

植物学报 ›› 2019, Vol. 54 ›› Issue (4): 522-530.DOI: 10.11983/CBB18229 cstr: 32102.14.CBB18229
收稿日期:2018-10-30
接受日期:2019-02-11
出版日期:2019-07-01
发布日期:2020-01-08
通讯作者:
阮继伟
基金资助:Received:2018-10-30
Accepted:2019-02-11
Online:2019-07-01
Published:2020-01-08
Contact:
Jiwei Ruan
摘要: 正向遗传学突变体筛选被广泛用于揭示减数分裂中涉及的遗传基因, 如调控减数分裂II型交叉形成途径的重组抑制基因。该研究利用拟南芥(Arabidopsis thaliana)花粉荧光标记系进行EMS突变体的正向遗传学筛选, 鉴定拟南芥野生型Col遗传背景下的重组抑制突变体, 共获得18个重组率显著提高3倍以上的重组抑制突变体, 其中包括显性和隐性遗传突变。研究表明, 基于荧光标记高通量鉴定重组抑制突变体是可行的, 可为植物减数分裂重组调控分子机制研究提供新方法和突变材料。
李帆,阮继伟. 利用荧光标记高通量鉴定减数分裂重组抑制突变体. 植物学报, 2019, 54(4): 522-530.
Fan Li,Jiwei Ruan. High-throughput Identification of Meiotic Anti-CO Mutants by Fluorescent Reporters. Chinese Bulletin of Botany, 2019, 54(4): 522-530.
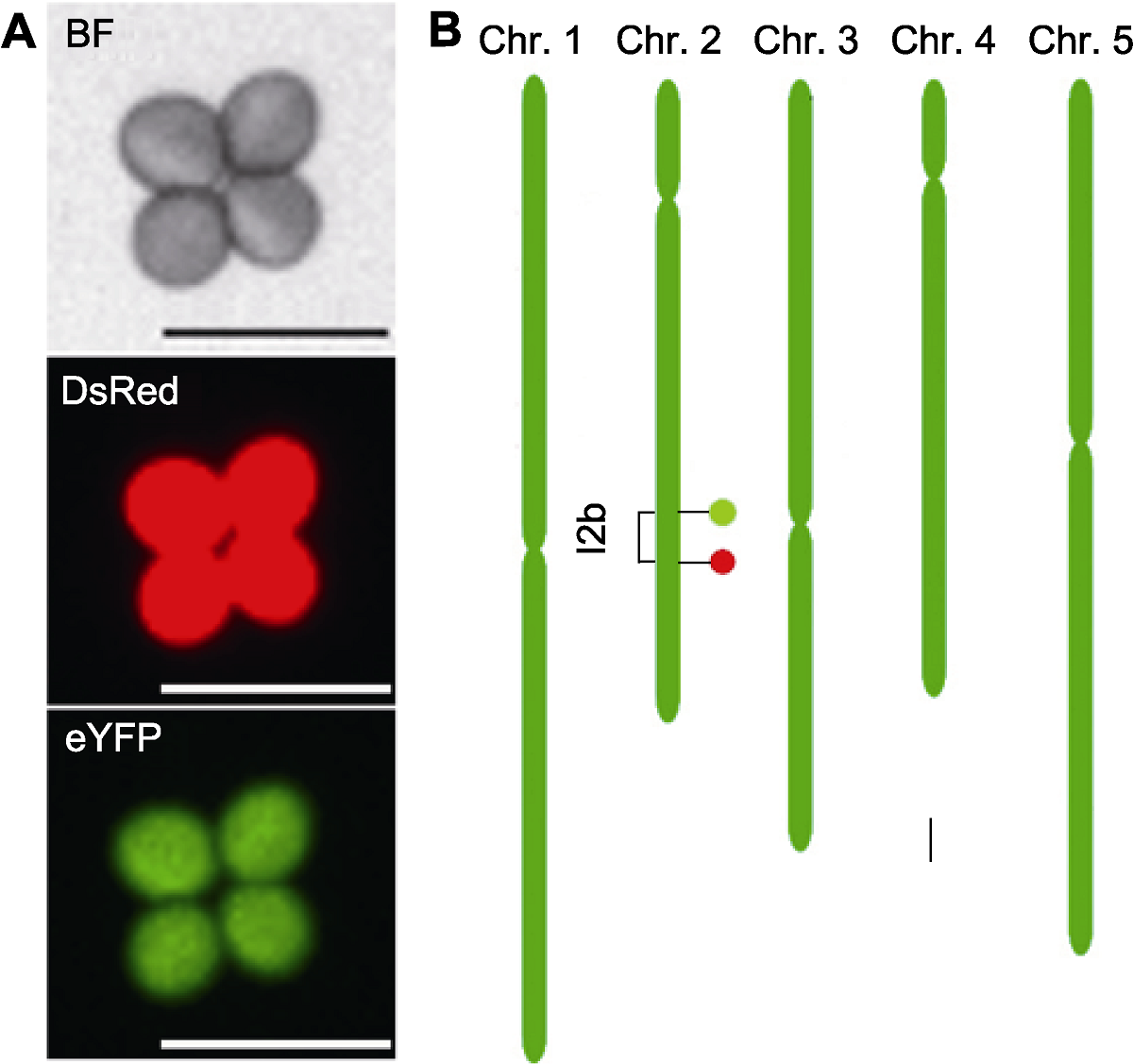
图1 拟南芥花粉荧光标记系FTL-I2b示意图 (A) 拟南芥花粉荧光标记系FTL-I2b四分体花粉荧光显微图, 其包含1个DsRed和1个eYFP标记, 能在不同的荧光激发下分别表达红荧光和绿荧光(Bars=50 μm); (B) 拟南芥花粉荧光标记系FTL-I2b在染色体上的位置示意图(Bar=1 Mb), 红色(下)和绿色(上)椭圆分别代表对应的荧光标记DsRed和eYFP, 并构建出1个I2b标记区间(1.45 Mb)。BF: 明场
Figure 1 A schematic illustration of Arabidopsis fluorescent tagged line FTL-I2b (A) Fluorescence micrograph of a tetrad pollen of FTL-I2b, which contains a DsRed and an eYFP fluorescent marker and expresses red fluorescence and green fluorescence under different fluorescence excitations respectively (Bars=50 μm); (B) The genomic location of FTL-I2b fluorescent markers on the chromosome (Bar=1 Mb), the DsRed and eYFP fluorescent markers are indicated by filled circles colored by red (down) and green (up), respectively, which constructing a I2b interval (1.45 Mb). BF: Bright field
