

植物学报 ›› 2018, Vol. 53 ›› Issue (1): 42-50.DOI: 10.11983/CBB17058 cstr: 32102.14.CBB17058
收稿日期:2017-03-22
接受日期:2017-08-30
出版日期:2018-01-01
发布日期:2018-08-10
通讯作者:
苏伟
基金资助:
Cheng Chen1,2, Aiwu Dong1,2, Wei Su1,2,*( )
)
Received:2017-03-22
Accepted:2017-08-30
Online:2018-01-01
Published:2018-08-10
Contact:
Wei Su
摘要: HIRA是组蛋白H3.3的特异分子伴侣, 在组蛋白H3.3掺入染色质的过程中发挥重要作用。研究表明, HIRA在哺乳动物胚胎发育和DNA损伤修复过程中不可或缺。而目前人们对于植物中HIRA同源基因功能的研究相对较少。该研究主要关注拟南芥(Arabidopsis thaliana) AtHIRA基因在植物体细胞同源重组以及减数分裂同源重组过程中的功能。将体细胞同源重组和减数分裂同源重组报告系统分别导入野生型和hira-1突变体后统计同源重组频率, 结果表明在正常生长条件下及在伯莱霉素(bleomycin)或UV-C处理条件下, hira-1突变体体细胞的分子内和分子间同源重组频率均低于野生型。而在正常生长条件下, 野生型与hira-1突变体花粉母细胞间的减数分裂同源重组频率没有明显差异, hira-1突变体的DNA损伤水平与野生型接近。qRT-PCR结果表明, DNA损伤修复相关基因RAD51和RAD54在hira-1突变体中的表达水平均高于野生型。此外, 盐胁迫处理实验表明, hira-1突变体对于高盐胁迫更加敏感。综上, AtHIRA在拟南芥体细胞同源重组及盐胁迫响应过程中发挥了一定作用。
陈成, 董爱武, 苏伟. 拟南芥组蛋白分子伴侣AtHIRA参与体细胞同源重组及盐胁迫响应. 植物学报, 2018, 53(1): 42-50.
Cheng Chen, Aiwu Dong, Wei Su. Histone Chaperone AtHIRA is Involved in Somatic Homologous Recombination and Salinity Response in Arabidopsis. Chinese Bulletin of Botany, 2018, 53(1): 42-50.
| Primer name | Primer sequence (5′-3′) |
|---|---|
| ACTIN2-F | GGCGATGAAGCTCAATCCAAA |
| ACTIN2-R | GGTCACGACCAGCAAGATCAAG |
| GUS-F | AAGTGGATTGATGTGATATCTC |
| GUS-R | TTCGCGCTGATACCAGACG |
| ATM-F | TGCAGCTGCGTCTCTGCATGA |
| ATM-R | CTTCATGCCGCCCTTGGGCA |
| BRCA1-F | TGCTCAGGGCTCACAGTTGAAGA |
| BRCA1-R | TGCAGGCTCCGTTTTCATTGATTG |
| PARP1-F | TGCTCGCGCGAACTCACTTCT |
| PARP1-R | AGCCTCTCCACCAGAACGGCT |
