

植物学报 ›› 2025, Vol. 60 ›› Issue (3): 393-406.DOI: 10.11983/CBB24118 cstr: 32102.14.CBB24118
徐田甜1,2, 杨培建1,2, 周晓茜1,2, 曹怡1,2, 陈艳红1,2, 刘国元1,2, 张健1,2, 魏辉1,2,*( )
)
收稿日期:2024-08-04
接受日期:2024-11-12
出版日期:2025-05-10
发布日期:2024-11-26
通讯作者:
*E-mail: 15850682752@163.com
基金资助:
Xu Tiantian1,2, Yang Peijian1,2, Zhou Xiaoxi1,2, Cao Yi1,2, Chen Yanhong1,2, Liu Guoyuan1,2, Zhang Jian1,2, Wei Hui1,2,*( )
)
Received:2024-08-04
Accepted:2024-11-12
Online:2025-05-10
Published:2024-11-26
Contact:
*E-mail: 15850682752@163.com
摘要: 植物肌醇半乳糖苷合成酶(GolS)是棉子糖家族寡糖(RFOs)生物合成途径中的关键酶, 为棉子糖系列寡糖提供活化的半乳糖基, 调控植物体内棉子糖系列寡糖的生物合成与积累, 在植物响应非生物胁迫中发挥重要作用。然而, 关于紫薇(Lagerstroemia indica) GolS (LiGolS)家族基因的分子结构特征未见报道。该研究在紫薇全基因组水平鉴定了13个LiGolS基因, 并对其理化性质、染色体定位、进化关系、基因结构、保守基序以及盐胁迫下的表达进行分析。结果表明, 13个LiGolS基因不均匀地分布在10条染色体上, 13个LiGolS蛋白的等电点为4.75-9.45, 分子量为37.69-46.12 kDa, 氨基酸数量为327-404 aa。亚细胞定位预测结果表明, 6个蛋白定位在叶绿体, 1个蛋白定位在线粒体, 5个蛋白定位在细胞质, 1个蛋白定位在液泡。13个基因含有的外显子数目为0-4。盐胁迫下LiGolS的表达分析表明, LiGolS家族基因均表现出不同程度的表达上调, 表明这些基因可能参与紫薇的盐胁迫响应。研究结果为解析紫薇GolS基因功能奠定了基础。
徐田甜, 杨培建, 周晓茜, 曹怡, 陈艳红, 刘国元, 张健, 魏辉. 紫薇GolS家族基因的理化特性与表达特征. 植物学报, 2025, 60(3): 393-406.
Xu Tiantian, Yang Peijian, Zhou Xiaoxi, Cao Yi, Chen Yanhong, Liu Guoyuan, Zhang Jian, Wei Hui. Analysis of Physicochemical Characteristics and Expression Characteristics of Lagerstroemia indica GolS Family Genes. Chinese Bulletin of Botany, 2025, 60(3): 393-406.
| Gene name | Gene ID | Length (bp) | Number of amino acids (aa) | Molecular weight (kDa) | Theoretical pI | Aliphatic index | Instability index | GRAVY | Subcelluar location |
|---|---|---|---|---|---|---|---|---|---|
| LiGolS1 | evm.model.Chr1.208 | 1215 | 404 | 46.00 | 9.45 | 92.20 | 57.70 | -0.086 | C |
| LiGolS2 | evm.model.Chr3.1524 | 1146 | 381 | 42.91 | 8.71 | 82.10 | 49.57 | -0.263 | CP |
| LiGolS3 | evm.model.Chr4.803 | 990 | 329 | 37.72 | 4.75 | 78.81 | 50.39 | -0.329 | C |
| LiGolS4 | evm.model.Chr4.806 | 990 | 329 | 37.85 | 4.78 | 78.51 | 48.28 | -0.338 | C |
| LiGolS5 | evm.model.Chr5.968 | 984 | 327 | 37.69 | 4.76 | 80.76 | 53.39 | -0.318 | C |
| LiGolS6 | evm.model.Chr8.101 | 1050 | 349 | 39.71 | 8.16 | 90.32 | 47.76 | -0.148 | CP |
| LiGolS7 | evm.model.Chr8.200 | 1164 | 387 | 43.06 | 7.63 | 87.93 | 44.71 | -0.095 | CP |
| LiGolS8 | evm.model.Chr10.348 | 1125 | 374 | 42.24 | 9.11 | 93.56 | 49.52 | -0.062 | M |
