽培品种中所占的比例。
Figure 6 Haplotype analysis of PHY genes in wild, landrace and cultivar The left figure represents the clustering of different materials, and the right figure represents the proportion of different haplotypes in wild, landrace and cultivar.
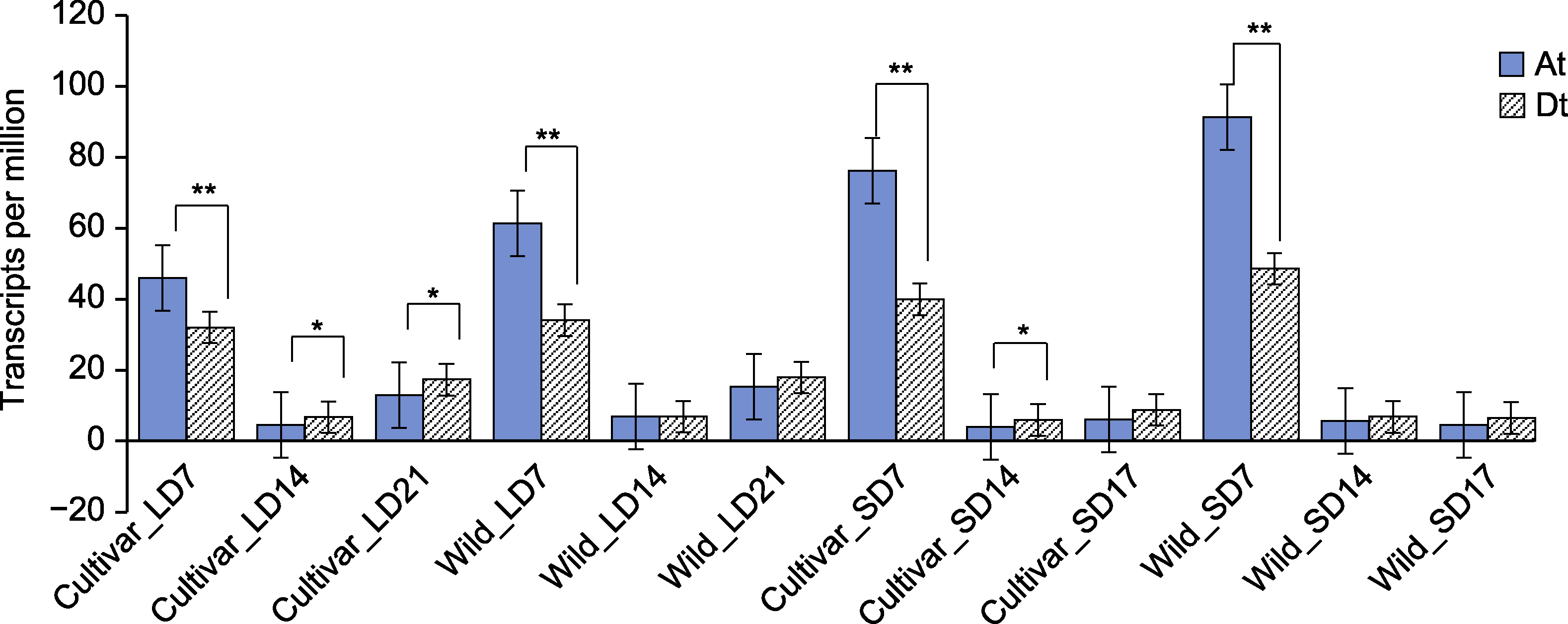
图7 陆地棉GhPHYE1基因在不同亚基因组中的表达 LD: 长日照; SD: 短日照; *P<0.05; **P<0.01
Figure 7 Expression of GhPHYE1 genes in different subgenomes of Gossypium hirsutum LD: Long-day; SD: Short-day; *P<0.05; **P<0.01
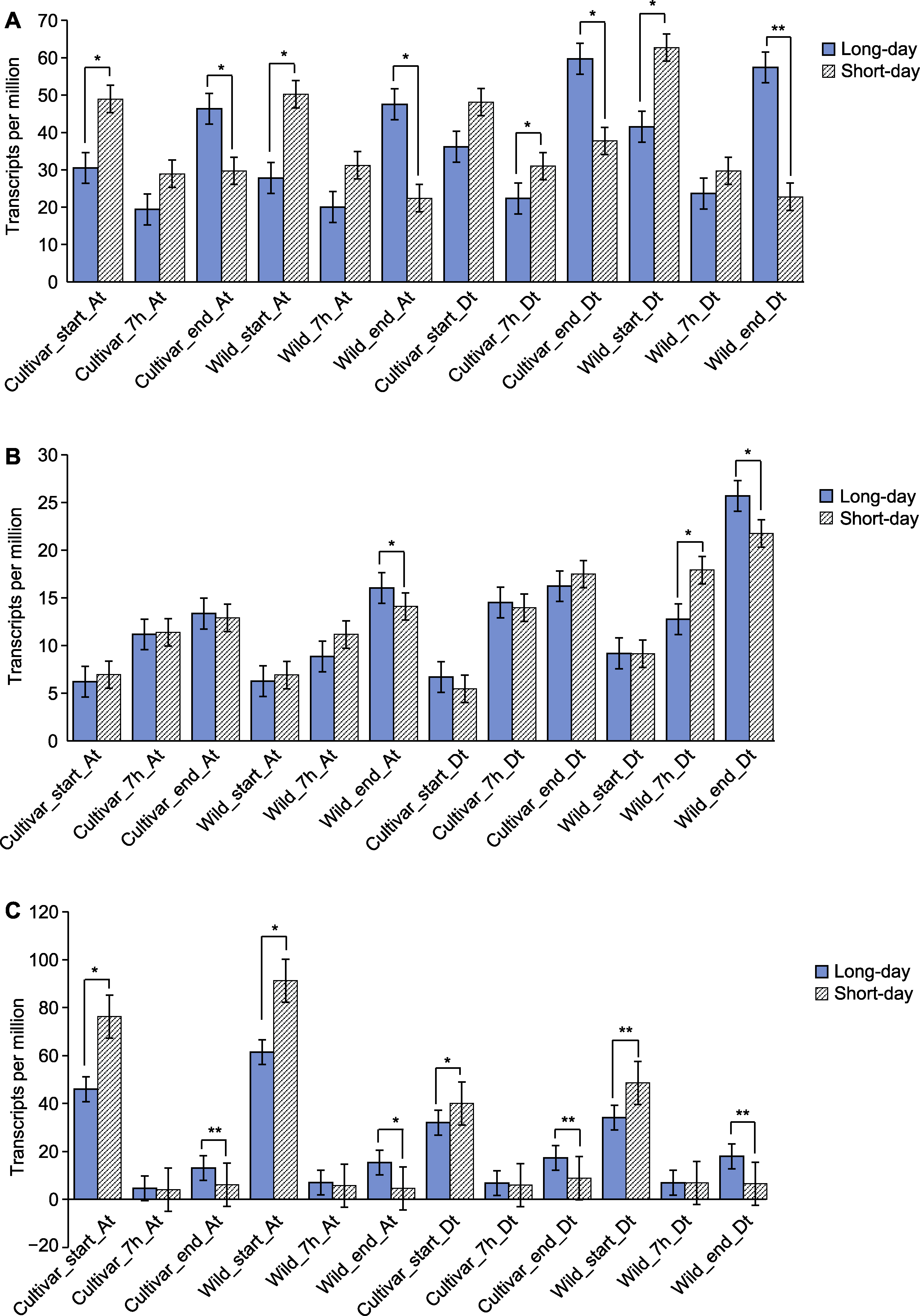
图8 陆地棉PHY基因在不同光照处理条件下的表达 (A) PHYA1基因的表达; (B) PHYB1基因的表达; (C) PHYE1基因的表达。*P<0.05; **P<0.01