| PARP2-F | AGCCTGAAGGCCCGGGTAACA |
| PARP2-R | GCTGTCTCAGTTTTGGCTGCCG |
| RAD51-F | CGCCATTTCCCTCCACTCTCAAGC |
| RAD51-R | ACCTGCTGCCTGAAGCTGTTCG |
| RAD54-F | TGAGAGACAGGTGGGCACTCC |
| RAD54-R | ACGTCACCTCGTCACCTGCTGA |
表1 本研究所用引物
Table 1 Primers used in this study
| Primer name | Primer sequence (5′-3′) |
|---|---|
| ACTIN2-F | GGCGATGAAGCTCAATCCAAA |
| ACTIN2-R | GGTCACGACCAGCAAGATCAAG |
| GUS-F | AAGTGGATTGATGTGATATCTC |
| GUS-R | TTCGCGCTGATACCAGACG |
| ATM-F | TGCAGCTGCGTCTCTGCATGA |
| ATM-R | CTTCATGCCGCCCTTGGGCA |
| BRCA1-F | TGCTCAGGGCTCACAGTTGAAGA |
| BRCA1-R | TGCAGGCTCCGTTTTCATTGATTG |
| PARP1-F | TGCTCGCGCGAACTCACTTCT |
| PARP1-R | AGCCTCTCCACCAGAACGGCT |
| PARP2-F | AGCCTGAAGGCCCGGGTAACA |
| PARP2-R | GCTGTCTCAGTTTTGGCTGCCG |
| RAD51-F | CGCCATTTCCCTCCACTCTCAAGC |
| RAD51-R | ACCTGCTGCCTGAAGCTGTTCG |
| RAD54-F | TGAGAGACAGGTGGGCACTCC |
| RAD54-R | ACGTCACCTCGTCACCTGCTGA |
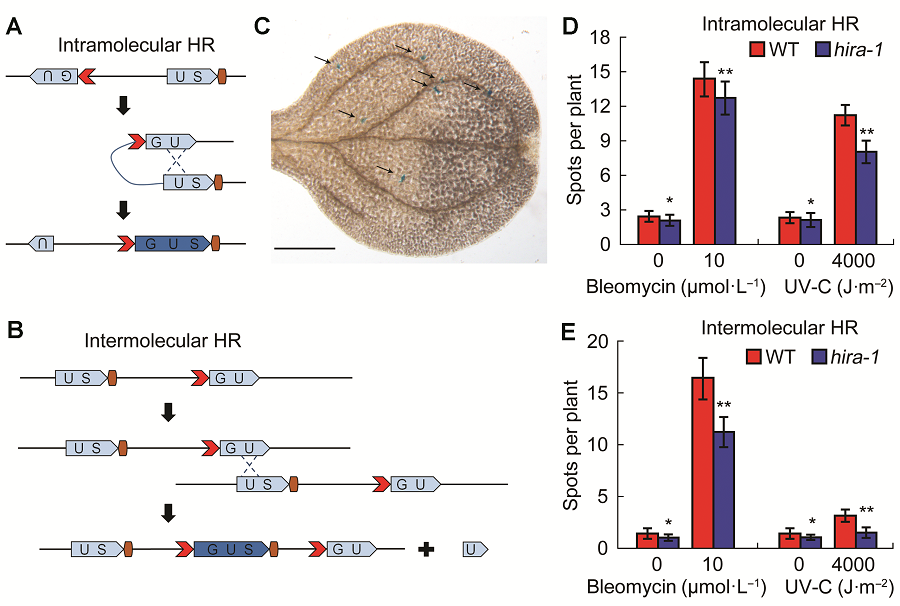
图1 拟南芥hira-1突变体的体细胞同源重组水平低于野生型(A) 1445株系中的分子内同源重组事件示意, 在经过1次同源重组事件后, GUS基因的两个分离片段可以重新组成1个有功能的GUS基因; (B) IC9C株系中的分子间同源重组事件示意, 不同分子间的片段经过同源重组事件重新形成有功能的GUS基因; (C) 组织化学染色后出现在拟南芥叶片中的每个箭头指示的蓝点代表1个有功能的GUS基因, 表明发生了1次同源重组事件(Bar=500 μm); (D) 野生型和hira-1突变体的分子内同源重组水平比较; (E) 野生型和hira-1突变体的分子间同源重组水平比较。WT: 野生型; HR: 同源重组。* P<0.05; ** P<0.01
Figure 1 hira1 mutant shows reduced homologus recombination frequency compared with Col-0 of Arabidopsis(A) Scheme of homologus recombination (HR) event in intramolecular line 1445. The two fragments of GUS gene can recombine to form a function