

Chinese Bulletin of Botany ›› 2023, Vol. 58 ›› Issue (1): 140-149.DOI: 10.11983/CBB22177 cstr: 32102.14.CBB22177
Special Issue: 杂粮生物学专辑 (2023年58卷1期)
• EXPERIMENTAL COMMUNICATIONS • Previous Articles Next Articles
Feifei Wang, Zhenxiang Zhou, Yi Hong, Yangyang Gu, Chao Lü, Baojian Guo, Juan Zhu, Rugen Xu*( )
)
Received:2022-07-31
Accepted:2022-10-24
Online:2023-01-01
Published:2023-01-05
Contact:
*E-mail: rgxu@yzu.edu.cn
About author:†These authors contributed equally to this paper
Feifei Wang, Zhenxiang Zhou, Yi Hong, Yangyang Gu, Chao Lü, Baojian Guo, Juan Zhu, Rugen Xu. Identification of the NF-YC Genes in Hordeum vulgare and Expression Analysis Under Salt Stress[J]. Chinese Bulletin of Botany, 2023, 58(1): 140-149.
| Gene name | Gene ID | Chromo- some | Location | Amino acid length (aa) | Protein molecular weight (kDa) | pI | Instability index | Aliphatic index | Subcellular location |
|---|---|---|---|---|---|---|---|---|---|
| HvNF-YC1 | HORVU.MOREX.- r3.1HG0069390.1 | 1 | 447067437- 447072149 | 282 | 31.44 | 5.4 | 48.23 | 75.07 | Nucleus |
| HvNF-YC2 | HORVU.MOREX.- r3.1HG0081770.1 | 1 | 489164568- 489164978 | 136 | 14.07 | 9.5 | 38.39 | 79.78 | Nucleus |
| HvNF-YC3 | HORVU.MOREX.- r3.3HG0237950.1 | 3 | 54989815- 54991052 | 371 | 39.09 | 4.74 | 52.22 | 66.66 | Nucleus |
| HvNF-YC4 | HORVU.MOREX.- r3.4HG0364520.1 | 4 | 231786893- 231791804 | 255 | 28.09 | 5.93 | 59.67 | 65.76 | Nucleus |
| HvNF-YC5 | HORVU.MOREX.- r3.4HG0391570.1 | 4 | 499791697- 499793529 | 241 | 25.63 | 5.32 | 60.13 | 66.18 | Nucleus |
| HvNF-YC6 | HORVU.MOREX.- r3.5HG0489580.1 | 5 | 474074907- 474075856 | 203 | 21.78 | 5.39 | 59.79 | 72.91 | Nucleus |
| HvNF-YC7 | HORVU.MOREX.- r3.6HG0571700.1 | 6 | 135835186- 135838701 | 260 | 28.59 | 4.87 | 81.41 | 71.81 | Nucleus |
| HvNF-YC8 | HORVU.MOREX.- r3.7HG0677850.1 | 7 | 160202157- 160203122 | 199 | 21.57 | 5.52 | 65.36 | 71.86 | Nucleus |
| HvNF-YC9 | HORVU.MOREX.- r3.7HG0737530.1 | 7 | 597862664- 597863461 | 265 | 29.76 | 4.79 | 69.77 | 63.06 | Nucleus |