Figure 8 Expression of PHY genes in Gossypium hirsutum under different light treatment conditions (A) Expression of PHYA1 genes; (B) Expression of PHYB1 genes; (C) Expression of PHYE1 genes. *P<0.05; **P<0.01
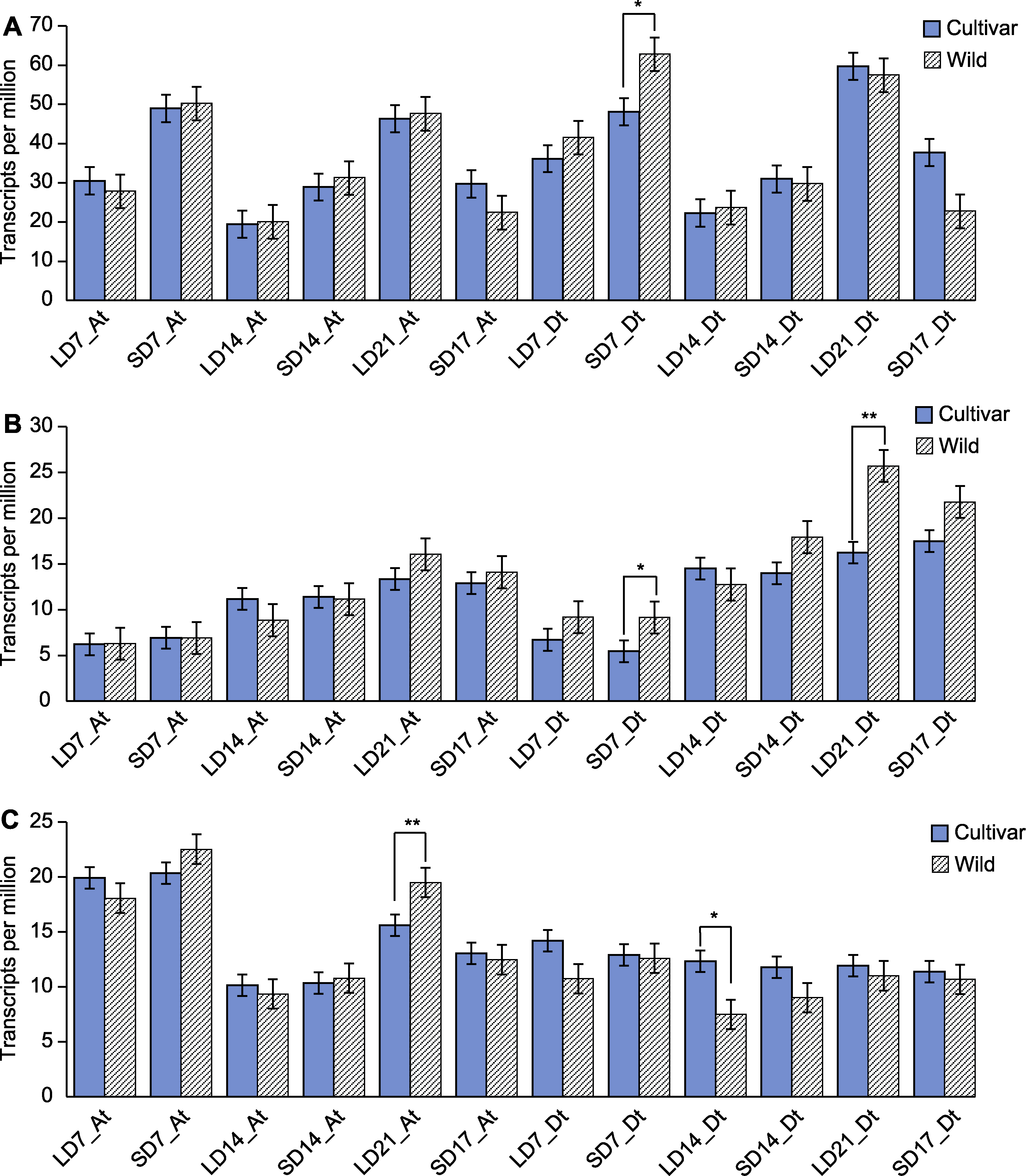
图9 陆地棉PHY基因在栽培品种与野生种中的表达 (A) PHYA1基因的表达; (B) PHYB1基因的表达; (C) PHYC1基因的表达。*P<0.05; **P<0.01
Figure 9 Expression of PHY genes in cultivated and wild Gossypium hirsutum (A) Expression of PHYA1 gene; (B) Expression of PHYB1 gene; (C) Expression of PHYC1 gene. *P<0.05; **P<0.01