| LiGolS9 | evm.model.Chr18.633 | 1215 | 404 | 46.12 | 9.22 | 94.36 | 51.32 | -0.071 | CP |
| LiGolS10 | evm.model.Chr22.379 | 1143 | 380 | 42.83 | 8.32 | 90.79 | 49.38 | -0.138 | V |
| LiGolS11 | evm.model.Chr23.402 | 1167 | 388 | 43.97 | 9.36 | 88.97 | 57.29 | -0.151 | C |
| LiGolS12 | evm.model.Chr23.618 | 1149 | 382 | 43.30 | 8.85 | 94.40 | 48.35 | -0.109 | CP |
| LiGolS13 | evm.model.Chr24.816 | 1038 | 345 | 38.60 | 7.69 | 92.75 | 43.61 | -0.027 | CP |
表1 LiGolS基因的分子特征
Table 1 Molecular characterization of the LiGolS genes
| Gene name | Gene ID | Length (bp) | Number of amino acids (aa) | Molecular weight (kDa) | Theoretical pI | Aliphatic index | Instability index | GRAVY | Subcelluar location |
|---|---|---|---|---|---|---|---|---|---|
| LiGolS1 | evm.model.Chr1.208 | 1215 | 404 | 46.00 | 9.45 | 92.20 | 57.70 | -0.086 | C |
| LiGolS2 | evm.model.Chr3.1524 | 1146 | 381 | 42.91 | 8.71 | 82.10 | 49.57 | -0.263 | CP |
| LiGolS3 | evm.model.Chr4.803 | 990 | 329 | 37.72 | 4.75 | 78.81 | 50.39 | -0.329 | C |
| LiGolS4 | evm.model.Chr4.806 | 990 | 329 | 37.85 | 4.78 | 78.51 | 48.28 | -0.338 | C |
| LiGolS5 | evm.model.Chr5.968 | 984 | 327 | 37.69 | 4.76 | 80.76 | 53.39 | -0.318 | C |
| LiGolS6 | evm.model.Chr8.101 | 1050 | 349 | 39.71 | 8.16 | 90.32 | 47.76 | -0.148 | CP |
| LiGolS7 | evm.model.Chr8.200 | 1164 | 387 | 43.06 | 7.63 | 87.93 | 44.71 | -0.095 | CP |
| LiGolS8 | evm.model.Chr10.348 | 1125 | 374 | 42.24 | 9.11 | 93.56 | 49.52 | -0.062 | M |
| LiGolS9 | evm.model.Chr18.633 | 1215 | 404 | 46.12 | 9.22 | 94.36 | 51.32 | -0.071 | CP |
| LiGolS10 | evm.model.Chr22.379 | 1143 | 380 | 42.83 | 8.32 | 90.79 | 49.38 | -0.138 | V |
| LiGolS11 | evm.model.Chr23.402 | 1167 | 388 | 43.97 | 9.36 | 88.97 | 57.29 | -0.151 | C |
| LiGolS12 | evm.model.Chr23.618 | 1149 | 382 | 43.30 | 8.85 | 94.40 | 48.35 | -0.109 | CP |
| LiGolS13 | evm.model.Chr24.816 | 1038 | 345 | 38.60 | 7.69 | 92.75 | 43.61 | -0.027 | CP |
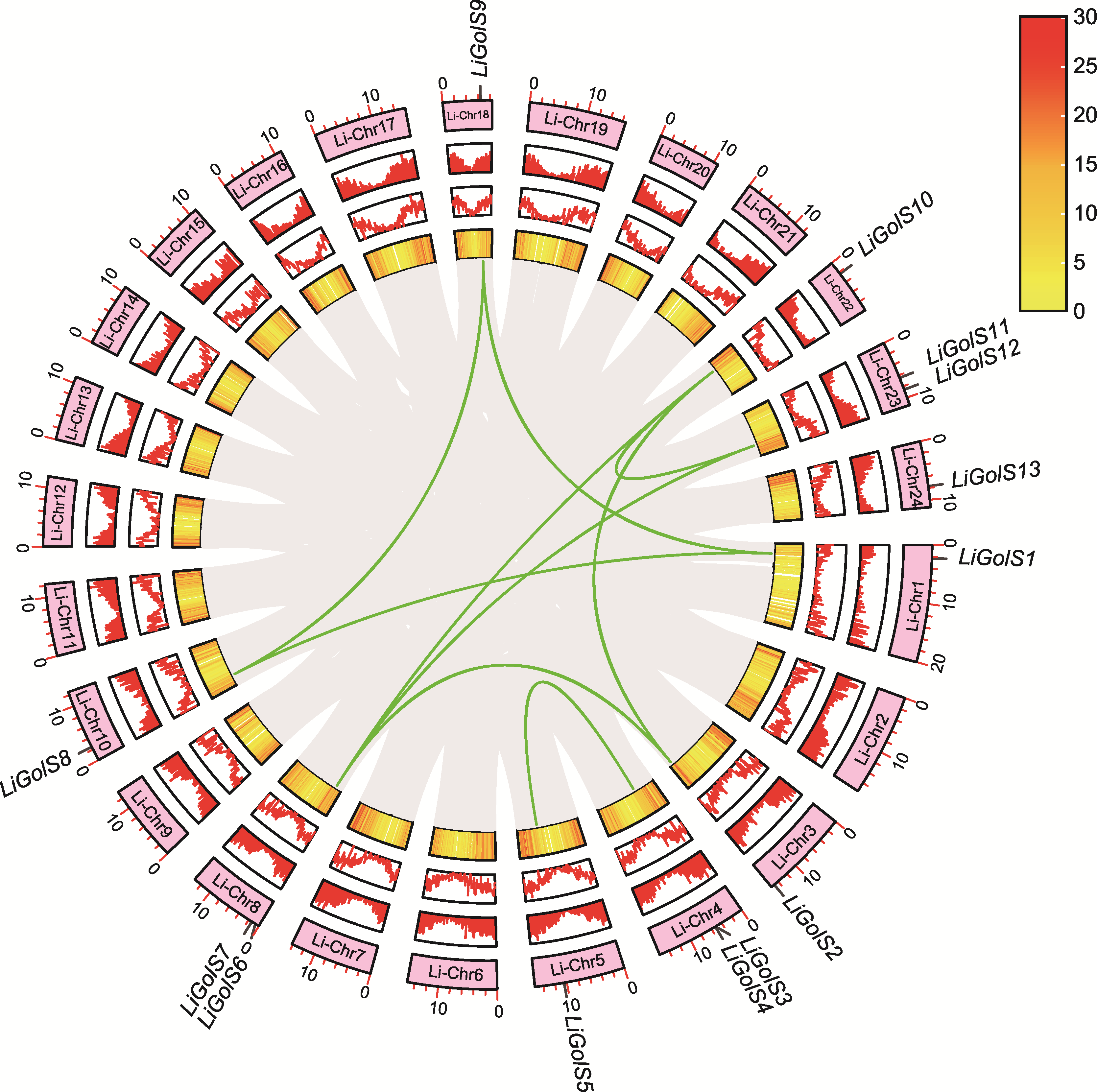
图2 LiGolS基因的共线性分析 圆环从外到内依次代表基因在染色体上的定位、基因密度和共线性区域(绿色连线为LiGolS基因)。
Figure 2 Collinearity analysis of LiGolS genes The ring, from the outside to the inside, represents the location of genes on chromosomes, gene density and collinear regions (the green lines represent LiGolS genes).