| HvNF-YC10 | HORVU.MOREX.- r3.7HG0737680.1 | 7 | 598086832- 598087587 | 251 | 27.85 | 6.22 | 68.44 | 73.19 | Nucleus |
| HvNF-YC11 | HORVU.MOREX.- r3.7HG0737720.1 | 7 | 598116971- 598117753 | 260 | 28.62 | 4.66 | 80.61 | 68.81 | Nucleus |
Table 1 Sequence information of HvNF-YCs gene family
| Gene name | Gene ID | Chromo- some | Location | Amino acid length (aa) | Protein molecular weight (kDa) | pI | Instability index | Aliphatic index | Subcellular location |
|---|---|---|---|---|---|---|---|---|---|
| HvNF-YC1 | HORVU.MOREX.- r3.1HG0069390.1 | 1 | 447067437- 447072149 | 282 | 31.44 | 5.4 | 48.23 | 75.07 | Nucleus |
| HvNF-YC2 | HORVU.MOREX.- r3.1HG0081770.1 | 1 | 489164568- 489164978 | 136 | 14.07 | 9.5 | 38.39 | 79.78 | Nucleus |
| HvNF-YC3 | HORVU.MOREX.- r3.3HG0237950.1 | 3 | 54989815- 54991052 | 371 | 39.09 | 4.74 | 52.22 | 66.66 | Nucleus |
| HvNF-YC4 | HORVU.MOREX.- r3.4HG0364520.1 | 4 | 231786893- 231791804 | 255 | 28.09 | 5.93 | 59.67 | 65.76 | Nucleus |
| HvNF-YC5 | HORVU.MOREX.- r3.4HG0391570.1 | 4 | 499791697- 499793529 | 241 | 25.63 | 5.32 | 60.13 | 66.18 | Nucleus |
| HvNF-YC6 | HORVU.MOREX.- r3.5HG0489580.1 | 5 | 474074907- 474075856 | 203 | 21.78 | 5.39 | 59.79 | 72.91 | Nucleus |
| HvNF-YC7 | HORVU.MOREX.- r3.6HG0571700.1 | 6 | 135835186- 135838701 | 260 | 28.59 | 4.87 | 81.41 | 71.81 | Nucleus |
| HvNF-YC8 | HORVU.MOREX.- r3.7HG0677850.1 | 7 | 160202157- 160203122 | 199 | 21.57 | 5.52 | 65.36 | 71.86 | Nucleus |
| HvNF-YC9 | HORVU.MOREX.- r3.7HG0737530.1 | 7 | 597862664- 597863461 | 265 | 29.76 | 4.79 | 69.77 | 63.06 | Nucleus |
| HvNF-YC10 | HORVU.MOREX.- r3.7HG0737680.1 | 7 | 598086832- 598087587 | 251 | 27.85 | 6.22 | 68.44 | 73.19 | Nucleus |
| HvNF-YC11 | HORVU.MOREX.- r3.7HG0737720.1 | 7 | 598116971- 598117753 | 260 | 28.62 | 4.66 | 80.61 | 68.81 | Nucleus |
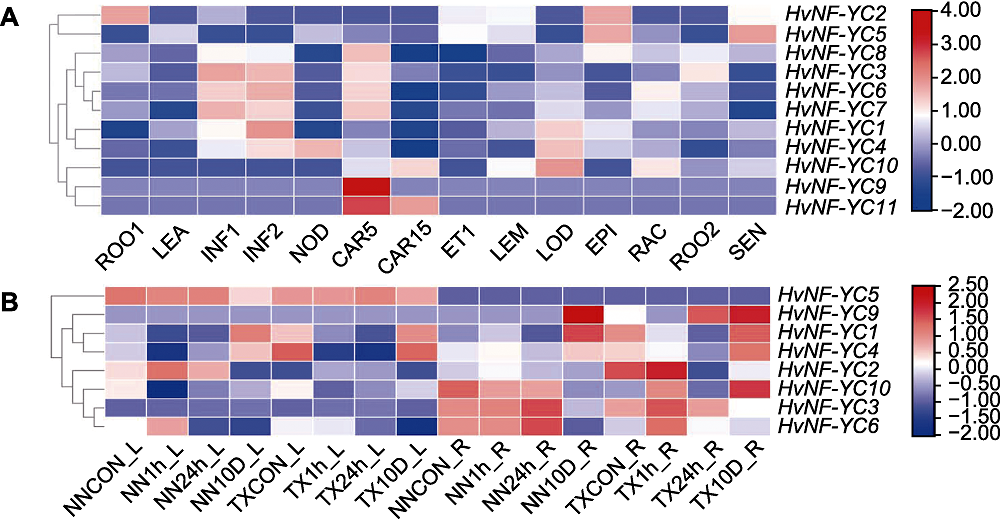
