

植物学报 ›› 2017, Vol. 52 ›› Issue (6): 713-722.DOI: 10.11983/CBB16239 cstr: 32102.14.CBB16239
刘凯歌, 齐双慧, 段绍伟, 李东, 金倡宇, 高晨浩, 刘绚霞, 陈明训*( )
)
收稿日期:2016-12-05
接受日期:2017-04-03
出版日期:2017-11-01
发布日期:2017-11-22
通讯作者:
陈明训
基金资助:
Liu Kaige, Qi Shuanghui, Duan Shaowei, Li Dong, Jin Changyu, Gao Chenhao, Liu Mingxun Chen Xuanxia*( )
)
Received:2016-12-05
Accepted:2017-04-03
Online:2017-11-01
Published:2017-11-22
Contact:
Liu Mingxun Chen Xuanxia
摘要: 拟南芥(Arabidopsis thaliana) AtTTG1作为WD40重复转录因子存在于细胞核中, 对表皮毛形成、花青素合成和储藏物质积累等具有重要调节作用。该研究从甘蓝型油菜(Brassica napus)品种秦优7号中克隆获得了BnTTG1-1基因的全长CDS序列, 对其进行了烟草(Nicotiana benthamiana)叶片细胞的亚细胞定位研究, 检测了BnTTG1-1在油菜(B. campestris)中的时空表达模式, 并比较分析了BnTTG1-1对多个生物学过程的影响作用。结果表明, BnTTG1-1定位于烟草叶片细胞的细胞核中, 推测其作为转录因子发挥调节作用。BnTTG1-1广泛存在于油菜营养组织和发育的种子中。在突变体ttg1-13背景下, 异源表达BnTTG1-1基因能够完全恢复该突变体的多个表型, 如无表皮毛形成和花青素合成、种皮呈黄色、种子脂肪酸和储藏蛋白含量高以及在种子萌发和幼苗形态建成过程中对高葡萄糖和高盐胁迫耐受力差等。由此可知, 甘蓝型油菜BnTTG1-1与拟南芥AtTTG1在植物生长发育的多个生物学过程中具有类似的功能。
刘凯歌, 齐双慧, 段绍伟, 李东, 金倡宇, 高晨浩, 刘绚霞, 陈明训. 甘蓝型油菜BnTTG1-1基因的功能分析. 植物学报, 2017, 52(6): 713-722.
Liu Kaige, Qi Shuanghui, Duan Shaowei, Li Dong, Jin Changyu, Gao Chenhao, Liu Mingxun Chen Xuanxia. Functional Analysis of Brassica napus BnTTG1-1 Gene. Chinese Bulletin of Botany, 2017, 52(6): 713-722.
| Primer name | Primer sequence (5′-3′) | Annotation |