GUS gene after a HR event; (B) Scheme of HR event in intermolecular line IC9C. The recombination of separated GUS fragments require intermolecular interaction to restore a functional GUS gene after a HR event; (C) Arabidopsis leaf with arrow labeled blue spots/sectors represent a functional GUS gene, which indicate an independent HR event (Bar=500 μm); (D) Comparison of intramolecular recombination frequency between Col-0 and hira-1; (E) Comparison of intermolecular recombination frequency between Col-0 and hira-1. WT: Wild type; HR: Homologus recombination. * P<0.05; ** P<0.01
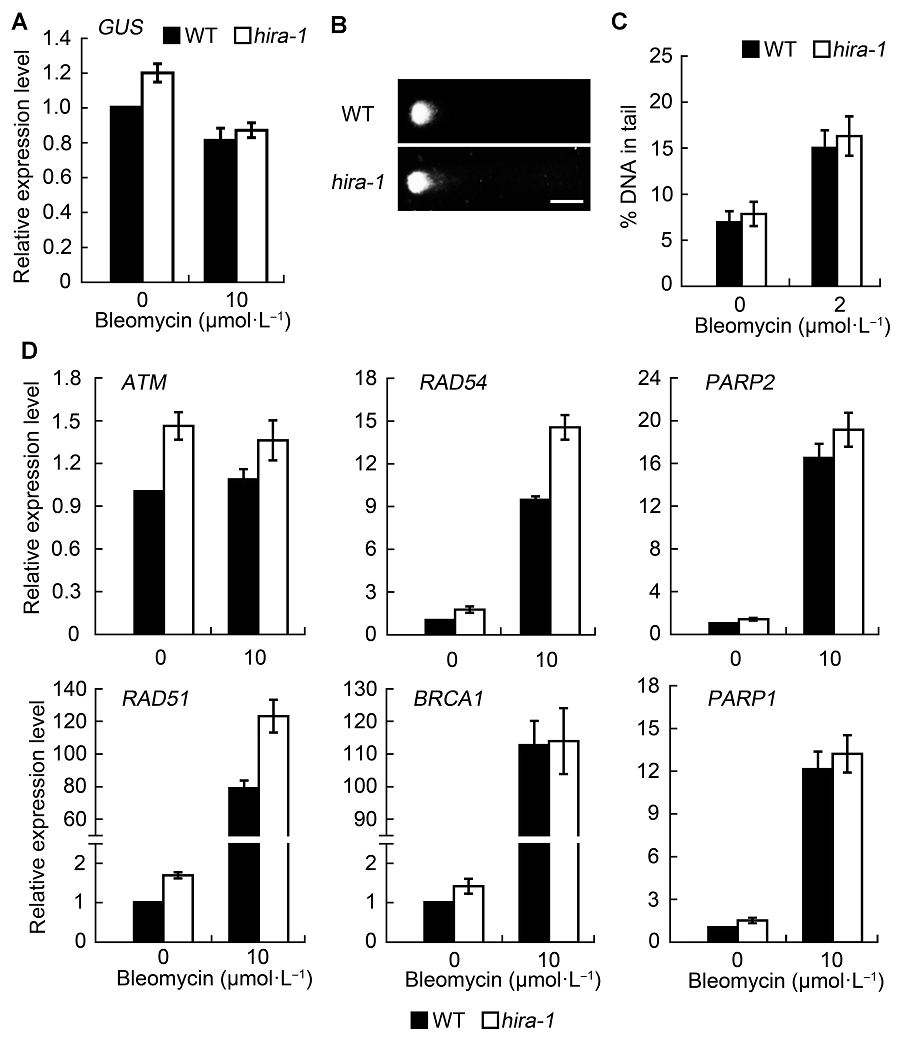
图2 AtHIRA功能缺失对DNA损伤水平的影响及DNA损伤修复相关基因的表达水平检测(平均值±标准差)(A) 在正常条件和bleomycin处理下, 拟南芥野生型和hira-1突变体中GUS基因的表达水平; (B) 损伤处理后, 野生型与hira-1突变体中细胞核彗星示意图(Bar=10 μm); (C) 野生型和hira-1突变体的细胞核彗尾中DNA百分比统计。统计结果均从超过100个细胞核中得出; (D) 实时荧光定量PCR检测野生型和hira-1突变体的DNA损伤修复相关基因的表达水平。实验经3次生物学重复。
Figure 2 Effect of AtHIRA mutation on DNA damage level and expression level of DNA repair genes (means±SD)(A) Expression level of GUS gene in wild type and hira-1 mutant of Arabidopsis under normal conditions and bleomycin treatment; (B) Representative comet images of wild-type and hira-1 nuclei after bleomycin treatment (Bar=10 μm); (C) The average percentage of DNA in comet tails of wild type and hira-1 mutant. More than 100 individual nuclei were recorded and calculated; (D) Relative expression level of DNA repair genes by RT-qPCR between wild type and hira-1. Three biological repeats were analyzed.