Figure 8 Tissue expression profile of NF-YC gene family members in Hordeum vulgare (A) and HvNF-YC expressions in the leaf and root of NN and TX varieties under salt stress (B) ROO1: Roots from seedlings (10 cm shoot stage); LEA: Shoots from seedlings (10 cm shoot stage); INF1: Young developing inflorescences (5 mm); INF2: Developing inflorescences (1-1.5 cm); NOD: Developing tillers (3rd internode); CAR5: Developing grain (5 days after pollination (DAP)); CAR15: Developing grain (15 DAP); ET1: Etiolated seedling (dark condition,10 days); LEM: Inflorescences (lemma, 42 DAP); LOD: Inflorescences (lodicule, 42 DAP); EPI: Epidermal strips (28 DAP); RAC: Inflorescences (rachis, 35 DAP); ROO2: Roots (28 DAP); SEN: Senescing leaves (56 DAP)
| [1] | 陈国户, 庞小可, 李广, 王浩, 吴思文, 温宏伟, 尹倩, 袁凌云, 侯金锋, 唐小燕, 汪承刚 (2022). 白菜NAC基因家族全基因组鉴定及其应答春化反应的表达分析. 南京农业大学学报 45, 656-665. |
| [2] | 黄俊文, 南建宗, 阳成伟 (2020). NF-Y转录因子调控植物生长发育及胁迫响应的研究进展. 植物生理学报 56, 2595-2605. |
| [3] | 李娟, 高凯, 安新民 (2019). 转录因子NF-Y在植物生长发育和逆境胁迫响应中的作用. 中国细胞生物学学报 41, 2434-2442. |
| [4] | 李世贵, 马瑞, 王芳芳, 刘维刚, 杨江伟, 唐勋, 张宁, 司怀军 (2021). 植物NF-Y转录因子研究进展. 植物生理学报 57, 248-256. |
| [5] |
马鑫磊, 许瑞琪, 索晓曼, 李婧实, 顾鹏鹏, 姚锐, 林小虎, 高慧 (2022). 谷子III型PRX基因家族全基因组鉴定及干旱胁迫下表达分析. 作物学报 48, 2517-2532.
DOI |
| [6] | 桑璐曼, 汤沙, 张仁梁, 贾小平, 刁现民 (2022). 谷子热激蛋白HSP90基因家族鉴定及分析. 植物遗传资源学报 23, 1085-1097. |
| [7] |
Alam MM, Tanaka T, Nakamura H, Ichikawa H, Kobayashi K, Yaeno T, Yamaoka N, Shimomoto K, Takayama K, Nishina H, Nishiguchi M (2015). Overexpression of a rice heme activator protein gene (OsHAP2E) confers resistance to pathogens, salinity and drought, and increases photosynthesis and tiller number. Plant Biotechnol J 13, 85-96.
DOI URL |
| [8] |
Dolfini D, Gatta R, Mantovani R (2012). NF-Y and the transcriptional activation of CCAAT promoters. Crit Rev Biochem Mol Biol 47, 29-49.
DOI URL |
| [9] |
Forsburg SL, Guarente L (1989). Identification and characterization of HAP4: a third component of the CCAAT- bound HAP2/HAP3 heteromer. Genes Dev 3, 1166-1178.
DOI URL |
| [10] |
Frontini M, Imbriano C, Manni I, Mantovani R (2004). Cell cycle regulation of NF-YC nuclear localization. Cell Cycle 3, 217-222.