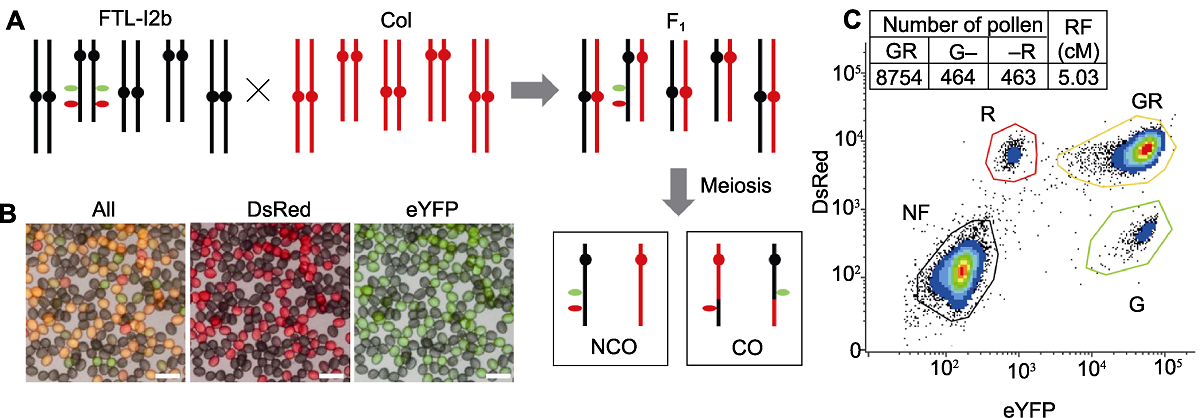
图2 拟南芥FTL-I2b半合子荧光标记(I2b/++) (A) FTL-I2b与Col杂交示意图, F1代经过杂交后具有半合子荧光标记, 通过减数分裂形成重组和未重组的配子; (B) 杂交F1代花粉半合子荧光标记显微图, 图中黄色花粉(GR)为未重组配子, 单红色(R)和单绿色(G)花粉为重组配子(Bars=50 μm); (C) 利用流式细胞仪统计花粉中荧光标记可计算该标记区间I2b的重组率, 计算公式为重组率(RF)=(G+R)/(2GR+G+R)×100。
Figure 2 The Arabidopsis hemizygous fluorescent markers (I2b/++) of FTL-I2b (A) A schematic illustration of FTL-I2b cross with Col, the hybrid F1 contains a hemizygous fluorescent markers after hybridization, the recombined and unrecombined gametes are formed through meiosis; (B) Fluorescence micrograph of pollen in hybrid F1 generation, in which yellow pollen (GR) is unrecombined gamete, and monochromatic red (R) and green (G) pollen is recombined gametes (Bars=50 μm); (C) The recombination frequency (RF) of I2b interval can be calculated by counting the fluorescent marker pattern in the gametes via flow cytometry, the formula calculating recombination frequency=(G+R)/(2GR+G+R)×100.
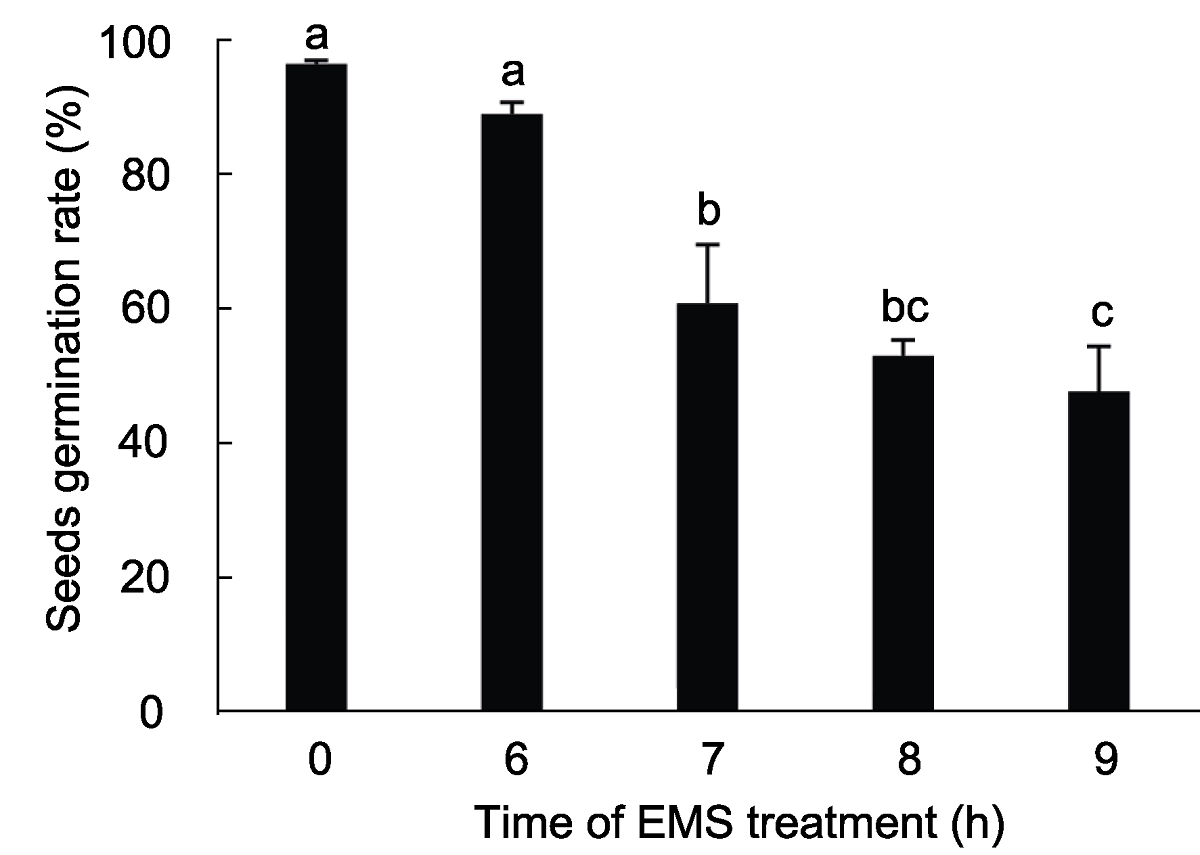
图3 不同时间的EMS (75 mmol∙L-1)处理下M1代拟南芥种子的萌发率 种子萌发率为3次重复的平均发芽率, 不同小写字母表示差异显著(P<0.05)。
Figure 3 The germination rate of M1 Arabidopsis seeds by different treat time using 75 mmol∙L-1 EMS The average germination rate was derived from three repeats, different lowercase letters indicate significant differences (P<0.05).
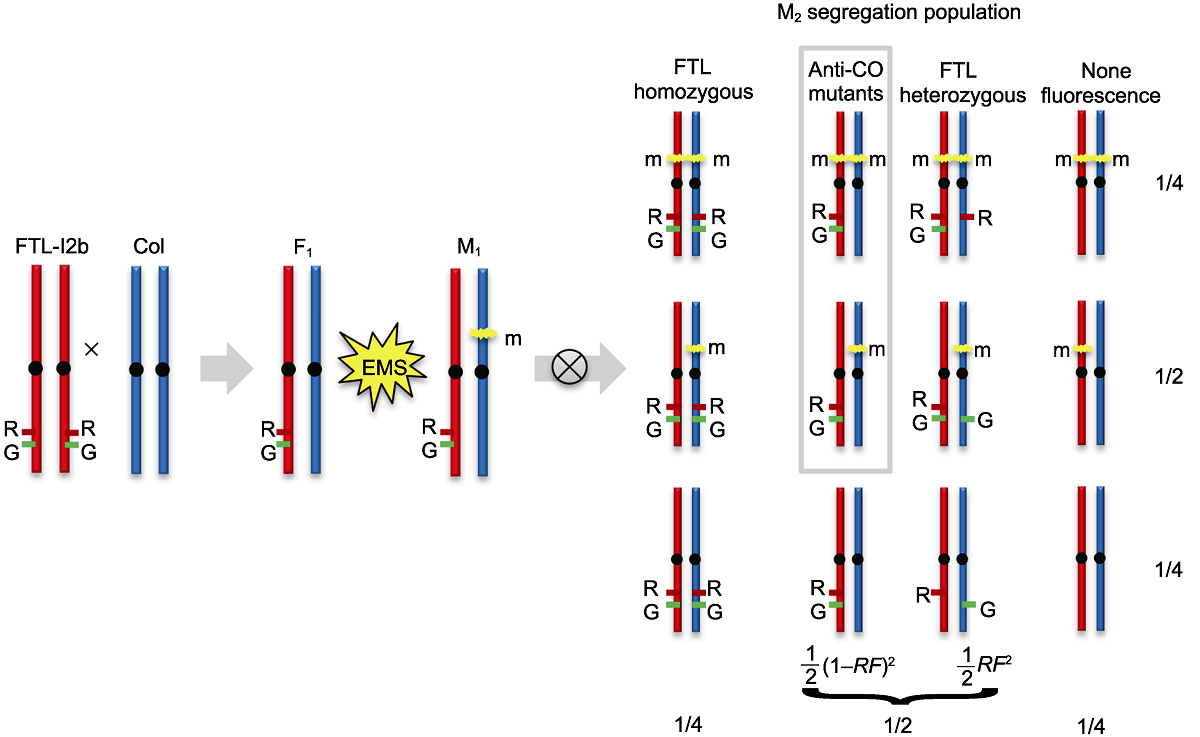
图4 拟南芥减数分裂重组抑制突变体正向遗传学筛选示意图 突变体正向遗传学的筛选建立在拟南芥野生型(WT)遗传背景基础上, 利用花粉荧光标记系FTL-I2b半合子荧光标记(I2b/++)进行高通量重组率检测, 筛选获得重组率提高的突变体。图中FTL-I2b中的DsRed和eYFP荧光标记分别由R和G表示。m代表基因的显性或隐性突变。RF: 重组率