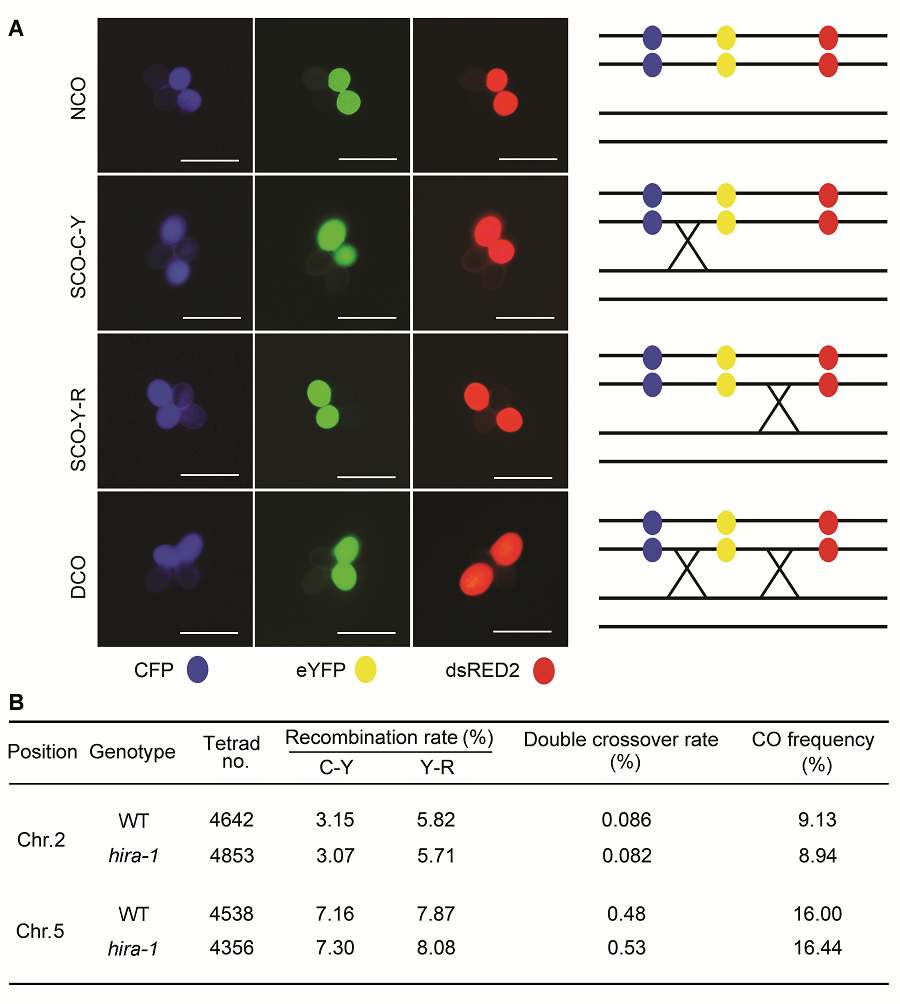
图3 利用荧光标记花粉四分体系统比较拟南芥野生型(WT)和hira-1突变体的减数分裂同源重组频率(A) 荧光四分体类型示意图: 不交换(NCO)、2种单交换(SCO-C-Y和SCO-Y-R)和1种双交换(DCO)。右侧是每种荧光类型对应的交换示意图(Bars=10 μm); (B) 在野生型和hira-1突变体中观察到的各类型荧光四分体数目。
Figure 3 Comparison of meiotic recombination frequency between wild type (WT) and hira-1 mutant of Arabidopsis using the fluorescent tagged line tetrad analysis system(A) Examples of tetrad fluorescent patterns including no cross over (NCO), two types of single cross overs (SCO-C-Y and SCO- Y-R) and one type of double cross over (DCO). The schematic representation of corresponding CO events is shown at right of each tetrad class (Bars=10 μm); (B) Number of each tetrad fluorescent patterns observed in wild type and hira-1.