|---|---|---|
| AtACTIN7-F | GCCCCTGAGGAGCACCCAGTT | RT-PCR |
| AtACTIN7-R | CCGGTTGTACGACCACTGGCA | |
| BnTTG1-1-F | GCCAGTATCCGTCCTCAACA | RT-PCR |
| BnTTG1-1-R | CTCCCAGATAAGAGCCTGCG | |
| BnACTIN7-F | GGAGCTGAGAGATTCCGTTG | qRT-PCR |
| BnACTIN7-R | GAACCACCACTGAGGACGAT | |
| BnTTG1-1-F | CTGCAGTGGTCTTCTTCGTT | qRT-PCR |
| BnTTG1-1-R | GTTACAATCACATAGATGCAGAGAC | |
| BnTTG1-1-Xma1-F | TATTcccgggATGGACAACTCAGCTCCAGACTC | 35S:BnTTG1-1-GFP and 35S:BnTTG1-1 |
| BnTTG1-1-Spe1-R | GGactagtAACTCTAAGGAGCTGCATTTTGTTAGC |
表1 引物序列
Table 1 Sequences of primers
| Primer name | Primer sequence (5′-3′) | Annotation |
|---|---|---|
| AtACTIN7-F | GCCCCTGAGGAGCACCCAGTT | RT-PCR |
| AtACTIN7-R | CCGGTTGTACGACCACTGGCA | |
| BnTTG1-1-F | GCCAGTATCCGTCCTCAACA | RT-PCR |
| BnTTG1-1-R | CTCCCAGATAAGAGCCTGCG | |
| BnACTIN7-F | GGAGCTGAGAGATTCCGTTG | qRT-PCR |
| BnACTIN7-R | GAACCACCACTGAGGACGAT | |
| BnTTG1-1-F | CTGCAGTGGTCTTCTTCGTT | qRT-PCR |
| BnTTG1-1-R | GTTACAATCACATAGATGCAGAGAC | |
| BnTTG1-1-Xma1-F | TATTcccgggATGGACAACTCAGCTCCAGACTC | 35S:BnTTG1-1-GFP and 35S:BnTTG1-1 |
| BnTTG1-1-Spe1-R | GGactagtAACTCTAAGGAGCTGCATTTTGTTAGC |
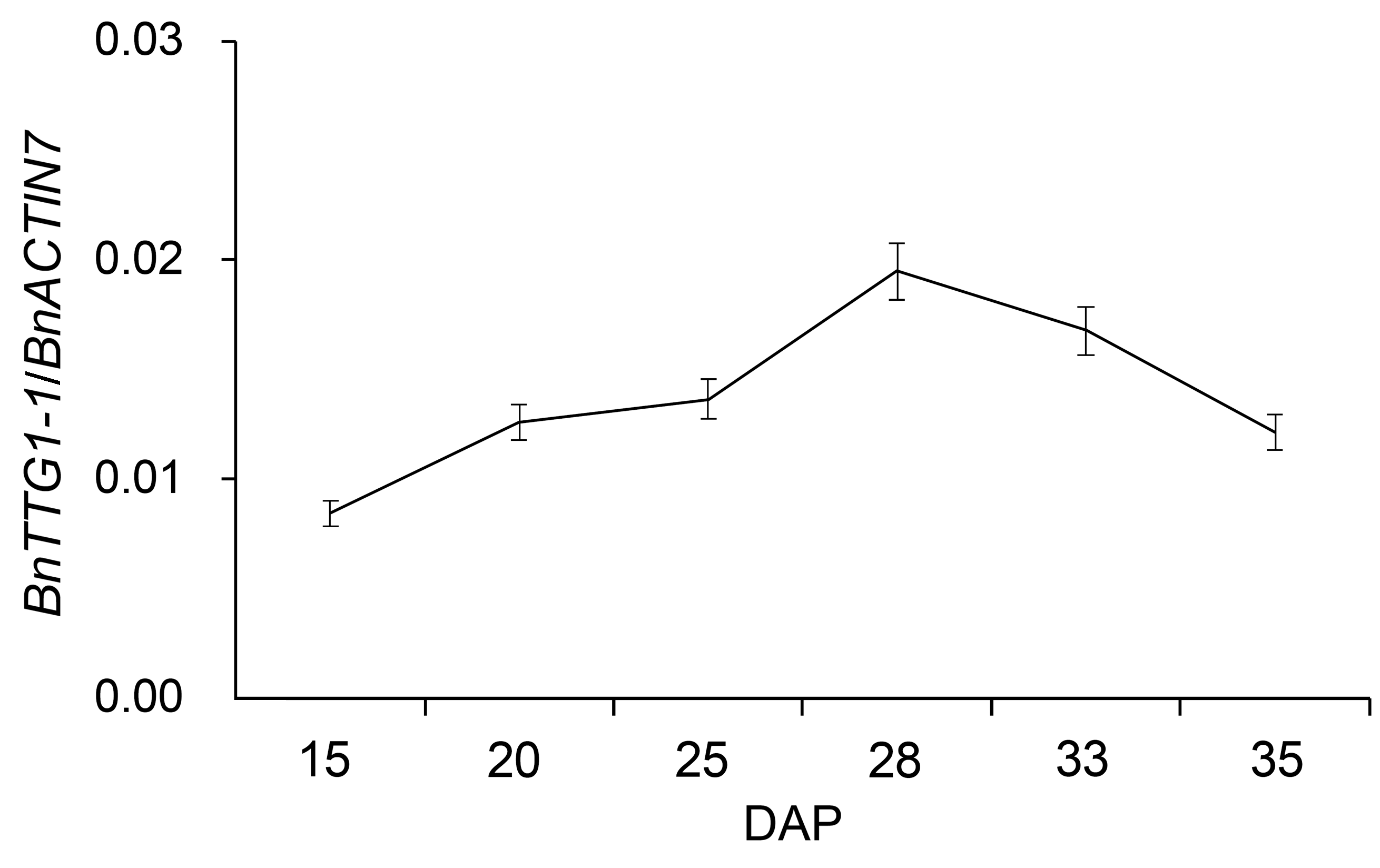
图1 qRT-PCR分析BnTTG1-1基因在甘蓝型油菜秦优7号种子不同发育时期的表达模式(平均值±标准差)DAP: 授粉后的天数。BnACTIN7为内参基因。