| [1] | 丁武思, 陈士瞻, 刘磊, 樊晓聪, 丁梦月, 孙广华, 王立建, 杨建平 (2021). 2个玉米光敏色素C基因的克隆及功能验证. 河南农业科学 50, 16-26. |
| [2] | 房迈莼, 王小菁, 李洪清 (2005). 光对植物生物钟的调节. 植物学通报 22, 207-214. |
| [3] | 惠婕, 黄丛林, 吴忠义, 张秀海 (2011). 拟南芥光敏色素基因PHYA转化菊花的研究. 江苏农业科学 39(2), 51-54. |
| [4] | 李建平 (2013). 光敏色素D在拟南芥生长和发育中的功能与机理研究. 博士论文. 北京: 中国农业科学院. pp. 1-75. |
| [5] | 李建平, 杨建平, 宋梅芳, 苏亮, 侯佩, 郝晓燕, 邵琳, 陈果, 足木热木, 黄全生 (2014). 拟南芥光敏色素D (AtphyD)在不同光质下的表达特性分析. 新疆农业科学 51, 305-310. |
| [6] | 李全权 (2020). 玉米光敏色素C在光形态建成与开花中的功能解析. 博士论文. 泰安: 山东农业大学. pp. 1-131. |
| [7] |
李壮, 马燕斌, 蔡应繁, 吴锁伟, 肖阳, 孟凡华, 付风铃, 黄玉碧, 杨建平 (2010). 小麦光敏色素基因TaPhyB3的克隆和表达分析. 作物学报 36, 779-787.