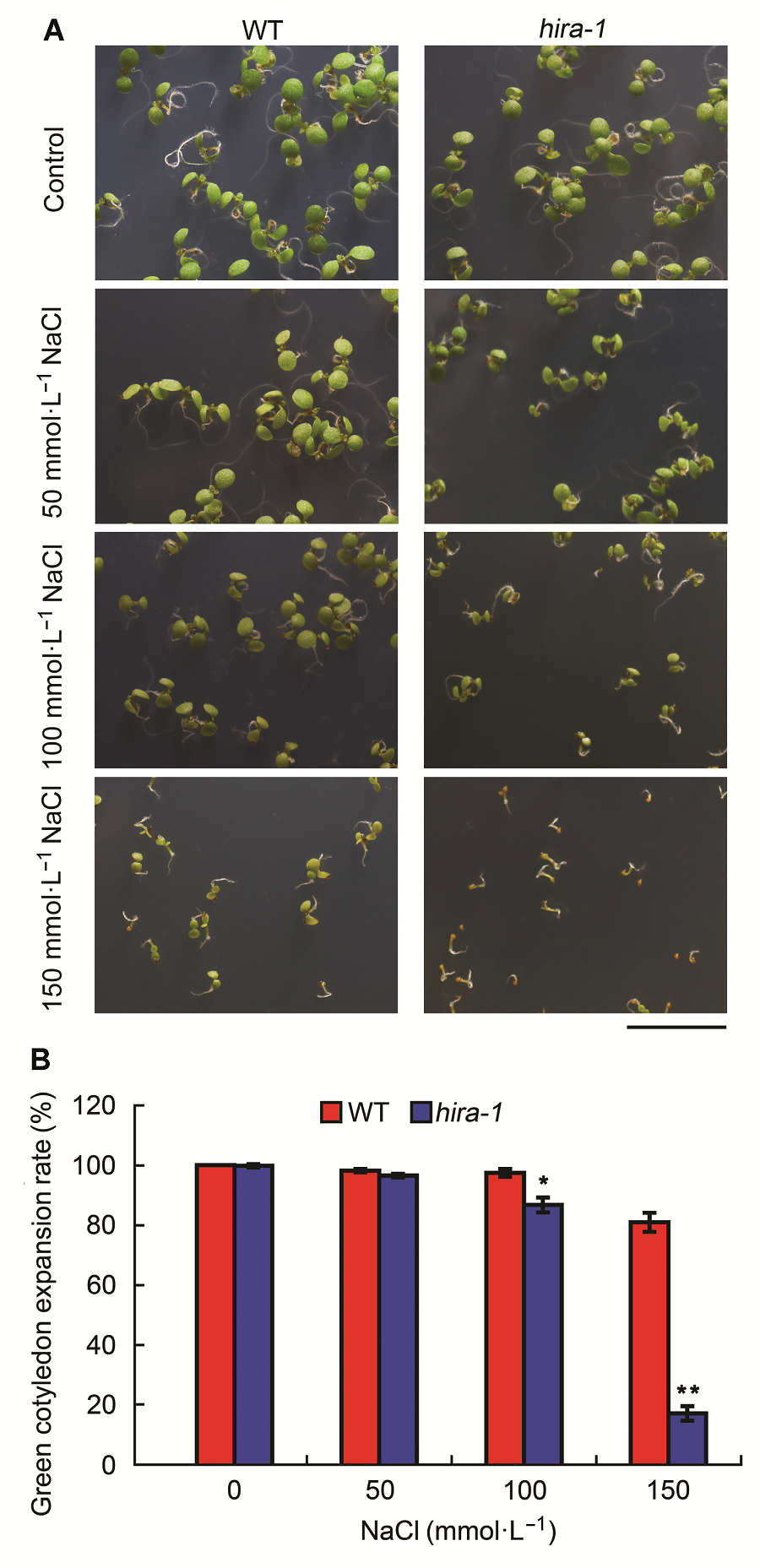
图4 拟南芥hira-1突变体对盐胁迫敏感(平均值±标准差)(A) 在含0、50、100和500 mmol·L-1 NaCl培养基上生长7天的野生型(WT)与hira-1突变体幼苗的表型(Bar=1 cm); (B) 生长7天的野生型与hira-1突变体幼苗绿色子叶展开率。每种基因型统计数目超过100株。实验经3次生物学重复。
Figure 4 Arabidopsis mutant hira-1 exhibited hypersensitivi- ty to salt stress (means±SD)(A) Phenotypes of 7-day-old seedlings of wild type (WT) and hira-1 grown on media containing 0, 50, 100, and 150 mmol·L-1 NaCl (Bar=1 cm); (B) Comparison of green cotyledon expansion rates of 7-day-old seedlings between wild type and hira-1. Over 100 seeds each genotype were recorded. Three biological repeats were analyzed.
| [1] | 夏志强, 何奕昆, 鲍时来, 种康 (2007). 植物开花的组蛋白甲基化调控分子机理. 植物学通报 24, 275-283. |
| [2] | Amin AD, Vishnoi N, Prochasson P (2012). A global requirement for the HIR complex in the assembly of chromatin.Biochim Biophys Acta 1819, 264-276. |
| [3] | Andersen SL, Sekelsky J (2010). Meiotic versus mitotic recombination: two different routes for double-strand b- reak repair: the different functions of meiotic versus mi- totic DSB repair are reflected in different pathway usage and different outcomes. Bioessays 32, 1058-1066. |
| [4] | Francis KE, Lam SY, Harrison BD, Bey AL, Berchowitz LE, Copenhaver GP (2007). Pollen tetrad-based visual assay for meiotic recombination in Arabidopsis. Proc Natl Acad Sci USA 104, 3913-3918. |
| [5] | Fritsch O, Benvenuto G, Bowler C, Molinier J, Hohn B (2004). The INO80 protein controls homologous recombination in Arabidopsis thaliana. Mol Cell 16, 479-485. |
| [6] | Gao J, Zhu Y, Zhou WB, Molinier J, Dong AW, Shen WH (2012). NAP1 family histone chaperones are required for somatic homologous recombination in Arabidopsis.Plant Cell 24, 1437-1447. |