Figure 1 qRT-PCR analysis of BnTTG1-1 expression in de- veloping seeds at different developmental stages in Brassica napus cv. ‘QINYOU Seven’ (means±SD)DAP: Days after pollination. The qRT-PCR result was normaliz- ed against the expression of BnACTIN7 as an internal control.
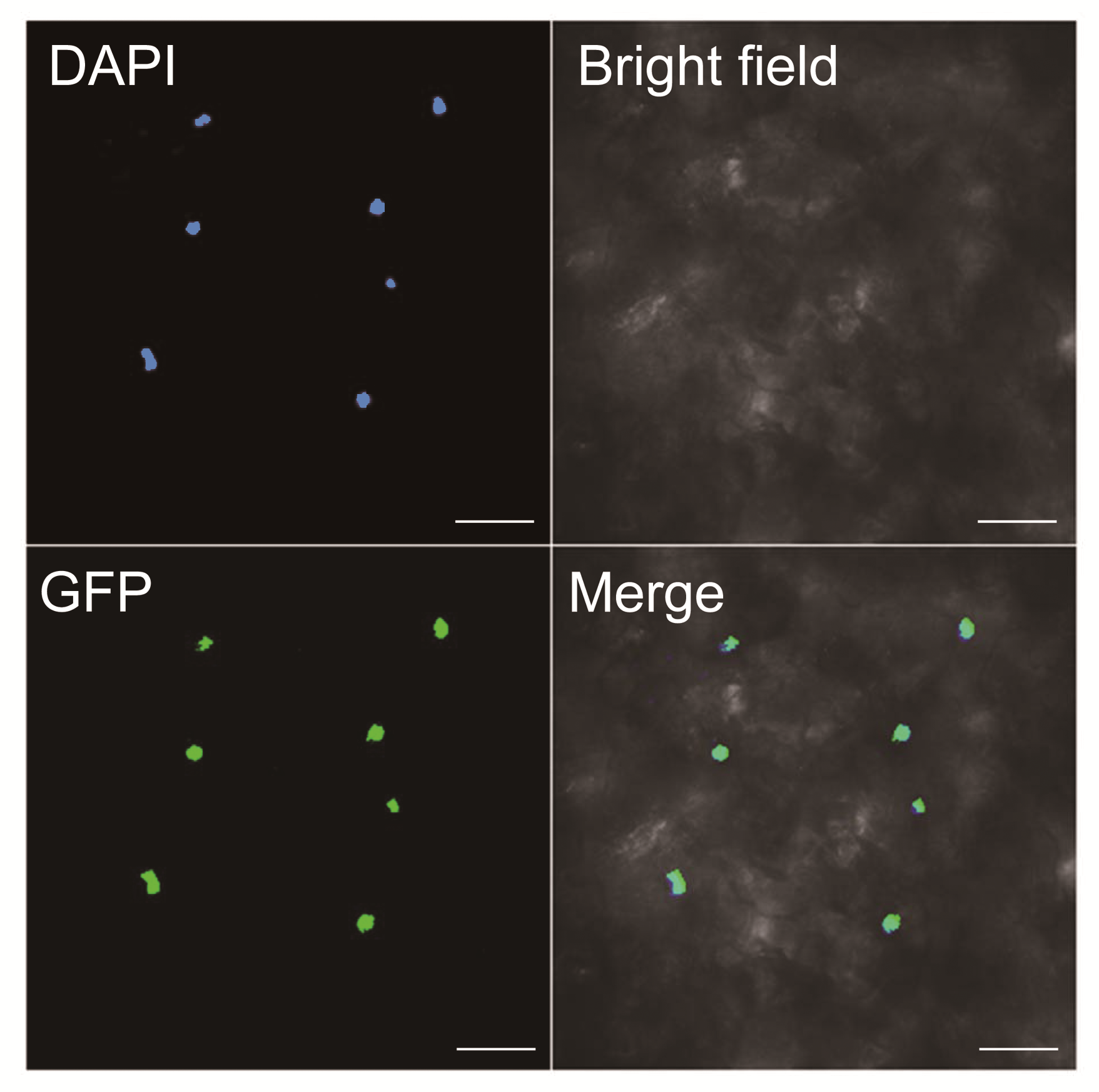
图2 BnTTG1-1在烟草叶片细胞中的亚细胞定位DAPI: 4', 6-二脒基-2-苯基吲哚; GFP: 绿色荧光蛋白; Merge: DAPI、GFP和亮场3个图像的合并图像。Bars=5 μm
Figure 2 Subcellular localization of BnTTG1-1 protein fus- ed with GFP (35S:BnTTG1-1-GFP) in tobacco (Nicotiana ben- thamiana) leave cellsDAPI: 4’, 6-diamidino-2-phenylindole dihydrochloride; GFP: Green fluorescent protein; Merge: Merged picture of bright, DAPI, and GFP fields. Bars=5 μm
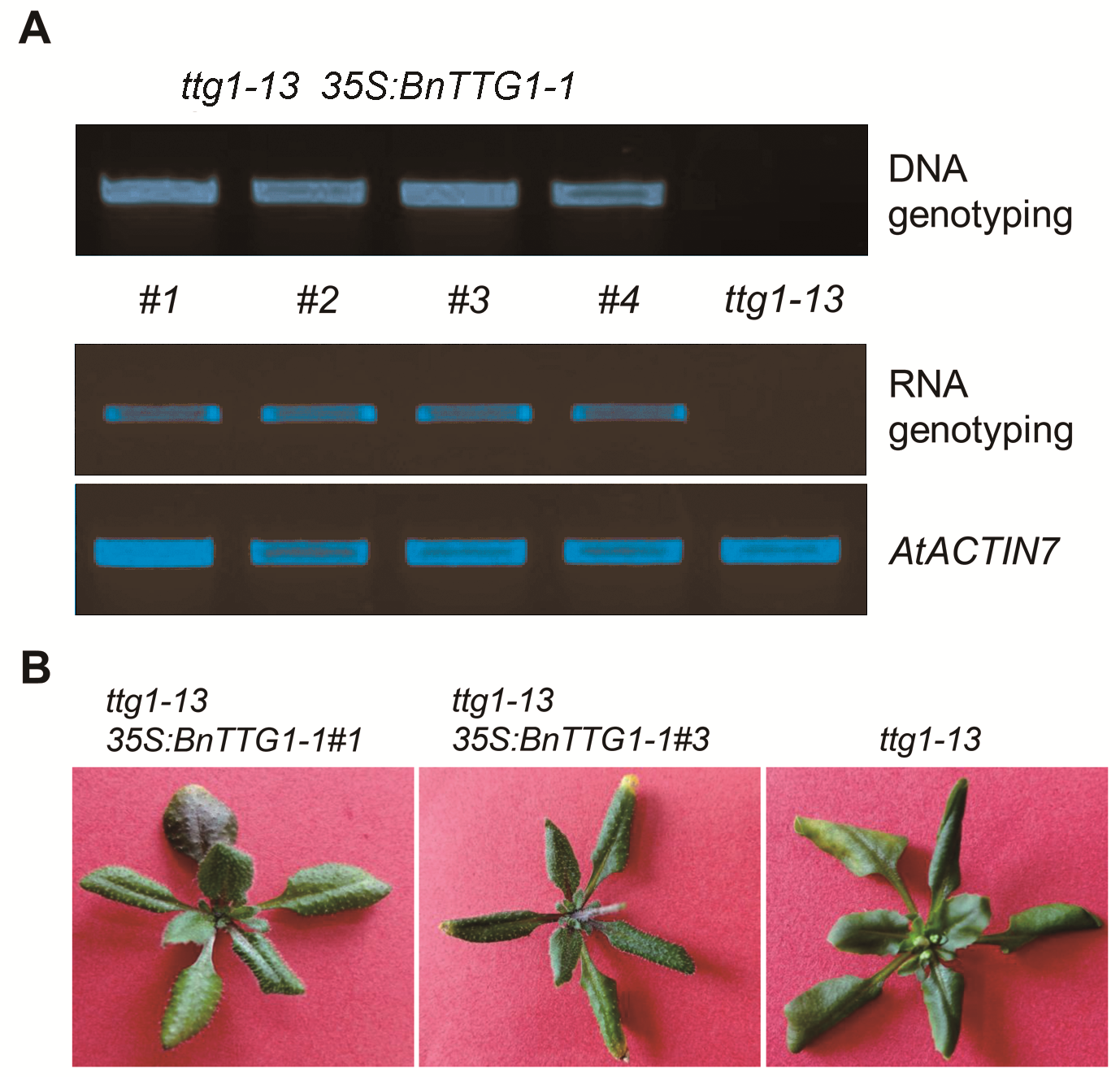
图3 ttg1-13 35S:BnTTG1-1转基因植株的鉴定(A) 在DNA和RNA水平鉴定ttg1-13 35S:BnTTG1-1转基因植株, AtACTIN7为内参基因; (B) 在突变体ttg1-13背景下异源表达BnTTG1-1能够完全恢复突变体的表型, 如无表皮毛和花青素等。
Figure 3 Identification of ttg1-13 35S:BnTTG1-1 transgenic plants(A) PCR-based DNA and RNA genotyping of ttg1-13 35S: BnTTG1-1 transgenic plants, AtACTIN7 was regarded as an internal control; (B) Heterologous expression of BnTTG1-1 in the ttg1-13 background fully rescued no trichomes and anthocyanins phenotypes of ttg1-13.