DOI |
| [8] | 连欣 (2020). 棉花开花时间相关基因的挖掘与功能分析. 硕士论文. 泰安: 山东农业大学. pp. 1-78. |
| [9] | 申慧敏, 吴年隆, 王亚敏, 王兴春 (2022). 谷子光敏色素家族基因的生物信息学及表达模式分析. 山西农业科学 50, 1-8. |
| [10] | 魏俊梅 (2018). 棉花开花时间相关基因的挖掘鉴定与功能分析. 硕士论文. 泰安: 山东农业大学. pp. 1-57. |
| [11] |
岳晶, 管利萍, 孟思远, 张静, 侯岁稳 (2015). 光敏色素信号通路中磷酸化修饰研究进展. 植物学报 50, 241-254.
DOI |
| [12] |
詹克慧, 李志勇, 侯佩, 习雨琳, 肖阳, 孟凡华, 杨建平 (2012). 利用修饰光敏色素信号途径进行作物改良的可行性. 中国农业科学 45, 3249-3255.
DOI |
| [13] |
赵翔, 赵青平, 杨煦, 慕世超, 张骁 (2015). 向光素调节植物向光性及其与光敏色素/隐花色素的相互关系. 植物学报 50, 122-132.
DOI |
| [14] |
Abdellatif KF, Khidr YA, El-Mansy YM, El-Lawendey MM, Soliman YA (2012). Molecular diversity of Egyptian cotton (Gossypium barbadense L.) and its relation to varietal development. J Crop Sci Biotechnol 15, 93-99.
DOI URL |
| [15] |
Argout X, Martin G, Droc G, Fouet O, Labadie K, Rivals E, Aury JM, Lanaud C (2017). The cacao Criollo genome v2.0: an improved version of the genome for genetic and functional genomic studies. BMC Genomics 18, 730.
DOI PMID |
| [16] |
Ballaré CL, Pierik R (2017). The shade-avoidance syndrome: multiple signals and ecological consequences. Plant Cell Environ 40, 2530-2543.
DOI URL |
| [17] |
Bao Y, Hu GJ, Grover CE, Conover J, Yuan DJ, Wendel JF (2019). Unraveling cis and trans regulatory evolution during cotton domestication. Nat Commun 10, 5399.
DOI |
| [18] |
Bolger AM, Lohse M, Usadel B (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114-2120.
DOI PMID |
| [19] |
Briggs WR, Olney MA (2001). Photoreceptors in plant photomorphogenesis to date. Five phytochromes, two cryptochromes, one phototropin, and one superchrome. Plant Physiol 125, 85-88.
PMID |
| [20] |
Cai YF, Cai XY, Wang QL, Wang P, Zhang Y, Cai CW, Xu YC, Wang KB, Zhou ZL, Wang CX, Geng SP, Li B, Dong Q, Hou YQ, Wang H, Ai P, Liu Z, Yi FF, Sun MS, An GY, Cheng JR, Zhang YY, Shi Q, Xie YH, Shi XY, Chang Y, Huang FF, Chen Y, Hong SM, Mi LY, Sun Q, Zhang L, Zhou BL, Peng R, Zhang X, Liu F (2020). Genome sequencing of the Australian wild diploid species Gossypium australe highlights disease resistance and de-a)layed gland morphogenesis. Plant Biotechnol J 18, 814-828.
DOI URL |
| [1] |
Chen CJ, Chen H, Zhang Y, Thomas HR, Frank MH, He YH, Xia R (2020a). TBtools: an integrative Toolkit developed for interactive analyses of big biological data. Mol Plant 13, 1194-1202.
DOI URL |
| [2] |
Chen M, Chory J (2011). Phytochrome signaling mechanisms and the control of plant development. Trends Cell Biol 21, 664-671.
DOI PMID |
| [3] | Chen ZJ, Sreedasyam A, Ando A, Song QX, De Santiago LM, Hulse-Kemp AM, Ding MQ, Ye WX, Kirkbride RC, Jenkins J, Plott C, Lovell J, Lin YM, Vaughn R, Liu B, Simpson S, Scheffler BE, Wen L, Saski CA, Grover CE, Hu GJ, Conover JL, Carlson JW, Shu SQ, Boston LB, Williams M, Peterson DG, McGee K, Jones DC, Wendel JF, Stelly DM, Grimwood J, Schmutz J (2020b). Genomic diversifications of five Gossypium allopolyploid species and their impact on cotton improvement. Nat Genet 52, 525-533. |
| [4] |
Chun L, Kawakami A, Christopher DA (2001). Phytochrome A mediates blue light and UV-A-dependent chloroplast gene transcription in green leaves. Plant Physiol 125, 1957-1966.