| [7] | Gherbi H, Gallego ME, Jalut N, Lucht JM, Hohn B, White CI (2001). Homologous recombination in planta is stimulated in the absence of Rad50.EMBO Rep 2, 287-291. |
| [8] | Giraut L, Falque M, Drouaud J, Pereira L, Martin OC, Mézard C (2011). Genome-wide crossover distribution in Arabidopsis thaliana meiosis reveals sex-specific patterns along chromosomes. PLoS Genet 7, e1002354. |
| [9] | Heyer WD, Ehmsen KT, Liu J (2010). Regulation of homologous recombination in eukaryotes.Annu Rev Genet 44, 113-139. |
| [10] | Ingouff M, Rademacher S, Holec S, Šoljić L, Xin N, Readshaw A, Foo SH, Lahouze B, Sprunck S, Berger F (2010). Zygotic resetting of the HISTONE 3 variant re- pertoire participates in epigenetic reprogramming in Arabi- dopsis.Curr Biol 20, 2137-2143. |
| [11] | Krejci L, Altmannova V, Spirek M, Zhao XL (2012). Homologous recombination and its regulation.Nucleic Acids Res 40, 5795-5818. |
| [12] | Loppin B, Bonnefoy E, Anselme C, Laurencon A, Karr TL, Couble P (2005). The histone H3.3 chaperone HIRA is essential for chromatin assembly in the male pronucleus.Nature 437, 1386-1390. |
| [13] | Melamed-Bessudo C, Levy AA (2012). Deficiency in DNA methylation increases meiotic crossover rates in euchro- matic but not in heterochromatic regions in Arabidopsis.Proc Natl Acad Sci USA 109, E981-E988. |
| [14] | Molinier J, Ries G, Bonhoeffer S, Hohn B (2004). Interchromatid and interhomolog recombination in Arabidopsis thaliana. Plant Cell 16, 342-352. |
| [15] | Nie X, Wang HF, Li J, Holec S, Berger F (2014). The HIRA complex that deposits the histone H3.3 is conserved in Arabidopsis and facilitates transcriptional dynamics.Biol Open 3, 794-802. |