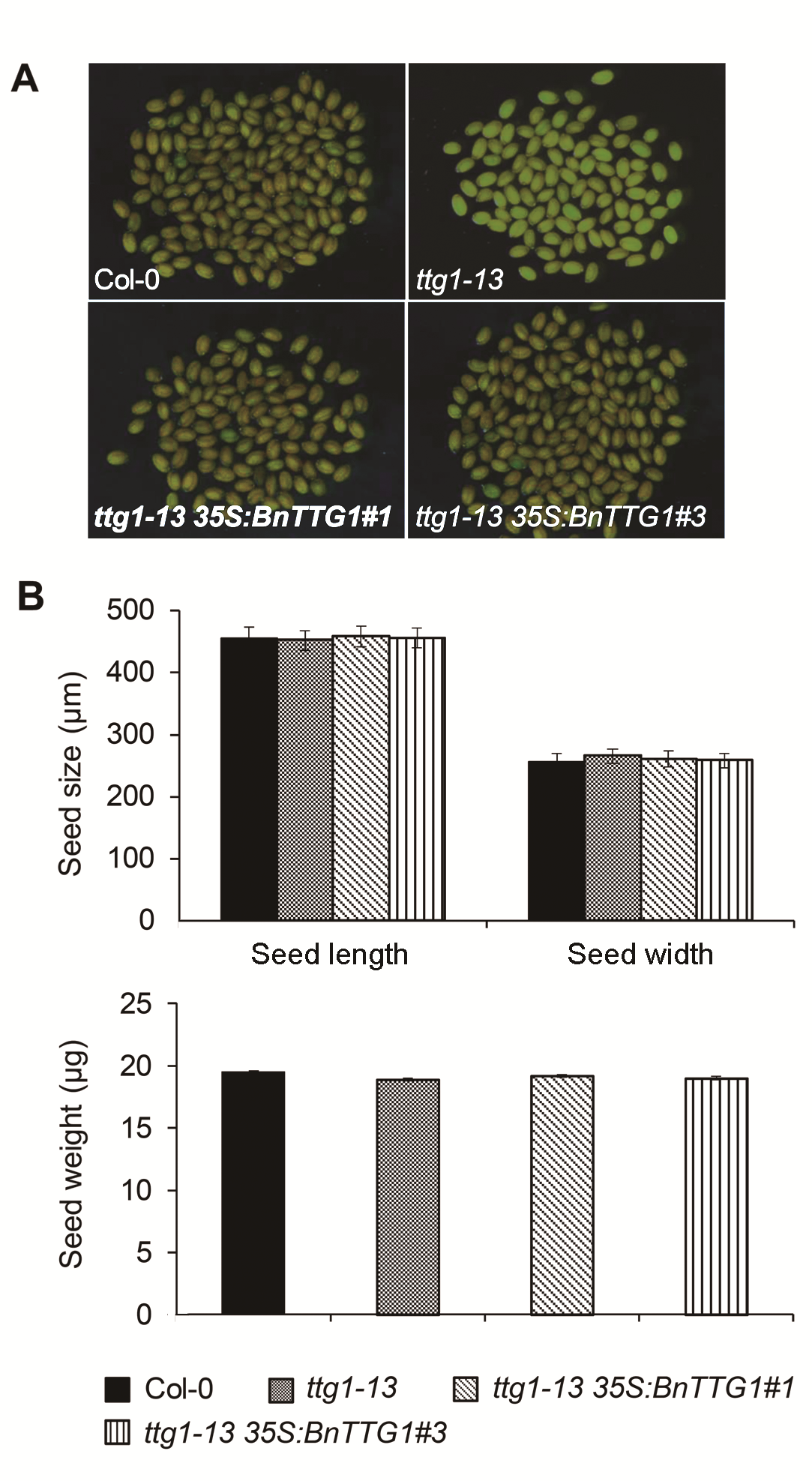
图4 比较拟南芥野生型(Col-0)、突变体ttg1-13和转基因植株ttg1-13 35S:BnTTG1-1的种皮颜色、种子大小和重量(平均值±标准差)(A) 成熟种子的显微观察; (B) 成熟种子的大小和重量比较
Figure 4 Comparison of seed coat color, seed size and se- ed weight among the wild-type (Col-0), ttg1-13, and ttg1-13 35S:BnTTG1-1 transgenic plants of Arabidopsis (means±SD)(A) Microscopic observation of mature seeds; (B) Comparison of seed size and weight of mature seeds
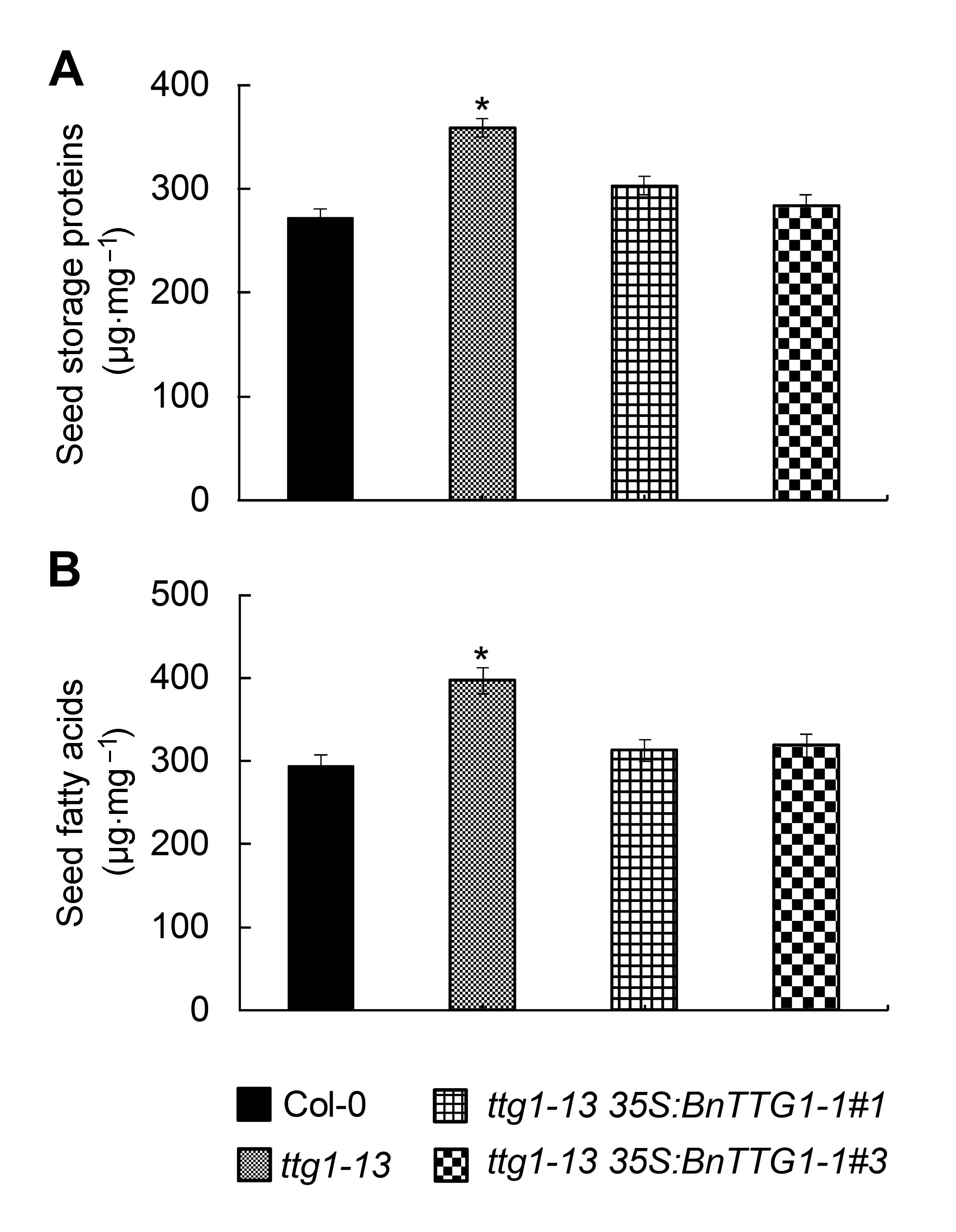
图5 比较拟南芥野生型(Col-0)、突变体ttg1-13和转基因植株ttg1-13 35S:BnTTG1-1种子的储藏蛋白与脂肪酸含量(平均值±标准差)(A) 种子储藏蛋白含量; (B) 种子脂肪酸含量。*表示在P<0.05水平上差异显著。
Figure 5 Comparison of seed storage compounds among the wild-type (Col-0), ttg1-13, and ttg1-13 35S:BnTTG1-1 trans- genic plants of Arabidopsis (means±SD)(A) The content of seed storage proteins in different lines; (B) The content of seed fatty acids in different lines. Asterisks de- note statistically signi?cant differences between the wild-type and ttg1-13 mutant (Student’s t test, P<0.05).