DOI PMID |
| [5] |
Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, McVean G, Durbin R, 1000 Genomes Project Analysis Group (2011). The variant call format and VCFtools. Bioinformatics 27, 2156-2158.
DOI PMID |
| [6] |
Doebley JF, Gaut BS, Smith BD (2006). The molecular genetics of crop domestication. Cell 127, 1309-1321.
DOI PMID |
| [7] |
Fang L, Gong H, Hu Y, Liu CX, Zhou BL, Huang T, Wang YK, Chen SQ, Fang DD, Du XM, Chen H, Chen JD, Wang S, Wang Q, Wan Q, Liu BL, Pan MQ, Chang LJ, Wu HT, Mei GF, Xiang D, Li XH, Cai CP, Zhu XF, Chen ZJ, Han B, Chen XY, Guo WZ, Zhang TZ, Huang XH (2017a). Genomic insights into divergence and dual domestication of cultivated allotetraploid cottons. Genome Biol 18, 33.
DOI URL |
| [8] |
Fang L, Wang Q, Hu Y, Jia YH, Chen JD, Liu BL, Zhang ZY, Guan XY, Chen SQ, Zhou BL, Mei GF, Sun JL, Pan ZE, He SP, Xiao SH, Shi WJ, Gong WF, Liu JG, Ma J, Cai CP, Zhu XF, Guo WZ, Du XM, Zhang TZ (2017b). Genomic analyses in cotton identify signatures of selection and loci associated with fiber quality and yield traits. Nat Genet 49, 1089-1098.
DOI URL |
| [9] |
Grover CE, Arick II MA, Thrash A, Conover JL, Sanders WS, Peterson DG, Frelichowski JE, Scheffler JA, Scheffler BE, Wendel JF (2019). Insights into the evolution of the new world diploid cottons (Gossypium, Subgenus Houzingenia) based on genome sequencing. Genome Biol Evol 11, 53-71.
DOI URL |
| [10] |
Grover CE, Pan MQ, Yuan DJ, Arick MA, Hu GJ, Brase L, Stelly DM, Lu ZF, Schmitz RJ, Peterson DG, Wendel JF, Udall JA (2020). The Gossypium longicalyx genome as a resource for cotton breeding and evolution. G3 10, 1457-1467.
DOI URL |
| [11] |
Hu Y, Chen JD, Fang L, Zhang ZY, Ma W, Niu YC, Ju LZ, Deng JQ, Zhao T, Lian JM, Baruch K, Fang D, Liu X, Ruan YL, Rahman MU, Han JL, Wang K, Wang Q, Wu HT, Mei GF, Zang YH, Han ZG, Xu CY, Shen WJ, Yang DF, Si ZF, Dai F, Zou LF, Huang F, Bai YL, Zhang YG, Brodt A, Ben-Hamo H, Zhu XF, Zhou BL, Guan XY, Zhu SJ, Chen XY, Zhang TZ (2019). Gossypium barbadense and Gossypium hirsutum genomes provide insights into the origin and evolution of allotetraploid cotton. Nat Genet 51, 739-748.
DOI PMID |
| [12] |
Huang G, Wu ZG, Percy RG, Bai MZ, Li Y, Frelichowski JE, Hu J, Wang K, Yu JZ, Zhu YX (2020). Genome sequence of Gossypium herbaceum and genome updates of Gossypium arboreum and Gossypium hirsutum provide insights into cotton A-genome evolution. Nat Genet 52, 516-524.
DOI PMID |
| [13] |
Hughes J (2013). Phytochrome cytoplasmic signaling. Annu Rev Plant Biol 64, 377-402.
DOI PMID |
| [14] |
Johnson E, Bradley M, Harberd NP, Whitelam GC (1994). Photoresponses of light-grown phyA mutants of Arabidopsis (Phytochrome A is required for the perception of daylength extensions). Plant Physiol 105, 141-149.
PMID |
| [15] |
Katoh K, Standley DM (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30, 772-780.
DOI PMID |
| [16] |
Kim D, Paggi JM, Park C, Bennett C, Salzberg SL (2019). Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol 37, 907-915.