PMID |
| [11] |
Kong HZ, Landherr LL, Frohlich MW, Leebens-Mack J, Ma H, DePamphilis CW (2007). Patterns of gene duplication in the plant SKP1 gene family in angiosperms: evidence for multiple mechanisms of rapid gene birth. Plant J 50, 873-885.
DOI URL |
| [12] |
Laloum T, De Mita S, Gamas P, Baudin M, Niebel A (2013). CCAAT-box binding transcription factors in plants: Y so many? Trends Plant Sci 18, 157-166.
DOI PMID |
| [13] |
Li QP, Yan WH, Chen HX, Tan C, Han ZM, Yao W, Li GW, Yuan MQ, Xing YZ (2016). Duplication of OsHAP family genes and their association with heading date in rice. J Exp Bot 67, 1759-1768.
DOI URL |
| [14] |
Li YJ, Fang Y, Fu YR, Huang JG, Wu CA, Zheng CC (2013). NFYA1 is involved in regulation of postgermina-tion growth arrest under salt stress in Arabidopsis. PLoS One 8, e61289.
DOI URL |
| [15] | Liang MX, Hole D, Wu JX, Blake T, Wu YJ (2012). Expres-sion and functional analysis of NUCLEAR FACTORY, subunit B genes in barley. Planta 235, 779-791. |
| [16] |
Mantovani R (1999). The molecular biology of the CCAAT- binding factor NF-Y. Gene 239, 15-27.
DOI PMID |
| [17] |
Nelson DE, Repetti PP, Adams TR, Creelman RA, Wu JR, Warner DC, Anstrom DC, Bensen RJ, Castiglioni PP, Donnarummo MG, Hinchey BS, Kumimoto RW, Maszle DR, Canales RD, Krolikowski KA, Dotson SB, Gutterson N, Ratcliffe OJ, Heard JE (2007). Plant nuclear factor Y (NF-Y) B subunits confer drought tolerance and lead to improved corn yields on water-limited acres. Proc Natl Acad Sci USA 104, 16450-16455.
DOI PMID |
| [18] |
Niu BX, Zhang ZY, Zhang J, Zhou Y, Chen C (2021). The rice LEC1-like transcription factor OsNF-YB9 interacts with SPK, an endosperm specific sucrose synthase pro-tein kinase, and functions in seed development. Plant J 106, 1233-1246.
DOI URL |
| [19] |
Panahi B, Mohammadi SA, Ruzicka K, Abbasi Holaso H, Zare Mehrjerdi M (2019). Genome-wide identification and co-expression network analysis of nuclear factor-Y in barley revealed potential functions in salt stress. Physiol Mol Biol Plants 25, 485-495.
DOI |
| [20] | Pelletier JM, Kwong RW, Park S, Le BH, Baden R, Cagliari A, Hashimoto M, Munoz MD, Fischer RL, Goldberg RB, Harada JJ (2017). LEC1 sequentially regulates the transcription of genes involved in diverse developmental processes during seed development. Proc Natl Acad Sci USA 114, E6710-E6719. |
| [21] |
Petroni K, Kumimoto RW, Gnesutta N, Calvenzani V, Fornari M, Tonelli C, Holt III BF, Mantovani R (2012). The promiscuous life of plant NUCLEAR FACTOR Y transcription factors. Plant Cell 24, 4777-4792.
DOI URL |
| [22] |
Qu BY, He X, Wang J, Zhao YY, Teng W, Shao A, Zhao XQ, Ma WY, Wang JY, Li B, Li ZS, Tong YP (2015). A wheat CCAAT box-binding transcription factor increases the grain yield of wheat with less fertilizer input. Plant Physiol 167, 411-423.
DOI PMID |
| [23] |
Rogozin IB, Wolf YI, Sorokin AV, Mirkin BG, Koonin EV (2003). Remarkable interkingdom conservation of intron positions and massive, lineage-specific intron loss and gain in eukaryotic evolution. Curr Biol 13, 1512-1517.