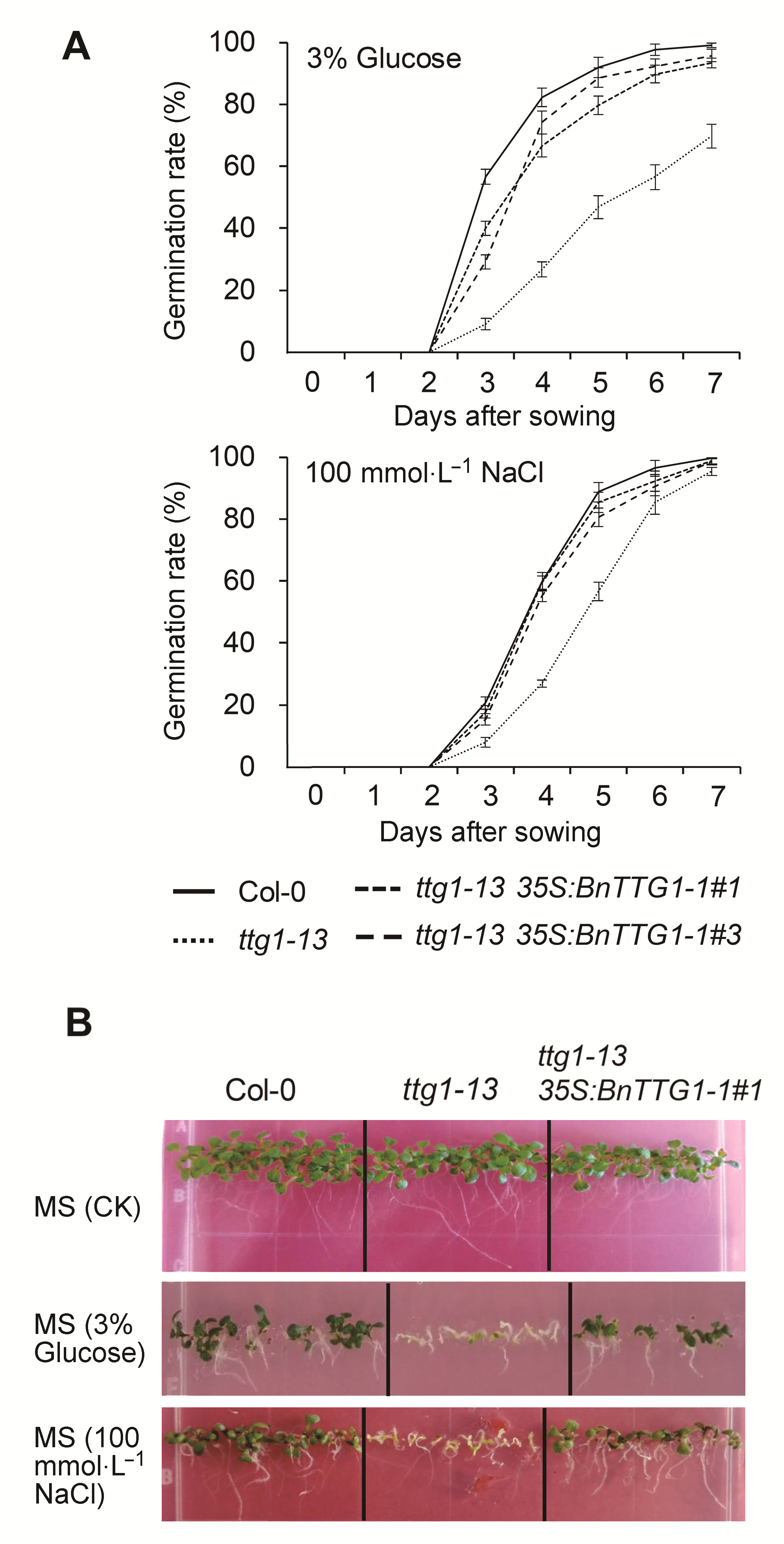
图6 非生物胁迫条件下(含有3%葡萄糖和100 mmol·L-1 NaCl)拟南芥野生型Col-0、突变体ttg1-13和转基因植株ttg1-13 35S: BnTTG1-1的发芽率和幼苗形态建成(A) 种子发芽率; (B) 幼苗的形态建成。数据为3个生物学重复的平均值±标准差, 每个生物学重复统计100粒种子。
Figure 6 Seed germination rate and seedling establishment on MS agar medium containing 3% (w/v) Glucose and containing 100 mmol·L-1 NaCl among the wild-type (Col-0), ttg1- 13, and ttg1-13 35S:BnTTG1-1 transgenic plants of Arabidopsis(A) Seed germination rate; (B) Seedling establishment. Values are the means±SD from three independent experiments evaluating 100 seeds.
| [1] | 刘后利, 傅廷栋, 陈怀庆, 易淑梅, 熊双娥 (1979). 甘蓝型黄籽油菜的发现及其遗传行为的初步研究. 遗传学报 6, 54. |
| [2] |
张子龙, 李加纳 (2001). 甘蓝型黄籽油菜粒色遗传及其育种研究进展. 作物杂志 (6), 37-40.
DOI URL |
| [3] | Bradford MM (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the |
| [4] | principle of protein-dye binding.Anal Biochem 72, 248-254. |
| [5] | Cavell AC, Lydiate DJ, Parkin IAP, Dean C, Trick M (1998). Collinearity between a 30-centimorgan segment of Arabidopsis thaliana chromosome 4 and duplicated regions within the Brassica napus genome. Genome 41, 62-69. |
| [6] |
Cernac A, Andre C, Hoffmann-Benning S, Benning C (2006). WRI1 is required for seed germination and seedling establishment.Plant Physiol 141, 745-757.
DOI URL |
| [7] |
Chen MX, Du X, Zhu Y, Wang Z, Hua SJ, Li ZL, Guo WL, Zhang GP, Peng JR, Jiang LX (2012a).Seed Fatty Acid Reducer acts downstream of gibberellin signaling pathway to lower seed fatty acid storage in Arabidopsis. Plant Cell Environ 35, 2155-2169.
DOI URL PMID |
| [8] | Chen MX, Wang Z, Zhu YN, Li ZL, Hussain N, Xuan LJ, Guo WL, Zhang GP, Jiang LX (2012b). The effect of TRANSPARENT TESTA2 on seed fatty acid biosynthesis and tolerance to environmental stresses during young seedling establishment in Arabidopsis. Plant Physiol 160, 1023-1036. |
| [9] |
Chen MX, Xuan LJ, Wang Z, Zhou LH, Li ZL, Du X, Ali E, Zhang GP, Jiang LX (2014). TRANSPARENT TESTA8 inhibits seed fatty acid accumulation by targeting several seed development regulators in Arabidopsis.Plant Phy- siol 165, 905-916.
DOI URL PMID |
| [10] | Chen MX, Zhang B, Li CX, Kulaveerasingam H, Chew FT, Yu H (2015). TRANSPARENT TESTA GLABRA1 regulates the accumulation of seed storage reserves in Ara- bidopsis. Plant Physiol 169, 391-402. |
| [11] | Clough SJ, Bent AF (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16, 735-743. |
| [12] |
Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR (2010). Abscisic acid: emergence of a core signaling network.Annu Rev Plant Biol 61, 651-679.