| [16] | Okada T, Endo M, Singh MB, Bhalla PL (2005). Analysis of the histoneH3 gene family in Arabidopsis and identification of the male-gamete-specific variant AtMGH3. Plant J 44, 557-568. |
| [17] | Otero S, Desvoyes B, Gutierrez C (2014). Histone H3 dynamics in plant cell cycle and development.Cytogenet Genome Res 143, 114-124. |
| [18] | Roberts C, Sutherland HF, Farmer H, Kimber W, Halford S, Carey A, Brickman JM, Wynshaw-Boris A, Scambler PJ (2002). Targeted mutagenesis of theHira gene results in gastrulation defects and patterning abnormalities of mesoendodermal derivatives prior to early embryo- nic lethality. Mol Cell Biol 22, 2318-2328. |
| [19] | Schuermann D, Fritsch O, Lucht JM, Hohn B (2009). Replication stress leads to genome instabilities in Arabidopsis DNA polymerase δ mutants.Plant Cell 21, 2700-2714. |
| [20] | Schuermann D, Molinier J, Fritsch O, Hohn B (2005). The dual nature of homologous recombination in plants.Tren- ds Genet 21, 172-181. |
| [21] | Sherwood PW, Osley MA (1991). Histone regulatory (hir) mutations suppress δ insertion alleles in Saccharomyces cerevisiae. Genetics 128, 729-738. |
| [22] | Szenker E, Lacoste N, Almouzni G (2012). A developmental requirement for HIRA-dependent H3.3 deposition revealed at gastrulation in Xenopus. Cell Rep 1, 730-740. |
| [23] | Tang Y, Poustovoitov MV, Zhao KH, Garfinkel M, Canu- tescu A, Dunbrack R, Adams PD, Marmorstein R (2006). Structure of a human ASF1a-HIRA complex and insights into specificity of histone chaperone complex assembly.Nat Struct Mol Biol 13, 921-929. |
| [24] | Wijeratne AJ, Ma H (2007). Genetic analyses of meiotic recombination in Arabidopsis. J Integr Plant Biol 49, 1199-1207. |
| [25] | Xu Y, Price BD (2011). Chromatin dynamics and the repair of DNA double strand breaks.Cell Cycle 10, 261-267. |