PMID |
| [24] |
Romier C, Cocchiarella F, Mantovani R, Moras D (2003). The NF-YB/NF-YC structure gives insight into DNA binding and transcription regulation by CCAAT factor NF-Y. J Biol Chem 278, 1336-1345.
DOI PMID |
| [25] |
Siefers N, Dang KK, Kumimoto RW, Bynum WE, Tayrose G, Holt III BF (2009). Tissue-specific expression patterns of Arabidopsis NF-Y transcription factors suggest potential for extensive combinatorial complexity. Plant Physiol 149, 625-641.
DOI URL |
| [26] |
Stephenson TJ, McIntyre CL, Collet C, Xue GP (2010). TaNF-YC11, one of the light-upregulated NF-YC members in Triticum aestivum, is co-regulated with photosynthesis-related genes. Funct Integr Genomics 10, 265-276.
DOI URL |
| [27] |
Stephenson TJ, McIntyre CL, Collet C, Xue GP (2007). Genome-wide identification and expression analysis of the NF-Y family of transcription factors in Triticum aesti-vum. Plant Mol Biol 65, 77-92.
DOI PMID |
| [28] |
Thirumurugan T, Ito Y, Kubo T, Serizawa A, Kurata N (2008). Identification, characterization and interaction of HAP family genes in rice. Mol Genet Genomics 279, 279-289.
DOI URL |
| [29] |
Xing Y, Fikes JD, Guarente L (1993). Mutations in yeast HAP2/HAP3 define a hybrid CCAAT box binding domain. EMBO J 12, 4647-4655.
DOI PMID |
| [30] |
Yang WJ, Lu ZH, Xiong YF, Yao JL (2017). Genome-wide identification and co-expression network analysis of the OsNF-Y gene family in rice. Crop J 5, 21-31.
DOI URL |
| [31] | Yu TF, Liu Y, Fu JD, Ma J, Fang ZW, Chen J, Zheng L, Lu ZW, Zhou YB, Chen M, Xu ZS, Ma YZ (2021). The NF-Y-PYR module integrates the abscisic acid signal pathway to regulate plant stress tolerance. Plant Biotech-nol J 19, 2589-2605. |
| [32] |
Zhang ZB, Li XL, Zhang C, Zou HW, Wu ZY (2016). Isola-tion, structural analysis, and expression characteristics of the maize nuclear factor Y gene families. Biochem Bio-phys Res Commun 478, 752-758.
DOI URL |
| [1] | Ziyun Wang, Yanwen Lv, Yu Xiao, Chao Wu, Xinsheng Hu. Advances in Regulation and Evolutionary Mechanisms of Plant Gene Expression [J]. Chinese Bulletin of Botany, 2025, 60(4): 1-0. |
| [2] | Xu Tiantian, Yang Peijian, Zhou Xiaoxi, Cao Yi, Chen Yanhong, Liu Guoyuan, Zhang Jian, Wei Hui. Analysis of Physicochemical Characteristics and Expression Characteristics of Lagerstroemia indica GolS Family Genes [J]. Chinese Bulletin of Botany, 2025, 60(3): 393-406. |
| [3] | Laipeng Zhao, Baike Wang, Tao Yang, Ning Li, Haitao Yang, Juan Wang, Huizhuan Yan. Investigation of the Regulation of Drought Tolerance by the SlHVA22l Gene in Tomato [J]. Chinese Bulletin of Botany, 2024, 59(4): 558-573. |
| [4] | Jinyu Du, Zhen Sun, Yanlong Su, Heping Wang, Yaling Liu, Zhenying Wu, Feng He, Yan Zhao, Chunxiang Fu. Identification and Functional Analysis of an Agropyron mongolicum Caffeic Acid 3-O-methyltransferase Gene AmCOMT1 [J]. Chinese Bulletin of Botany, 2024, 59(3): 383-396. |