Figure 4 A schematic illustration of Arabidopsis meiotic anti-CO mutants forward genetics screen The forward genetics mutants screen was used the FTL-I2b containing linked hemizygous fluorescent reporters (I2b/++) on the genetic background of Arabidopsis wild type (WT). High-throughput detecting the recombination frequency by flow cytometry could identify the mutants with increased recombination rate. The DsRed and eYFP fluorescent reporters of FTL-I2b are indicated by R and G, respectively. The m refers to a dominant or recessive mutation. RF: Recombination frequency
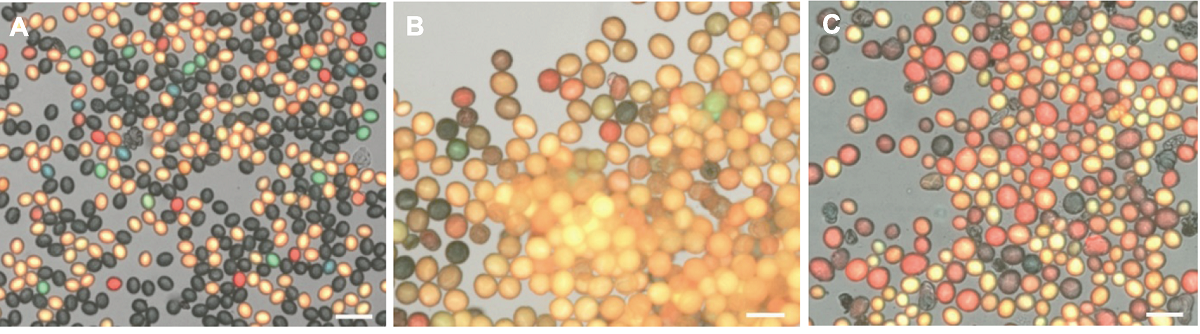
图5 拟南芥野生型与大花粉突变体花粉显微图 (A) 拟南芥野生型(WT)花粉大小为(21.82±0.98) µm (n=10); (B) 大花粉突变体1 (big pollen 1)的大花粉率为100%, 花粉大小为(38.36±1.37) µm (n=10); (C) 大花粉突变体2 (big pollen 2)的大花粉率为50%, 花粉大小为(36.00±2.70) µm (n=10)。Bars=50 µm
Figure 5 The pollen micrograph of Arabidopsis wild type and big pollen mutants (A) The pollen size of Arabidopsis wild type (WT) is (21.82±0.98) µm (n=10); (B) The large pollen rate of big pollen 1 mutant is 100%, pollen size is (38.36±1.37) µm (n=10); (C) The large pollen rate of big pollen 2 mutant is 50%, pollen size is (36.00±2.70) µm (n=10). Bars=50 µm

图6 拟南芥野生型与重组抑制突变体 (A) 拟南芥野生型(WT)、显性重组抑制突变体(drs)和隐性重组抑制突变体(rrs1)植株表型(Bars=1 cm); (B) 拟南芥野生型(WT)、显性重组抑制突变体(drs)和隐性重组抑制突变体(rrs1)花粉荧光标记显微图(Bars=50 μm)。
Figure 6 Arabidopsis wild type and recombination suppressor mutants (A) The plant phenotype of Arabidopsis wild type (WT), dominant recombination suppressor mutant (drs) and recessive recombination suppressor mutant (rrs1) (Bars=1 cm); (B) The fluorescent pollen micrograph of Arabidopsis wild type (WT), dominant recombination suppressor mutant (drs) and recessive recombination suppressor mutant (rrs1) (Bars=50 μm)