DOI PMID |
| [17] | Kim YM, Kim S, Koo N, Shin AY, Yeom SI, Seo E, Park SJ, Kang WH, Kim MS, Park J, Jang I, Kim PG, Byeon I, Kim MS, Choi J, Ko G, Hwang J, Yang TJ, Choi SB, Lee JM, Lim KB, Lee J, Choi IY, Park BS, Kwon SY, Choi D, Kim RW (2017). Genome analysis of Hibiscus syriacus provides insights of polyploidization and indeterminate flowering in woody plants. DNA Res 24, 71-80. |
| [18] |
Korunes KL, Samuk K (2021). Pixy: unbiased estimation of nucleotide diversity and divergence in the presence of missing data. Mol Ecol Res 21, 1359-1368.
DOI URL |
| [19] | Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouzé P, Rombauts S (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 30, 325-327. |
| [20] |
Li JG, Li G, Wang HY, Deng X (2011). Phytochrome signaling mechanisms. Arabidopsis Book 9, e0148.
DOI URL |
| [21] |
Liao Y, Smyth GK, Shi W (2014). FeatureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923-930.
DOI PMID |
| [22] |
Ma ZY, He SP, Wang XF, Sun JL, Zhang Y, Zhang GY, Wu LQ, Li ZK, Liu ZH, Sun GF, Yan YY, Jia YH, Yang J, Pan ZE, Gu QS, Li XY, Sun ZW, Dai PH, Liu ZW, Gong WF, Wu JH, Wang M, Liu HW, Feng KY, Ke HF, Wang JD, Lan HY, Wang GN, Peng J, Wang N, Wang LR, Pang BY, Peng Z, Li RQ, Tian SL, Du XM (2018). Resequencing a core collection of upland cotton identifies genomic variation and loci influencing fiber quality and yield. Nat Genet 50, 803-813.
DOI PMID |
| [23] |
Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ (2015). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32, 268-274.
DOI URL |
| [24] |
Pearce S, Kippes N, Chen A, Debernardi JM, Dubcovsky J (2016). RNA-seq studies using wheat PHYTOCHROME B and PHYTOCHROME C mutants reveal shared and specific functions in the regulation of flowering and shade- avoidance pathways. BMC Plant Biol 16, 141.
DOI URL |
| [25] |
Peng H, Phung J, Zhai Y, Neff MM (2020). Self-transcriptional repression of the Arabidopsis NAC transcription factor ATAF2 and its genetic interaction with phytochrome A in modulating seedling photomorphogenesis. Planta 252, 48.
DOI PMID |
| [26] |
Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL (2015). StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol 33, 290-295.
DOI PMID |
| [27] | Procter JB, Carstairs GM, Soares B, Mourão K, Ofoegbu TC, Barton D, Lui L, Menard A, Sherstnev N, Roldan- Martinez D, Duce S, Martin DMA, Barton GJ (2021). Alignment of biological sequences with Jalview. In: KatohK,ed. Multiple Sequence Alignment:Methods and Protocols. New York: Humana. pp. 203-224. |
| [28] |
Purugganan MD, Fuller DQ (2009). The nature of selection during plant domestication. Nature 457, 843-848.
DOI |
| [29] | Rao AQ, Irfan M, Saleem Z, Nasir IA, Riazuddin S, Husnain T (2011). Overexpression of the phytochrome B gene from Arabidopsis thaliana increases plant growth and yield of cotton (Gossypium hirsutum). J Zhejiang Univ Sci B Biomed Biotechnol 12, 326-334. |
| [30] |
Reed JW, Nagpal P, Poole DS, Furuya M, Chory J (1993). Mutations in the gene for the red/far-red light receptor phytochrome B alter cell elongation and physiological responses throughout Arabidopsis development. Plant Cell 5, 147-157.
PMID |
| [31] |
Saski CA, Scheffler BE, Hulse-Kemp AM, Liu B, Song QX, Ando A, Stelly DM, Scheffler JA, Grimwood J, Jones DC, Peterson DG, Schmutz J, Chen ZJ (2017). Sub genome anchored physical frameworks of the allotetraploid Upland cotton (Gossypium hirsutum L.) genome, and an approach toward reference-grade assemblies of polyploids. Sci Rep 7, 15274.
DOI |
| [32] |
Tian Y, Zhang TZ (2021). MIXTAs and phytohormones orchestrate cotton fiber development. Curr Opin Plant Biol 59, 101975.
DOI URL |
| [33] |
Udall JA, Long E, Hanson C, Yuan DJ, Ramaraj T, Conover JL, Gong L, Arick MA, Grover CE, Peterson DG, Wendel JF (2019a). De novo genome sequence assemblies of Gossypium raimondii and Gossypium turneri. G3 9, 3079-3085.
DOI URL |
| [34] |
Udall JA, Long E, Ramaraj T, Conover JL, Yuan DJ, Grover CE, Gong L, Arick II MA, Masonbrink RE, Peterson DG, Wendel JF (2019b). The genome sequence of Gossypioides kirkii illustrates a descending dysploidy in plants. Front Plant Sci 10, 1541.