| [5] | Zhengyong Duan, Min Ding, Yuzhuo Wang, Yibing Ding, Ling Chen, Ruiyun Wang, Zhijun Qiao. Genome-wide Identification and Expression Analysis of SBP Genes in Panicum miliaceum [J]. Chinese Bulletin of Botany, 2024, 59(2): 231-244. |
| [6] | Hanqian Zhao, Jiayi Song, Jie Yang, Yongjing Zhao, Wennian Xia, Weizhuo Gu, Zhongyi Wang, Nan Yang, Huizhen Hu. Identification of XTH Family Genes in Antirrhinum majus and Screening of Genes Involoved in Sclerotinia sclerotiorum Resistance and Stamen Petalization [J]. Chinese Bulletin of Botany, 2024, 59(2): 188-203. |
| [7] | Nan Wu, Lei Qin, Kan Cui, Haiou Li, Zhongsong Liu, Shitou Xia. Cloning of Brassica napus EXA1 Gene and Its Regulation on Plant Disease Resistance [J]. Chinese Bulletin of Botany, 2023, 58(3): 385-393. |
| [8] | Qi Zhang, Wenjing Zhang, Xiankai Yuan, Ming Li, Qiang Zhao, Yanli Du, Jidao Du. The Regulatory Mechanism of Melatonin on Nucleic Acid Repairing of Common Bean (Phaseolus vulgaris) at the Sprout Stage Under Salt Stress [J]. Chinese Bulletin of Botany, 2023, 58(1): 108-121. |
| [9] | Kai Fan, Fangting Ye, Zhijun Mao, Xinfeng Pan, Zhaowei Li, Wenxiong Lin. Comparative Genomics of the Small Heat Shock Protein Family in Angiosperms [J]. Chinese Bulletin of Botany, 2021, 56(3): 245-261. |
| [10] | Lulu Xie, Qingqing Cui, Chunjuan Dong, Qingmao Shang. Recent Advances in Molecular Mechanisms of Plant Graft Healing Process [J]. Chinese Bulletin of Botany, 2020, 55(5): 634-643. |
| [11] | Mingzhe Che, Yajun Wang, Chuangxin Ma, Xiaoquan Qi. Methods for Identification and Resistance Evaluation of Barley Slow Rusting to Leaf Rust [J]. Chinese Bulletin of Botany, 2020, 55(5): 573-576. |
| [12] | Nan Zhang,Ziguang Liu,Shichen Sun,Shengyi Liu,Jianhui Lin,Yifang Peng,Xiaoxu Zhang,He Yang,Xi Cen,Juan Wu. Response of AtR8 lncRNA to Salt Stress and Its Regulation on Seed Germination in Arabidopsis [J]. Chinese Bulletin of Botany, 2020, 55(4): 421-429. |
| [13] | Dongdong Cao,Shanyu Chen,Yebo Qin,Huaping Wu,Guanhai Ruan,Yutao Huang. Regulatory Mechanism of Salicylic Acid on Seed Germination Under Salt Stress in Kale [J]. Chinese Bulletin of Botany, 2020, 55(1): 49-61. |
| [14] | Yegeng Fan,Lihang Qiu,Xing Huang,Huiwen Zhou,Chongkun Gan,Yangrui Li,Rongzhong Yang,Jianming Wu,Rongfa Chen. Expression Analysis of Key Genes in Gibberellin Biosynthesis and Related Phytohormonal Dynamics During Sugarcane Internode Elongation [J]. Chinese Bulletin of Botany, 2019, 54(4): 486-496. |
| [15] | Xiaolong Wang,Fengzhi Liu,Xiangbin Shi,Xiaodi Wang,Xiaohao Ji,Zhiqiang Wang,Baoliang Wang,Xiaocui Zheng,Haibo Wang. Evolution and Expression of NCED Family Genes in Vitis vinifera [J]. Chinese Bulletin of Botany, 2019, 54(4): 474-485. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||