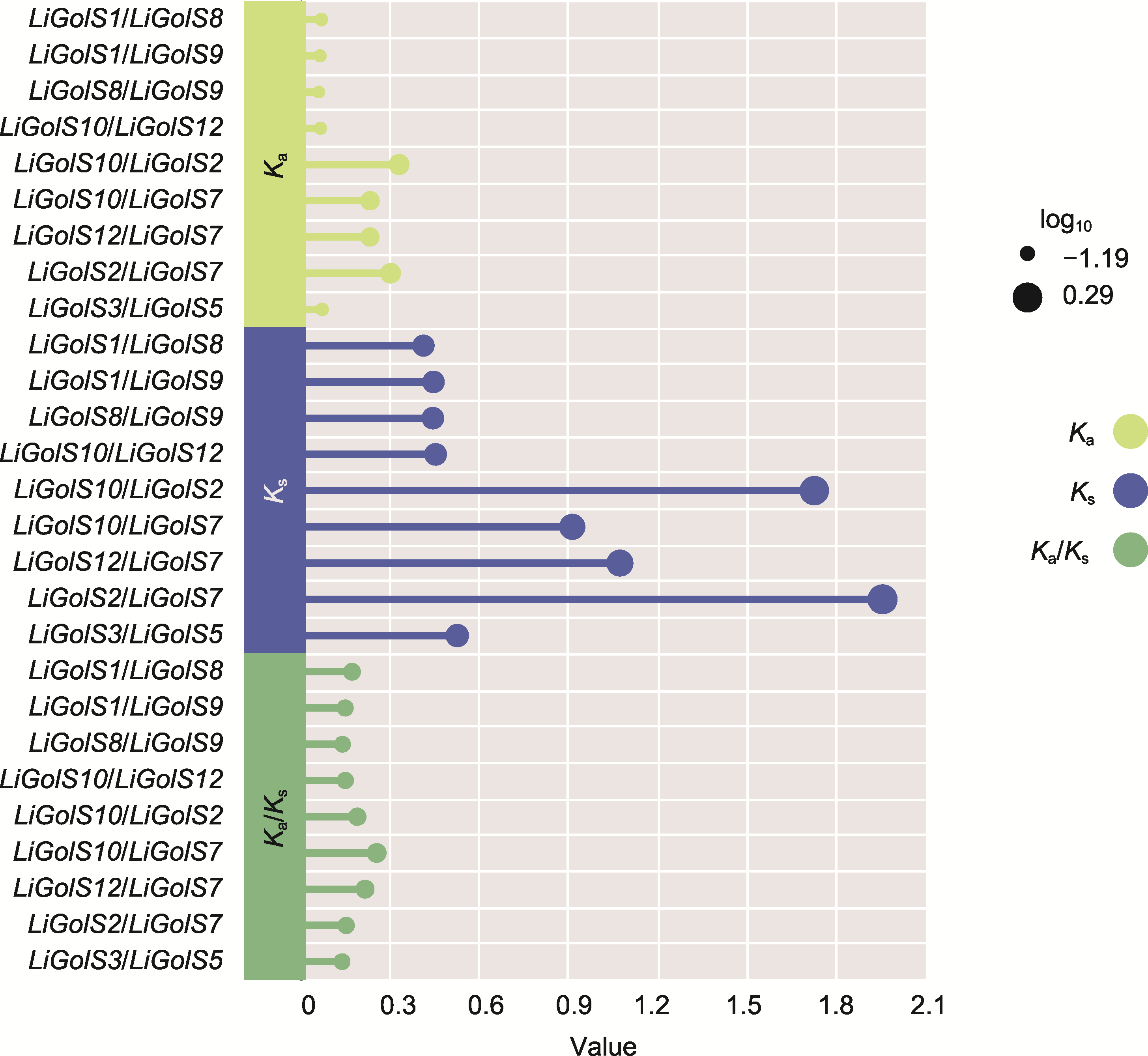
图3 LiGolS基因的非同义替换率(Ka)、同义替换率(Ks)和Ka/Ks 基因的进化压力可通过计算Ka、Ks及Ka/Ks来推断。使用TBtools v2.119的Simple Ka/Ks_Calculator程序, 计算LiGolS基因的Ka和Ks。每个圆圈代表1个共线性基因对, 杆长对应于特定位点的Ka、Ks和Ka/Ks。
Figure 3 The ratio of non-synonymous (Ka), synonymous (Ks) and Ka/Ks of LiGolS genes The evolutionary pressure of genes can be inferred by calculating the Ka, Ks, and Ka/Ks. The Ka and Ks values of LiGolS genes were calculated using the Simple Ka/Ks_Calculator program of TBtools v2.119. Each circle on the chart represents a collinearity gene pair, and the length of the rod corresponds to the Ka, Ks, and Ka/Ks values at a specific site.