| [1] | Berchowitz LE, Copenhaver GP ( 2008). Fluorescent Arabidopsis tetrads: a visual assay for quickly developing large crossover and crossover interference data sets. Nat Protoc 3, 41-50. |
| [2] | Berchowitz LE, Copenhaver GP ( 2010). Genetic interference: don't stand so close to me. Curr Genomics 11, 91-102. |
| [3] | Berchowitz LE, Francis KE, Bey AL, Copenhaver GP ( 2007). The role of AtMUS81 in interference-insensitive crossovers in A. thaliana. PLoS Genet 3, e132. |
| [4] | Crismani W, Girard C, Froger N, Pradillo M, Santos JL, Chelysheva L, Copenhaver GP, Horlow C, Mercier R ( 2012). FANCM limits meiotic crossovers. Science 336, 1588-1590. |
| [5] | Fernandes JB, Duhamel M, Seguéla-Arnaud M, Froger N, Girard C, Choinard S, Solier V, De Winne N, De Jaeger G, Gevaert K, Andrey P, Grelon M, Guerois R, Kumar R, Mercier R ( 2017). FIGL1 and its novel partner FLIP form a conserved complex that regulates homologous recombination. PLoS Genet 14, e1007317. |
| [6] | Fernandes JB, Séguéla-Arnaud M, Larchevêque C, Lloyd AH, Mercier R ( 2018). Unleashing meiotic crossovers in hybrid plants. Proc Natl Acad Sci USA 115, 2431-2436. |
| [7] | Francis KE, Lam SY, Harrison BD, Bey AL, Berchowitz LE, Copenhaver GP ( 2007). Pollen tetrad-based visual assay for meiotic recombination in Arabidopsis. Proc Natl Acad Sci USA 104, 3913-3918. |