DOI URL |
| [35] |
Wang MJ, Li JY, Wang PC, Liu F, Liu ZP, Zhao GN, Xu ZP, Pei LL, Grover CE, Wendel JF, Wang KB, Zhang XL (2021). Comparative genome analyses highlight transposon-mediated genome expansion and the evolutionary architecture of 3D genomic folding in cotton. Mol Biol Evol 38, 3621-3636.
DOI PMID |
| [36] |
Wang MJ, Tu LL, Yuan DJ, Zhu D, Shen C, Li JY, Liu FY, Pei LL, Wang PC, Zhao GN, Ye ZX, Huang H, Yan FL, Ma YZ, Zhang L, Liu M, You JQ, Yang YC, Liu ZP, Huang F, Li BQ, Qiu P, Zhang QH, Zhu LF, Jin SX, Yang XY, Min L, Li GL, Chen LL, Zheng HK, Lindsey K, Lin ZX, Udall JA, Zhang XL (2019). Reference genome sequences of two cultivated allotetraploid cottons, Gossypium hirsutum and Gossypium barbadense. Nat Genet 51, 224-229.
DOI |
| [37] |
Wang YP, Tang HB, DeBarry JD, Tan X, Li JP, Wang XY, Lee TH, Jin HZ, Marler B, Guo H, Kissinger JC, Paterson AH (2012). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res 40, e49.
DOI URL |
| [38] | Wendel JF, Brubaker C, Alvarez I, Cronn R, Stewart JM (2009). Evolution and natural history of the cotton genus. In: PatersonAH,ed. Genetics and Genomics of Cotton. New York: Springer. pp. 3-22. |
| [39] | Wendel JF, Cronn RC (2003). Polyploidy and the evolutionary history of cotton. Adv Agron 78, 139-186. |
| [40] |
Westengen OT, Huamán Z, Heun M (2005). Genetic diversity and geographic pattern in early South American cotton domestication. Theor Appl Genet 110, 392-402.
PMID |
| [41] |
Xie YR, Liu Y, Wang H, Ma XJ, Wang BB, Wu GX, Wang HY (2017). Phytochrome-interacting factors directly suppress MIR156 expression to enhance shade-avoidance syndrome in Arabidopsis. Nat Commun 8, 348.
DOI |
| [42] |
Yang ZE, Ge XY, Li WN, Jin YY, Liu LS, Hu W, Liu FY, Chen YL, Peng SL, Li FG (2021). Cotton D genome assemblies built with long-read data unveil mechanisms of centromere evolution and stress tolerance divergence. BMC Biol 19, 115.
DOI PMID |
| [43] |
Yang ZR, Qanmber G, Wang Z, Yang ZE, Li FG (2020). Gossypium genomics: trends, scope, and utilization for cotton improvement. Trends Plant Sci 25, 488-500.
DOI URL |
| [44] |
Yuan DJ, Grover CE, Hu GJ, Pan MQ, Miller ER, Conover JL, Hunt SP, Udall JA, Wendel JF (2021). Parallel and intertwining threads of domestication in allopolyploid cotton. Adv Sci (Weinh) 8, 2003634.
DOI URL |
| [45] |
Zhang HK, Gao SH, Lercher MJ, Hu SN, Chen WH (2012). EvolView, an online tool for visualizing, annotating and ma- naging phylogenetic trees. Nucleic Acids Res 40, W569-W572.
DOI URL |
| [46] |
Zhang LL, Ma XK, Zhang XT, Xu Y, Ibrahim AK, Yao JY, Huang HX, Chen S, Liao ZY, Zhang Q, Niyitanga S, Yu JX, Liu Y, Xu XM, Wang JJ, Tao AF, Xu JT, Chen SY, Yang X, He QY, Lin LH, Fang PP, Zhang LM, Ming R, Qi JM, Zhang LW (2021). Reference genomes of the two cultivated jute species. Plant Biotechnol J 19, 2235-2248.
DOI PMID |
| [47] |
Zhang M, Zheng XL, Song SQ, Zeng QW, Hou L, Li DM, Zhao J, Wei Y, Li XB, Luo M, Xiao YH, Luo XY, Zhang JF, Xiang CB, Pei Y (2011). Spatiotemporal manipulation of auxin biosynthesis in cotton ovule epidermal cells enhances fiber yield and quality. Nat Biotechnol 29, 453-458.