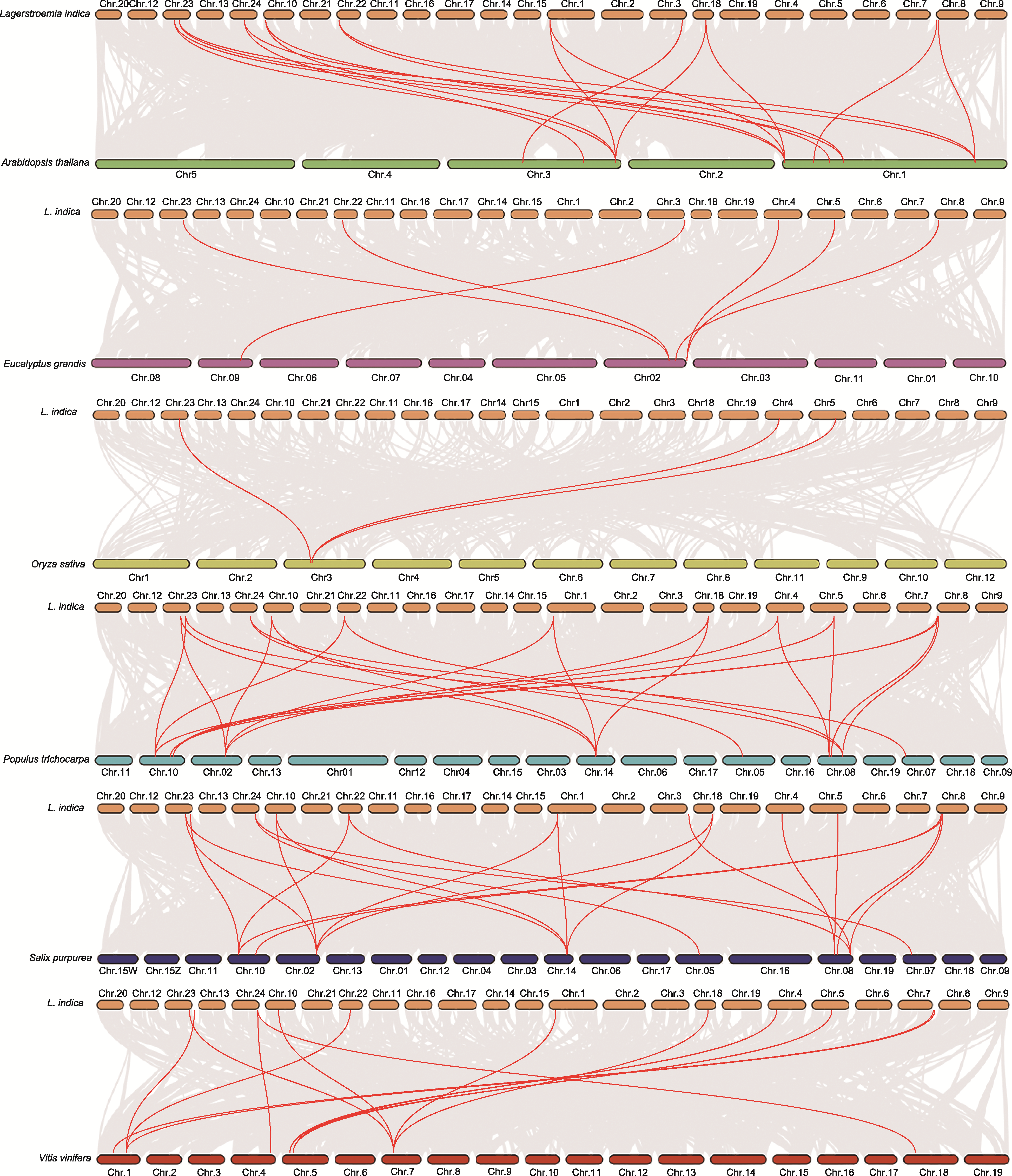
图4 不同物种GolS基因的共线性分析 灰线表示共线基因对, 红线表示共线GolS基因对。染色体用不同颜色的矩形表示。橙色: 紫薇; 绿色: 拟南芥; 紫色: 大桉; 黄色: 水稻; 天蓝色: 杨树; 湛蓝色: 红皮柳; 红色: 葡萄
Figure 4 Collinearity analysis of GolS genes among different species The gray lines represent collinear gene pairs, the red lines represent collinear GolS gene pairs. The chromosomes are represented by rectangles of different colors. Orange: Lagerstroemia indica; Green: Arabidopsis thaliana; Purple: Eucalyptus grandis; Yellow: Oryza sativa; Sky blue: Populus trichocarpa; Blue: Salix purpurea; Red: Vitis vinifera
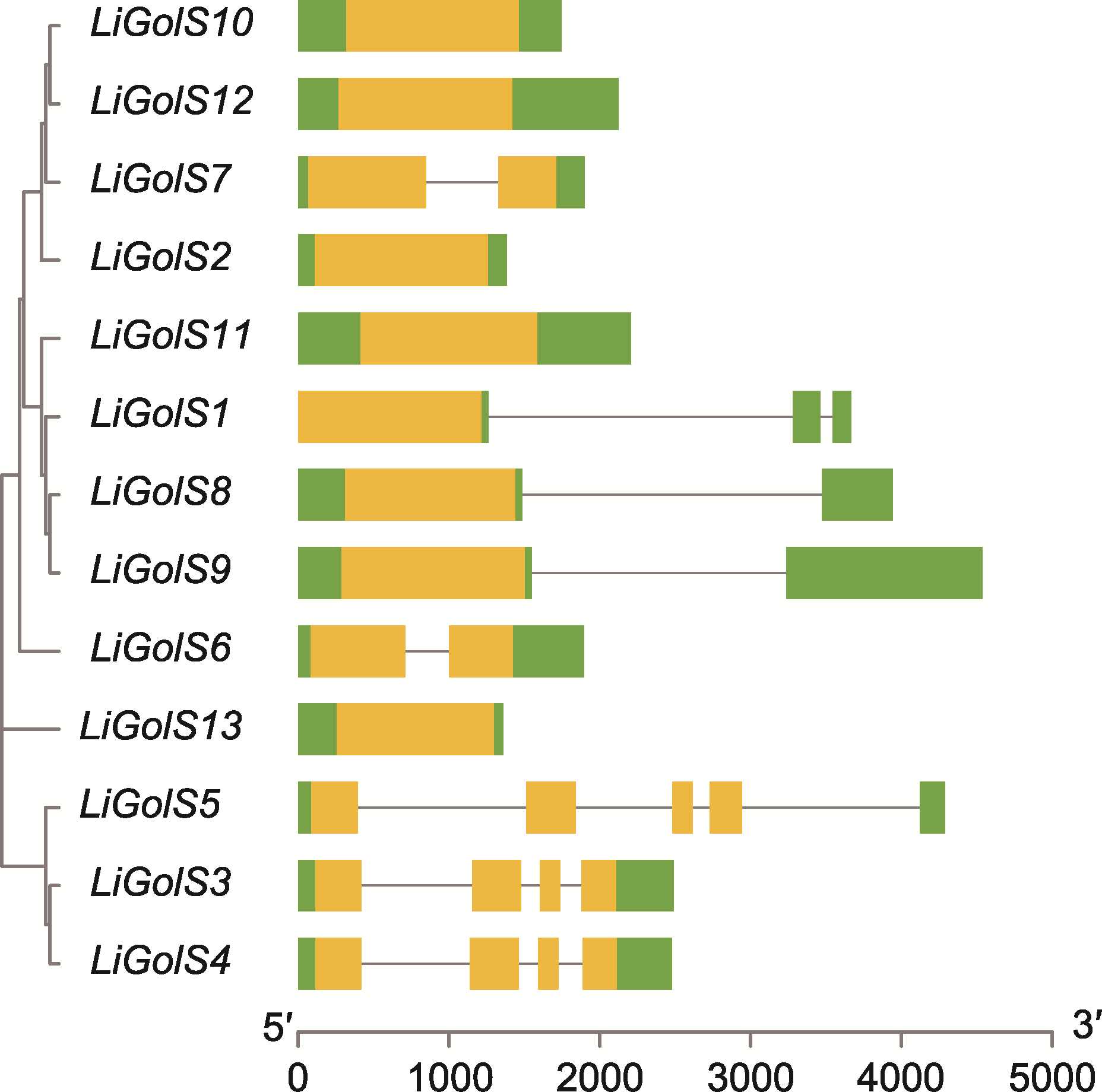
图5 13个LiGolS基因的结构 绿色矩形表示非翻译区(UTRs), 黄色矩形表示外显子, 黑色线条表示内含子。
Figure 5 Gene structure of 13 LiGolS genes Untranslated regions (UTRs) are represented by green rectangles, exons are represented by yellow rectangles, and introns are represented by black lines.