| [8] | Girard C, Chelysheva L, Choinard S, Froger N, Macaisne N, Lehmemdi A, Mazel J, Crismani W, Mercier R ( 2015). AAA-ATPase FIDGETIN-LIKE 1 and helicase FANCM antagonize meiotic crossovers by distinct mechanisms. PLoS Genet 11, e1005369. |
| [9] | Girard C, Crismani W, Froger N, Mazel J, Lemhemdi A, Horlow C, Mercier R ( 2014). FANCM-associated proteins MHF1 and MHF2, but not the other Fanconi anemia factors, limit meiotic crossovers. Nucleic Acids Res 42, 9087-9095. |
| [10] | Giraut L, Falque M, Drouaud J, Pereira L, Martin OC, Mézard C ( 2011). Genome-wide crossover distribution in Arabidopsis thaliana meiosis reveals sex-specific patterns along chromosomes. PLoS Genet 7, e1002354. |
| [11] | Hatkevich T, Kohl KP, McMahan S, Hartmann MA, Williams AM, Sekelsky J ( 2017). Bloom syndrome helicase promotes meiotic crossover patterning and homolog disjunction. Curr Biol 27, 96-102. |
| [12] | Higgins JD, Buckling EF, Franklin FC, Jones GH ( 2008). Expression and functional analysis of AtMUS81 in Arabidopsis meiosis reveals a role in the second pathway of crossing-over. Plant J 54, 152-162. |
| [13] | Hu Q, Li YF, Wang HJ, Shen Y, Zhang C, Du GJ, Tang D, Cheng ZK ( 2017). Meiotic chromosome association 1 interacts with TOP3α and regulates meiotic recombination in rice. Plant Cell 29, 1697-1708. |
| [14] | Huang JY, Cheng ZH, Wang C, Hong Y, Su H, Wang J, Copenhaver GP, Ma H, Wang YX ( 2015). Formation of interference-sensitive meiotic cross-overs requires sufficient DNA leading-strand elongation. Proc Natl Acad Sci USA 112, 12534-12539. |
| [15] | Jones GH, Franklin FCH ( 2006). Meiotic crossing-over: obligation and interference. Cell 126, 246-248. |