| [26] | Zhu Y, Dong AW, Meyer D, Pichon O, Renou JP, Cao KW, Shen WH (2006). Arabidopsis NRP1 and NRP2 encode histone chaperones and are required for maintaining postembryonic root growth. Plant Cell 18, 2879-2892. |
| [1] | 徐田甜, 杨培建, 周晓茜, 曹怡, 陈艳红, 刘国元, 张健, 魏辉. 紫薇GolS家族基因的理化特性与表达特征[J]. 植物学报, 2025, 60(3): 393-406. |
| [2] | 杜锦瑜, 孙震, 苏彦龙, 王贺萍, 刘亚玲, 吴振映, 何峰, 赵彦, 付春祥. 蒙古冰草咖啡酸氧甲基转移酶基因AmCOMT1的鉴定及功能分析[J]. 植物学报, 2024, 59(3): 383-396. |
| [3] | 张琦, 张文静, 袁宪凯, 李明, 赵强, 杜艳丽, 杜吉到. 褪黑素对盐胁迫下普通菜豆芽期核酸修复的调控机制[J]. 植物学报, 2023, 58(1): 108-121. |
| [4] | 王菲菲, 周振祥, 洪益, 谷洋洋, 吕超, 郭宝健, 朱娟, 许如根. 大麦NF-YC基因鉴定及在盐胁迫下的表达分析[J]. 植物学报, 2023, 58(1): 140-149. |
| [5] | 张楠,刘自广,孙世臣,刘圣怡,林建辉,彭疑芳,张晓旭,杨贺,岑曦,吴娟. 拟南芥AtR8 lncRNA对盐胁迫响应及其对种子萌发的调节作用[J]. 植物学报, 2020, 55(4): 421-429. |
| [6] | 曹栋栋,陈珊宇,秦叶波,吴华平,阮关海,黄玉韬. 水杨酸调控盐胁迫下羽衣甘蓝种子萌发的机理[J]. 植物学报, 2020, 55(1): 49-61. |
| [7] | 徐婉约,王应祥. 染色体展片法观察拟南芥雄性减数分裂过程中的染色体形态[J]. 植物学报, 2019, 54(5): 620-624. |
| [8] | 李帆,阮继伟. 利用荧光标记高通量鉴定减数分裂重组抑制突变体[J]. 植物学报, 2019, 54(4): 522-530. |
| [9] | 程新杰, 于恒秀, 程祝宽. 水稻减数分裂染色体分析方法[J]. 植物学报, 2019, 54(4): 503-508. |
| [10] | 栗露露,殷文超,牛梅,孟文静,张晓星,童红宁. 油菜素甾醇调控水稻盐胁迫应答的作用研究[J]. 植物学报, 2019, 54(2): 185-193. |
| [11] | 薛治慧, 种康. 中国科学家在杂种F1克隆繁殖研究领域取得突破性进展[J]. 植物学报, 2019, 54(1): 1-3. |
| [12] | 郭淑华, 孙永江, 牛彦杰, 韩宁, 翟衡, 杜远鹏. 碱性盐胁迫对葡萄种间杂交育种F1代光系统活性的影响[J]. 植物学报, 2018, 53(2): 196-202. |
| [13] | 徐晨, 刘晓龙, 李前, 凌凤楼, 武志海, 张治安. 供氮水平对盐胁迫下水稻叶片光合及叶绿素荧光特性的影响[J]. 植物学报, 2018, 53(2): 185-195. |
| [14] | 刘宝玲, 张莉, 孙岩, 薛金爱, 高昌勇, 苑丽霞, 王计平, 贾小云, 李润植. 谷子bZIP转录因子的全基因组鉴定及其在干旱和盐胁迫下的表达分析[J]. 植物学报, 2016, 51(4): 473-487. |
| [15] | 孔庆仙, 夏江宝, 赵自国, 屈凡柱. 不同地下水矿化度对柽柳光合特征及树干液流的影响[J]. 植物生态学报, 2016, 40(12): 1298-1309. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||