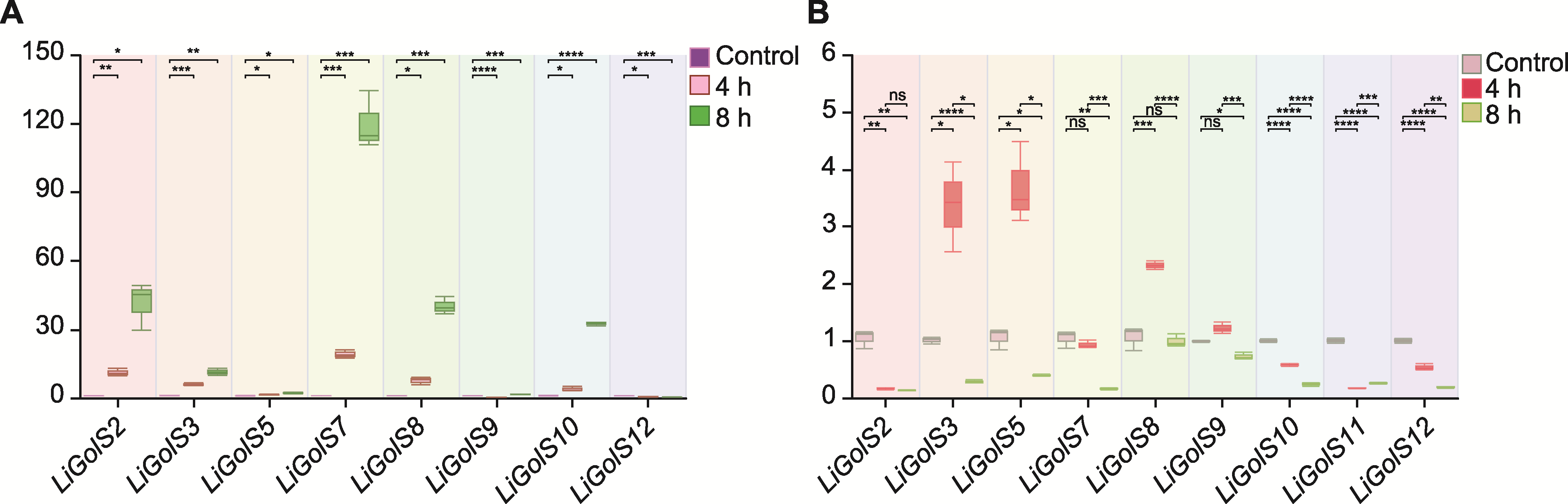
图9 盐胁迫下LiGolS基因的表达模式分析 (A) 叶片中LiGolS基因的表达水平; (B) 根中LiGolS基因的表达水平。ns表示无统计学意义。* P<0.05; ** P<0.01; *** P<0.001; **** P<0.0001
Figure 9 Analysis of LiGolS gene expression patterns under salt stress (A) Expression levels of LiGolS genes in the leaves; (B) Expression levels of LiGolS genes in the roots. ns means no statistical significance. * P<0.05; ** P<0.01; *** P<0.001; **** P<0.0001
| [1] | Artimo P, Jonnalagedda M, Arnold K, Baratin D, Csardi G, de Castro E, Duvaud S, Flegel V, Fortier A, Gasteiger E, Grosdidier A, Hernandez C, Ioannidis V, Kuznetsov D, Liechti R, Moretti S, Mostaguir K, Redaschi N, Rossier G, Xenarios I, Stockinger H (2012). ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res 40, W597-W603. |
| [2] | Bailey TL, Williams N, Misleh C, Li WW (2006). MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res 34, W369-W373. |
| [3] | Bateman A, Coin L, Durbin R, Finn RD, Hollich V, Griffiths-Jones S, Khanna A, Marshall M, Moxon S, Sonnhammer ELL, Studholme DJ, Yeats C, Eddy SR (2004). The Pfam protein families database. Nucleic Acids Res 32, D138-D141. |
| [4] | Chen CJ, Chen H, Zhang Y, Thomas HR, Frank MH, He YH, Xia R (2020). TBtools: an integrative toolkit develo- a)ped for interactive analyses of big biological data. Mol Plant 13, 1194-1202. |
| [5] | Chou KC, Shen HB (2010). Plant-mPLoc: a top-down strategy to augment the power for predicting plant protein subcellular localization. PLoS One 5, e11335. |
| [6] |
da Silveira Falavigna V, Porto DD, Miotto YE, dos Santos HP, de Oliveira PR, Margis-Pinheiro M, Pasquali G, Revers LF (2018). Evolutionary diversification of galactinol synthases in Rosaceae: adaptive roles of galactinol and raffinose during apple bud dormancy. J Exp Bot 69, 1247-1259.
DOI PMID |
| [7] | ElSayed AI, Rafudeen MS, Golldack D (2014). Physiological aspects of raffinose family oligosaccharides in plants: protection against abiotic stress. Plant Biol (Stuttg) 16, 1-8. |
| [8] | Fan YH, Yu MN, Liu M, Zhang R, Sun W, Qian MC, Duan HC, Chang W, Ma JQ, Qu CM, Zhang K, Lei B, Lu K (2017). Genome-wide identification, evolutionary and expression analyses of the GALACTINOL SYNTHASE gene family in rapeseed and tobacco. Int J Mol Sci 18, 2768. |
| [9] | Filiz E, Ozyigit II, Vatansever R (2015). Genome-wide identification of galactinol synthase (GolS) genes in Solanum lycopersicum and Brachypodium distachyon. Comput Biol Chem 58, 149-157. |
| [10] | Gawłowska M, Święcicki W, Lahuta L, Kaczmarek Z (2017). Raffinose family oligosaccharides in seeds of Pisum wild taxa, type lines for seed genes, domesticated and advanced breeding materials. Genet Resour Crop Evol 64, 569-578. |
| [11] | Gu L, Zhang YM, Zhang MS, Li T, Dirk LMA, Downie B, Zhao TY (2016). ZmGOLS2, a target of transcription factor ZmDREB2A, offers similar protection against abiotic stress as ZmDREB2A. Plant Mol Biol 90, 157-170. |
| [12] |
Guerra D, Crosatti C, Khoshro HH, Mastrangelo AM, Mica E, Mazzucotelli E (2015). Post-transcriptional and post-translational regulations of drought and heat response in plants: a spider's web of mechanisms. Front Plant Sci 6, 57.