| [16] | Kim Y, Schumaker KS, Zhu JK ( 2006). EMS mutagenesis of Arabidopsis. In: Salinas J, Sanchez-Serrano JJ, eds. Arabidopsis Protocols. Totowa: Humana Press. pp. 101-103. |
| [17] | Kurzbauer MT, Pradillo M, Kerzendorfer C, Sims J, Ladurner R, Oliver C, Janisiw MP, Mosiolek M, Schweizer D, Copenhaver GP, Schlögelhofer P ( 2018). Arabidopsis thaliana FANCD2 promotes meiotic crossover formation. Plant Cell 30, 415-428. |
| [18] | Li F, De Storme N, Geelen D ( 2017). Dynamics of male meiotic recombination frequency during plant development using fluorescent tagged lines in Arabidopsis thaliana. Sci Rep 7, 42535. |
| [19] | Lu PL, Han XW, Qi J, Yang JG, Wijeratne AJ, Li T, Ma H ( 2012). Analysis of Arabidopsis genome-wide variations before and after meiosis and meiotic recombination by resequencing Landsberg erecta and all four products of a single meiosis. Genome Res 22, 508-518. |
| [20] | Lu PL, Wijeratne AJ, Wang ZJ, Copenhaver GP, Ma H ( 2014). Arabidopsis PTD is required for type I crossover formation and affects recombination frequency in two different chromosomal regions. J Genet Genomics 41, 165-175. |
| [21] | Lukowitz W, Gillmor CS, Scheible WR ( 2000). Positional cloning in Arabidopsis. Why it feels good to have a genome initiative working for you. Plant Physiol 123, 795-806. |
| [22] | Macaisne N, Novatchkova M, Peirera L, Vezon D, Jolivet S, Froger N, Chelysheva L, Grelon M, Mercier R ( 2008). SHOC1, an XPF endonuclease-related protein, is essential for the formation of class I meiotic crossovers. Curr Biol 18, 1432-1437. |