DOI PMID |
| [48] |
Zhang TZ, Hu Y, Jiang WK, Fang L, Guan XY, Chen JD, Zhang JB, Saski CA, Scheffler BE, Stelly DM, Hulse- Kemp AM, Wan Q, Liu BL, Liu CX, Wang S, Pan MQ, Wang YK, Wang DW, Ye WX, Chang LJ, Zhang WP, Song QX, Kirkbride RC, Chen XY, Dennis E, Llewellyn DJ, Peterson DG, Thaxton P, Jones DC, Wang Q, Xu XY, Zhang H, Wu HT, Zhou L, Mei GF, Chen SQ, Tian Y, Xiang D, Li XH, Ding J, Zuo QY, Tao LN, Liu YC, Li J, Lin Y, Hui YY, Cao ZS, Cai CP, Zhu XF, Jiang Z, Zhou BL, Guo WZ, Li RQ, Chen ZJ (2015). Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat Biotechnol 33, 531-537.
DOI |
| [1] | 杨莉, 曲茜彤, 陈子航, 邹婷婷, 王全华, 王小丽. 菠菜AT-hook基因家族鉴定与表达谱分析[J]. 植物学报, 2025, 60(3): 377-392. |
| [2] | 姚祥坦, 张心怡, 陈阳, 袁晔, 程旺大, 王天瑞, 邱英雄. 基于基因组重测序揭示栽培欧菱遗传多样性及‘南湖菱’的起源驯化历史[J]. 生物多样性, 2024, 32(9): 24212-. |
| [3] | 罗燕, 刘奇源, 吕元兵, 吴越, 田耀宇, 安田, 李振华. 拟南芥光敏色素突变体种子萌发的光温敏感性[J]. 植物学报, 2024, 59(5): 752-762. |
| [4] | 康敏, 张美莹, 齐秀双, 佟宁宁, 李旸, 舒庆艳, 刘政安, 吕长平, 彭丽平. 伊藤杂种‘和谐’组培快繁体系的建立[J]. 植物学报, 2024, 59(3): 441-451. |
| [5] | 陈艳晓, 李亚萍, 周晋军, 解丽霞, 彭永彬, 孙伟, 和亚男, 蒋聪慧, 王增兰, 郑崇珂, 谢先芝. 拟南芥光敏色素B氨基酸位点突变对其结构与功能的影响[J]. 植物学报, 2024, 59(3): 481-494. |
| [6] | 胡楚婷, 杨柳依依, 石绍林, 周琰, 陈婷婷, 郑博瀚, 杨暘, 卢小玲, 王陈玲, 倪健. 浙江金华典型人工植被的植物功能性状[J]. 植物生态学报, 2024, 48(10): 1336-1350. |
| [7] | 于熙婷, 黄学辉. 现代玉米起源新见解——两类大刍草的混血[J]. 植物学报, 2023, 58(6): 857-860. |
| [8] | 许亚楠, 闫家榕, 孙鑫, 王晓梅, 刘玉凤, 孙周平, 齐明芳, 李天来, 王峰. 红光和远红光在调控植物生长发育及应答非生物胁迫中的作用[J]. 植物学报, 2023, 58(4): 622-637. |
| [9] | 孙福辉, 方慧仪, 温小蕙, 张亮生. 马银花MADS-box基因家族系统进化与表达分析[J]. 植物学报, 2023, 58(3): 404-416. |
| [10] | 冯旭飞, 雷长英, 张玉洁, 向导, 杨明凤, 张旺锋, 张亚黎. 棉花花铃期叶片氮分配对光合氮利用效率的影响[J]. 植物生态学报, 2023, 47(11): 1600-1610. |
| [11] | 张慧, 梁红凯, 智慧, 张林林, 刁现民, 贾冠清. 谷子β-胡萝卜素异构酶家族基因的表达与变异分析[J]. 植物学报, 2023, 58(1): 34-50. |
| [12] | 王菲菲, 周振祥, 洪益, 谷洋洋, 吕超, 郭宝健, 朱娟, 许如根. 大麦NF-YC基因鉴定及在盐胁迫下的表达分析[J]. 植物学报, 2023, 58(1): 140-149. |
| [13] | 黄宏文, 廖景平. 论我国国家植物园体系建设: 以任务带学科构建国家植物园迁地保护综合体系[J]. 生物多样性, 2022, 30(6): 22220-. |
| [14] | 李聪, 齐立娟, 谷晓峰, 李继刚. 植物光信号途径重要新调控因子TZP的研究进展[J]. 植物学报, 2022, 57(5): 579-587. |
| [15] | 蔡新宇, 毛晓伟, 赵毅强. 家养动物驯化起源的研究方法与进展[J]. 生物多样性, 2022, 30(4): 21457-. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||