DOI PMID |
| [13] | Guo ZJ, Ma DN, Li J, Wei MY, Zhang LD, Zhou LC, Zhou XX, He SS, Wang L, Shen YJ, Li QQ, Zheng HL (2022). Genome-wide identification and characterization of aquaporins in mangrove plant Kandelia obovata and its role in response to the intertidal environment. Plant Cell Environ 45, 1698-1718. |
| [14] | Huang TW, Luo XL, Fan ZP, Yang YN, Wan W (2021). Genome-wide identification and analysis of the sucrose synthase gene family in cassava (Manihot esculenta Crantz). Gene 769, 145191. |
| [15] |
Kollist H, Zandalinas SI, Sengupta S, Nuhkat M, Kangasjärvi J, Mittler R (2019). Rapid responses to abiotic stress: priming the landscape for the signal transduction network. Trends Plant Sci 24, 25-37.
DOI PMID |
| [16] | Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouzé P, Rombauts S (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 30, 325-327. |
| [17] | Letunic I, Doerks T, Bork P (2012). SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res 40, D302-D305. |
| [18] | Liu D, Wang KA, Ni P, Wang QY, Zhu K, Wei WL (2022). Identification of soybean GolS gene family and analysis of expression patterns under salt and drought stresses. Chin J Biotechnol 38, 3757-3772. (in Chinese) |
| 刘丹, 王柯蔼, 倪蓬, 王秋艳, 朱康, 危文亮 (2022). 大豆GolS基因家族鉴定及盐旱胁迫下的表达分析. 生物工程学报 38, 3757-3772. | |
| [19] | Liu YD, Zhang L, Chen LJ, Ma H, Ruan YY, Xu T, Xu CQ, He Y, Qi MF (2016). Molecular cloning and expression of an encoding galactinol synthase gene (AnGolS1) in seedling of Ammopiptanthus nanus. Sci Rep 6, 36113. |
| [20] |
Livak KJ, Schmittgen TD (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCTmethod. Methods 25, 402-408.
DOI PMID |
| [21] |
Montillet JL, Chamnongpol S, Rustérucci C, Dat J, van de Cotte B, Agnel JP, Battesti C, Inzé D, Van Breusegem F, Triantaphylidès C (2005). Fatty acid hydroperoxides and H2O2 in the execution of hypersensitive cell death in tobacco leaves. Plant Physiol 138, 1516-1526.
DOI PMID |
| [22] |
Obata T, Fernie AR (2012). The use of metabolomics to dissect plant responses to abiotic stresses. Cell Mol Life Sci 69, 3225-3243.
DOI PMID |
| [23] | Panikulangara TJ, Eggers-Schumacher G, Wunderlich M, Stransky H, Schöffl F (2004). Galactinol synthase1. A novel heat shock factor target gene responsible for heat- induced synthesis of raffinose family oligosaccharides in Arabidopsis. Plant Physiol 136, 3148-3158. |
| [24] | Sami F, Yusuf M, Faizan M, Faraz A, Hayat S (2016). Role of sugars under abiotic stress. Plant Physiol Biochem 109, 54-61. |
| [25] |
Saravitz DM, Pharr DM, Carter TE (1987). Galactinol synthase activity and soluble sugars in developing seeds of four soybean genotypes. Plant Physiol 83, 185-189.
DOI PMID |
| [26] |
Selvaraj MG, Ishizaki T, Valencia M, Ogawa S, Dedicova B, Ogata T, Yoshiwara K, Maruyama K, Kusano M, Saito K, Takahashi F, Shinozaki K, Nakashima K, Ishitani M (2017). Overexpression of an Arabidopsis thaliana galactinol synthase gene improves drought tolerance in transgenic rice and increased grain yield in the field. Plant Biotechnol J 15, 1465-1477.
DOI PMID |
| [27] |
Sengupta S, Mukherjee S, Parween S, Majumder AL (2012). Galactinol synthase across evolutionary diverse taxa: functional preference for higher plants? FEBS Lett 586, 1488-1496.
DOI PMID |
| [28] |
Sprenger N, Keller F (2000). Allocation of raffinose family oligosaccharides to transport and storage pools in Ajuga reptans: the roles of two distinct galactinol synthases. Plant J 21, 249-258.
DOI PMID |
| [29] | Taji T, Ohsumi C, Iuchi S, Seki M, Kasuga M, Kobayashi M, Yamaguchi-Shinozaki K, Shinozaki K (2002). Important roles of drought- and cold-inducible genes for galactinol synthase in stress tolerance in Arabidopsis thaliana. Plant J 29, 417-426. |
| [30] | Verma SS, Chinnusamy V, Bansa KC (2008). A simplified floral dip method for transformation of Brassica napus and B. carinata. J Plant Biochem Biotechnol 17, 197-200. |
| [31] | Vinson CC, Mota APZ, Porto BN, Oliveira TN, Sampaio I, Lacerda AL, Danchin EGJ, Guimaraes PM, Williams TCR, Brasileiro ACM (2020). Characterization of raffinose metabolism genes uncovers a wild Arachis galactinol synthase conferring tolerance to abiotic stresses. Sci Rep 10, 15258. |
| [32] | Wang LX, Lin YX, Hou GY, Yang M, Peng YT, Jiang YY, He CX, She MS, Chen Q, Li MY, Zhang Y, Zhang YT, Wang Y, He W, Wang XR, Tang HR, Luo Y (2024). A histone deacetylase, FaSRT1-2, plays multiple roles in regulating fruit ripening, plant growth and stresses resistance of cultivated strawberry. Plant Cell Environ 47, 2258-2273. |
| [33] |
You J, Wang YY, Zhang YJ, Dossa K, Li DH, Zhou R, Wang LH, Zhang XR (2018). Genome-wide identification and expression analyses of genes involved in raffinose accumulation in sesame. Sci Rep 8, 4331.