| [23] | Macaisne N, Vignard J, Mercier R ( 2011). SHOC1 and PTD form an XPF-ERCC1-like complex that is required for formation of class I crossovers. J Cell Sci 124, 2687-2691. |
| [24] | Mercier R, Mézard C, Jenczewski E, Macaisne N, Grelon M ( 2015). The molecular biology of meiosis in plants. Annu Rev Plant Biol 66, 297-327. |
| [25] | Mieulet D, Aubert G, Bres C, Klein A, Droc G, Vieille E, Rond-Coissieux C, Sanchez M, Dalmais M, Mauxion JP, Rothan C, Guiderdoni E, Mercier R ( 2018). Unleashing meiotic crossovers in crops. Nat Plants 4, 1010-1016. |
| [26] | Qi J, Chen YM, Copenhaver GP, Ma H ( 2014). Detection of genomic variations and DNA polymorphisms and impact on analysis of meiotic recombination and genetic mapping. Proc Natl Acad Sci USA 111, 10007-10012. |
| [27] | Qu LJ, Qin GJ ( 2014). Generation and identification of Arabidopsis EMS mutants. In: Sanchez-Serrano JJ, Salinas J, eds. Arabidopsis Protocols. Totowa: Humana Press. pp. 225-239. |
| [28] | Séguéla-Arnaud M, Choinard S, Larchevêque C, Girard C, Froger N, Crismani W, Mercier R ( 2017). RMI1 and TOP3α limit meiotic CO formation through their C-terminal domains. Nucleic Acids Res 4, 1860-1871. |
| [29] | Séguéla-Arnaud M, Crismani W, Larchevêque C, Mazel J, Froger N, Choinard S, Lemhemdi A, Macaisne N, Van Leene J, Gevaert K, De Jaeger G, Chelysheva L, Mercier R ( 2015). Multiple mechanisms limit meiotic crossovers: TOP3α and two BLM homologs antagonize crossovers in parallel to FANCM. Proc Natl Acad Sci USA 112, 4713-4718. |