DOI URL PMID |
| [13] |
Debeaujon I, Léon-Kloosterziel KM, Koornneef M (2000). Influence of the testa on seed dormancy, germination, and longevity in Arabidopsis.Plant Physiol 122, 403-414.
DOI URL |
| [14] |
Debeaujon I, Nesi N, Perez P, Devic M, Grandjean O, Caboche M, Lepiniec L (2003). Proanthocyanidin- accumulating cells in Arabidopsis testa: regulation of differentiation and role in seed development.Plant Cell 15, 2514-2531.
DOI URL PMID |
| [15] |
Finkelstein RR, Gampala SSL, Rock CD (2002). Abscisic acid signaling in seeds and seedlings.Plant Cell 14, S15-S45.
DOI URL PMID |
| [16] |
Gibson SI (2001). Plant sugar-response pathways. Part of a complex regulatory web.Plant Physiol 125, 2203-2203.
DOI URL PMID |
| [17] |
Hong JK, Choi HW, Hwang IS, Kim DS, Kim NH, Choi DS, Kim YJ, Hwang BK (2008). Function of a novel GDSL- type pepper lipase gene, CaGLIP1, in disease susceptibility and abiotic stress tolerance. Planta 227, 539-558.
DOI URL PMID |
| [18] |
Koes RE, Quattrocchio F, Mol JNM (1994). The flavonoid biosynthetic pathway in plants: function and evolution.BioEssays 16, 123-132.
DOI URL |
| [19] | Koornneef M (1981). The complex syndrome of ttg mutants. Arabidopsis Inf Serv 18, 45-51. |
| [20] |
Lepiniec L, Debeaujon I, Routaboul JM, Baudry A, Pourcel L, Nesi N, Caboche M (2006). Genetics and biochemistry of seed flavonoids.Annu Rev Plant Biol 57, 405-430.
DOI URL PMID |
| [21] |
Liu KG, Qi SH, Li D, Jin CY, Gao CH, Duan SW, Feng BL, Chen MX (2017). TRANSPARENT TESTA GLABRA 1 ubiquitously regulates plant growth and development from Arabidopsis to foxtail millet (Setaria italica). Plant Sci 254, 60-69.
DOI URL PMID |
| [22] |
Livak KJ, Schmittgen TD (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method.Methods 25, 402-408.
DOI URL |
| [23] |
Lu J, Li JN, Lei B, Wang SG, Chai YR (2009). Molecular cloning and characterization of two Brassica napus TTG1 genes reveal genus-specific nucleotide preference, extreme protein-level conservation and fast divergence of organ-specificity. Genes Genom 31, 129-142.
DOI URL |
| [24] |
Mol J, Grotewold E, Koes R (1998). How genes paint flowers and seeds.Trends Plant Sci 3, 212-217.
DOI URL |
| [25] | Mu JY, Tan HL, Zheng Q, Fu FY, Liang Y, Zhang J, Yang XH, Wang T, Chong K, Wang XJ, Zuo JR (2008). LEAFY COTYLEDON1 is a key regulator of fatty acid biosynthesis in Arabidopsis. Plant Physiol 148, 1042-1054. |
| [26] |
Nakabayashi R, Yonekura-Sakakibara K, Urano K, Suzuki M, Yamada Y, Nishizawa T, Matsuda F, Kojima M, Sakakibara H, Shinozaki K, Michael AJ, Tohge T, Yamazaki M, Saito K (2014). Enhancement of oxidative and drought tolerance in Arabidopsis by overaccumulation of antioxidant flavonoids.Plant J 77, 367-379.
DOI URL PMID |
| [27] |
Nesi N, Jond C, Debeaujon I, Caboche M, Lepiniec L (2001). The Arabidopsis TT2 gene encodes an R2R3 MYB domain protein that acts as a key determinant for proanthocyanidin accumulation in developing seed. Plant Cell 13, 2099-2114.
DOI URL PMID |
| [28] |
Nguyen HN, Kim JH, Hyun WY, Nguyen NT, Hong SW, Lee H (2013). TTG1-mediated flavonols biosynthesis alleviates root growth inhibition in response to ABA.Plant Cell Rep 32, 503-514.
DOI URL PMID |
| [29] | Osborn TC, Kole C, Parkin IAP, Sharpe AG, Kuiper M, Lydiate DJ, Trick M (1997). Comparison of flowering time genes inBrassica rapa, B. napus and Arabidopsis tha- liana. Genetics 146, 1123-1129. |
| [30] |
Peer WA, Murphy AS (2007). Flavonoids and auxin transport: modulators or regulators?Trends Plant Sci 12, 556-563.
DOI URL PMID |
| [31] |
Petrussa E, Braidot E, Zancani M, Peresson C, Bertolini A, Patui S, Vianello A (2013). Plant flavonoids-biosyn- thesis, transport and involvement in stress responses.Int J Mol Sci 14, 14950-14973.
DOI URL |
| [32] |
Shi L, Katavic V, Yu YY, Kunst L, Haughn G (2012). Arabidopsis glabra2 mutant seeds deficient in mucilage biosynthesis produce more oil. Plant J 69, 37-46.
DOI URL PMID |
| [33] |
Shirley BW (1996). Flavonoid biosynthesis: ‘new’ functions for an ‘old’ pathway.Trends Plant Sci 1, 377-382.
DOI URL |
| [34] |
Shirley BW, Kubasek WL, Storz G, Bruggemann E, Koornneef M, Ausubel FM, Goodman HM (1995). Analysis of Arabidopsis mutants deficient in flavonoid bio- synthesis.Plant J 8, 659-671.
DOI URL PMID |
| [35] |
Szymanski DB, Lloyd AM, Marks MD (2000). Progress in the molecular genetic analysis of trichome initiation and morphogenesis in Arabidopsis.Trends Plant Sci 5, 214-219.