DOI PMID |
| [34] | Yu CM, Liu GY, Qin J, Wan X, Guo AF, Wei H, Chen YH, Lian BL, Zhong F, Zhang J (2024). Genomic and transcriptomic studies on flavonoid biosynthesis in Lagerstroemia indica. BMC Plant Biol 24, 171. |
| [35] | Zhang Z, Li J, Zhao XQ, Wang J, Wong GKS, Yu J (2006). KaKs_calculator: calculating Ka and Ks through model selection and model averaging. Genomics Proteomics Bioinformatics 4, 259-263. |
| [36] | Zhou Y, Liu Y, Wang SS, Shi C, Zhang R, Rao J, Wang X, Gu XG, Wang YS, Li DX, Wei CL (2017). Molecular cloning and characterization of galactinol synthases in Camellia sinensis with different responses to biotic and abiotic stressors. J Agric Food Chem 65, 2751-2759. |
| [37] | Zhuo CL, Wang T, Lu SY, Zhao YQ, Li XG, Guo ZF (2013). A cold responsive galactinol synthase gene from Medicago falcata (MfGolS1) is induced by myo-inositol and confers multiple tolerances to abiotic stresses. Physiol Plant 149, 67-78. |
| [38] | Zuther E, Büchel K, Hundertmark M, Stitt M, Hincha DK, Heyer AG (2004). The role of raffinose in the cold acclimation response of Arabidopsis thaliana. FEBS Lett 576, 169-173. |
| [1] | 杜锦瑜, 孙震, 苏彦龙, 王贺萍, 刘亚玲, 吴振映, 何峰, 赵彦, 付春祥. 蒙古冰草咖啡酸氧甲基转移酶基因AmCOMT1的鉴定及功能分析[J]. 植物学报, 2024, 59(3): 383-396. |
| [2] | 王菲菲, 周振祥, 洪益, 谷洋洋, 吕超, 郭宝健, 朱娟, 许如根. 大麦NF-YC基因鉴定及在盐胁迫下的表达分析[J]. 植物学报, 2023, 58(1): 140-149. |
| [3] | 张琦, 张文静, 袁宪凯, 李明, 赵强, 杜艳丽, 杜吉到. 褪黑素对盐胁迫下普通菜豆芽期核酸修复的调控机制[J]. 植物学报, 2023, 58(1): 108-121. |
| [4] | 张楠,刘自广,孙世臣,刘圣怡,林建辉,彭疑芳,张晓旭,杨贺,岑曦,吴娟. 拟南芥AtR8 lncRNA对盐胁迫响应及其对种子萌发的调节作用[J]. 植物学报, 2020, 55(4): 421-429. |
| [5] | 曹栋栋,陈珊宇,秦叶波,吴华平,阮关海,黄玉韬. 水杨酸调控盐胁迫下羽衣甘蓝种子萌发的机理[J]. 植物学报, 2020, 55(1): 49-61. |
| [6] | 栗露露,殷文超,牛梅,孟文静,张晓星,童红宁. 油菜素甾醇调控水稻盐胁迫应答的作用研究[J]. 植物学报, 2019, 54(2): 185-193. |
| [7] | 郭淑华, 孙永江, 牛彦杰, 韩宁, 翟衡, 杜远鹏. 碱性盐胁迫对葡萄种间杂交育种F1代光系统活性的影响[J]. 植物学报, 2018, 53(2): 196-202. |
| [8] | 徐晨, 刘晓龙, 李前, 凌凤楼, 武志海, 张治安. 供氮水平对盐胁迫下水稻叶片光合及叶绿素荧光特性的影响[J]. 植物学报, 2018, 53(2): 185-195. |
| [9] | 陈成, 董爱武, 苏伟. 拟南芥组蛋白分子伴侣AtHIRA参与体细胞同源重组及盐胁迫响应[J]. 植物学报, 2018, 53(1): 42-50. |
| [10] | 刘宝玲, 张莉, 孙岩, 薛金爱, 高昌勇, 苑丽霞, 王计平, 贾小云, 李润植. 谷子bZIP转录因子的全基因组鉴定及其在干旱和盐胁迫下的表达分析[J]. 植物学报, 2016, 51(4): 473-487. |
| [11] | 祁琳, 柏新富, 牛玮浩, 张振华. 根际通气状况对盐胁迫下棉花幼苗生长的影响[J]. 植物学报, 2016, 51(1): 16-23. |
| [12] | 姜琼, 王幼宁, 王利祥, 孙政玺, 李霞. 盐胁迫下大豆根组织定量PCR分析中内参基因的选择[J]. 植物学报, 2015, 50(6): 754-764. |
| [13] | 陈莎莎, 贺转转, 姜生秀, 邢佳佳, 吕秀云, 兰海燕. 藜钙依赖磷酸激酶基因CaCPK的克隆及胁迫表达[J]. 植物学报, 2014, 49(2): 139-149. |
| [14] | 刘焱, 邢立静, 李俊华, 戴绍军. 水稻含有B-box锌指结构域的OsBBX25蛋白参与植物对非生物胁迫的响应[J]. 植物学报, 2012, 47(4): 366-378. |
| [15] | 杨利艳, 韩榕. Ca2+对小麦萌发及幼苗抗盐性的效应[J]. 植物学报, 2011, 46(2): 155-161. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||