| [30] | Wang YX, Cheng ZH, Huang JY, Shi Q, Hong Y, Copenhaver GP, Gong ZZ, Ma H ( 2012). The DNA replication factor RFC1 is required for interference-sensitive meiotic crossovers in Arabidopsis thaliana. PLoS Genet 8, e1003039. |
| [31] | Yelina NE, Lambing C, Hardcastle TJ, Zhao XH, Santos B, Henderson IR ( 2015). DNA methylation epigenetically silences crossover hot spots and controls chromosomal domains of meiotic recombination in Arabidopsis. Gene Dev 29, 2183-2202. |
| [32] | Yelina NE, Ziolkowski PA, Miller N, Zhao X, Kelly KA, Muñoz DF, Mann DJ, Copenhaver GP, Henderson IR ( 2013). High-throughput analysis of meiotic crossover frequency and interference via flow cytometry of fluorescent pollen in Arabidopsis thaliana. Nat Protoc 8, 2119-2134. |
| [1] | 徐婉约,王应祥. 染色体展片法观察拟南芥雄性减数分裂过程中的染色体形态[J]. 植物学报, 2019, 54(5): 620-624. |
| [2] | 程新杰, 于恒秀, 程祝宽. 水稻减数分裂染色体分析方法[J]. 植物学报, 2019, 54(4): 503-508. |
| [3] | 陶乃奇,张斌,刘信凯,周和达,钟乃盛,严丹峰,张敏,高继银,张文驹. 利用荧光标记SSR鉴别21个茶花新品种[J]. 植物学报, 2019, 54(1): 37-45. |
| [4] | 薛治慧, 种康. 中国科学家在杂种F1克隆繁殖研究领域取得突破性进展[J]. 植物学报, 2019, 54(1): 1-3. |
| [5] | 陈成, 董爱武, 苏伟. 拟南芥组蛋白分子伴侣AtHIRA参与体细胞同源重组及盐胁迫响应[J]. 植物学报, 2018, 53(1): 42-50. |
| [6] | 罗向东, 戴亮芳, 万勇, 胡标林, 李佛生, 李霞, 谢建坤. 东乡野生稻与栽培稻正反交种间杂种F1的雄配子发生与发育[J]. 植物学报, 2011, 46(4): 407-412. |
| [7] | 谭何新 文铁桥 张大兵. 水稻花粉发育的分子机理[J]. 植物学报, 2007, 24(03): 330-339. |
| [8] | 郭军洋;陈劲枫;钱春桃;曹清河. 植物减数分裂染色体配对与染色体组分析的研究进展[J]. 植物学报, 2004, 21(05): 513-520. |
| [9] | 李雅轩. 植物减数分裂研究进展[J]. 植物学报, 1999, 16(05): 526-529. |
| [10] | 曹雅娟 方瑾 刘宁 王慧. 连翅小孢子细胞减数分裂中期II的超微结构[J]. 植物学报, 1996, 13(专辑): 33-34. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