DOI URL PMID |
| [36] |
Tsuchiya Y, Nambara E, Naito S, McCourt P (2004). The FUS3 transcription factor functions through the epidermal regulator TTG1 during embryogenesis in Arabidopsis. Plant J 37, 73-81.
DOI URL PMID |
| [37] | Walker AR, Davison PA, Bolognesi-Winfield AC, James CM, Srinivasan N, Blundell TL, Esch JJ, Marks MD, Gray JC (1999). The TRANSPARENT TESTA GLABRA1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein. Plant Cell 11, 1337-1350. |
| [38] | Wang Z, Chen MX, Chen TL, Xuan LJ, Li ZL, Du X, Zhou LH, Zhang GP, Jiang LX (2014). TRANSPARENT TESTA2 regulates embryonic fatty acid biosynthesis by targeting FUSCA3 during the early developmental stage of Arabidopsis seeds. Plant J 77, 757-769. |
| [39] |
Western TL, Burn J, Tan WL, Skinner DJ, Martin- McCaffrey L, Moffatt BA, Haughn GW (2001). Isolation and characterization of mutants defective in seed coat mucilage secretory cell development in Arabidopsis.Plant Physiol 127, 998-1011.
DOI URL |
| [40] |
Winkel-Shirley B (2002). Biosynthesis of flavonoids and ef- fects of stress.Curr Opin Plant Biol 5, 218-223.
DOI URL PMID |
| [41] |
Xu WJ, Grain D, Bobet S, Le Gourrierec J, Thévenin J, Kelemen Z, Lepiniec L, Dubos C (2014). Complexity and robustness of the flavonoid transcriptional regulatory network revealed by comprehensive analyses of MYB- bHLH-WDR complexes and their targets in Arabidopsis seed.New Phytol 202, 132-144.
DOI URL PMID |
| [42] |
Zhang YM, Rock CO (2004). Evaluation of epigallocatechin gallate and related plant polyphenols as inhibitors of the FabG and FabI reductases of bacterial type II fatty-acid synthase.J Biol Chem 279, 30994-31001.
DOI URL |
| [1] | 刘雨函, 曹启江, 张诗晗, 李益慧, 王菁, 谭晓萌, 刘筱儒, 王显玲. 拟南芥AtFTCD-L参与根系响应土壤紧实度的机制研究[J]. 植物学报, 2025, 60(4): 1-0. |
| [2] | 杨柳卿, 王劲, 燕敬利, 陈芹芹, 程浩坤, 李春, 赵培玉, 杨博, 江元清. 甘蓝型油菜转录因子BnaABF2的表征分析及互作蛋白鉴定[J]. 植物学报, 2025, 60(1): 49-61. |
| [3] | 李青洋, 刘翠, 何李, 彭姗, 马嘉吟, 胡子祎, 刘宏波. 甘蓝型油菜BnaA02.CPSF6基因的克隆及功能分析(长英文摘要)[J]. 植物学报, 2025, 60(1): 62-73. |
| [4] | 景艳军, 林荣呈. 蓝光受体CRY2化身“暗黑舞者”[J]. 植物学报, 2024, 59(6): 878-882. |
| [5] | 罗燕, 刘奇源, 吕元兵, 吴越, 田耀宇, 安田, 李振华. 拟南芥光敏色素突变体种子萌发的光温敏感性[J]. 植物学报, 2024, 59(5): 752-762. |
| [6] | 陈艳晓, 李亚萍, 周晋军, 解丽霞, 彭永彬, 孙伟, 和亚男, 蒋聪慧, 王增兰, 郑崇珂, 谢先芝. 拟南芥光敏色素B氨基酸位点突变对其结构与功能的影响[J]. 植物学报, 2024, 59(3): 481-494. |
| [7] | 杨继轩, 王雪霏, 顾红雅. 西藏野生拟南芥开花时间变异的遗传基础[J]. 植物学报, 2024, 59(3): 373-382. |
| [8] | 张盈川, 吴晓明玉, 陶保龙, 陈丽, 鲁海琴, 赵伦, 文静, 易斌, 涂金星, 傅廷栋, 沈金雄. Bna-miR43介导甘蓝型油菜响应干旱胁迫[J]. 植物学报, 2023, 58(5): 701-711. |
| [9] | 王钢, 王二涛. “卫青不败由天幸”——WeiTsing的广谱抗根肿病机理被揭示[J]. 植物学报, 2023, 58(3): 356-358. |
| [10] | 吴楠, 覃磊, 崔看, 李海鸥, 刘忠松, 夏石头. 甘蓝型油菜EXA1的克隆及其对植物抗病的调控作用[J]. 植物学报, 2023, 58(3): 385-393. |
| [11] | 杨永青, 郭岩. 植物细胞质外体pH感受机制的解析[J]. 植物学报, 2022, 57(4): 409-411. |
| [12] | 支添添, 周舟, 韩成云, 任春梅. PAD4突变加速拟南芥酪氨酸降解缺陷突变体sscd1的程序性细胞死亡[J]. 植物学报, 2022, 57(3): 288-298. |
| [13] | 李艳艳, 齐艳华. 植物Aux/IAA基因家族生物学功能研究进展[J]. 植物学报, 2022, 57(1): 30-41. |
| [14] | 马炬峰, 辛敏, 徐陈超, 祝琬莹, 毛传澡, 陈欣, 程磊. 丛枝菌根真菌与氮添加对不同根形态基因型水稻氮吸收的影响[J]. 植物生态学报, 2021, 45(7): 728-737. |
| [15] | 车永梅, 孙艳君, 卢松冲, 侯丽霞, 范欣欣, 刘新. AtMYB77促进NO合成参与调控干旱胁迫下拟南芥侧根发育[J]. 植物学报, 2021, 56(4): 404-413. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||