

Chinese Bulletin of Botany ›› 2024, Vol. 59 ›› Issue (4): 558-573.DOI: 10.11983/CBB23129 cstr: 32102.14.CBB23129
• EXPERIMENTAL COMMUNICATIONS • Previous Articles Next Articles
Laipeng Zhao1,2, Baike Wang2, Tao Yang2, Ning Li2, Haitao Yang2, Juan Wang2,*( ), Huizhuan Yan1,*(
), Huizhuan Yan1,*( )
)
Received:2023-09-15
Accepted:2023-12-19
Online:2024-07-10
Published:2024-07-10
Contact:
*E-mail: drjuanwang@126.com; hzhyan1118@163.com
Laipeng Zhao, Baike Wang, Tao Yang, Ning Li, Haitao Yang, Juan Wang, Huizhuan Yan. Investigation of the Regulation of Drought Tolerance by the SlHVA22l Gene in Tomato[J]. Chinese Bulletin of Botany, 2024, 59(4): 558-573.
| Primer name | Forward primer (5'-3') | Reverse primer (5'-3') |
|---|---|---|
| SlHVA22l | GGCATGGCTAGTTTTTCCACATT | TGGGAGTGATGAAGTCCACAAA |
| SlPDS | GTGAACCCTGTCGGCCTTTA | AACTACGCTTGCTTCCGACA |
| Slactin | AGCAGGAACTTGAAACCGCT | CTCATGGATACCCGCAGCTT |
| SlDREB1A | GGTTTGTGTGCCGTTGGATT | AGGAACTTTACGCACTGGCT |
| SlHK1 | GCTCTGAGTGGGTTTGCTCT | GGCAAAGACTCTCTGGCCTT |
| SlAREB1 | AATGAGGGCAGGGATGGTTG | TCCATTGCTCTTCCCAAGTCC |
| SlPYL9 | CGGTTCATCCCGAGGTCATT | GTACCGCCAAACGCTCTGAT |
| SlAVP1 | CAACACCGCAAATATCGCCC | CGTCCATACCAGCCATCTCC |
| SlABI1 | AGTCAGCCGCAGTGTTTTTG | AGACAGTCCATCAAGCACGC |
| SlAAPK | TCTGGAGGGGAGCTGTTTGA | GTGCAGGACTTCCATCCAGT |
| SlMPK8 | CCAAGGTACCCAAGGCAACA | GCCTCGGACAAACAGGTTCT |
Table 1 Primers for qRT-PCR
| Primer name | Forward primer (5'-3') | Reverse primer (5'-3') |
|---|---|---|
| SlHVA22l | GGCATGGCTAGTTTTTCCACATT | TGGGAGTGATGAAGTCCACAAA |
| SlPDS | GTGAACCCTGTCGGCCTTTA | AACTACGCTTGCTTCCGACA |
| Slactin | AGCAGGAACTTGAAACCGCT | CTCATGGATACCCGCAGCTT |
| SlDREB1A | GGTTTGTGTGCCGTTGGATT | AGGAACTTTACGCACTGGCT |
| SlHK1 | GCTCTGAGTGGGTTTGCTCT | GGCAAAGACTCTCTGGCCTT |
| SlAREB1 | AATGAGGGCAGGGATGGTTG | TCCATTGCTCTTCCCAAGTCC |
| SlPYL9 | CGGTTCATCCCGAGGTCATT | GTACCGCCAAACGCTCTGAT |
| SlAVP1 | CAACACCGCAAATATCGCCC | CGTCCATACCAGCCATCTCC |
| SlABI1 | AGTCAGCCGCAGTGTTTTTG | AGACAGTCCATCAAGCACGC |
| SlAAPK | TCTGGAGGGGAGCTGTTTGA | GTGCAGGACTTCCATCCAGT |
| SlMPK8 | CCAAGGTACCCAAGGCAACA | GCCTCGGACAAACAGGTTCT |

Figure 1 Amino acid sequence analysis of SlHVA22l (A) Phylogenetic tree of the HVA22l protein in eukaryotic organisms; (B) Phylogenetic tree of HVA22l proteins in dicotyledonous and monocotyledonous plants; (C) Comparison of homologous proteins between SlHVA22l and other plant species through multiple sequence alignment; (D) The TB2/DP1 domain. Cs: Citrus sinensis; Gh: Gossypium hirsutum; Tc: Theobroma cacao; Vv: Vitis vinifera; Cas: Camellia sinensis; Ah: Arachis hypogaea; Coa: Coffea arabica; Nt: Nicotiana tomentosiformis; Ca: Capsicum annuum; Sl: Solanum lycopersicum; St: So. tuberosum; Mb: Musa balbisiana; Zo: Zingiber officinale; Os: Oryza sativa; Bd: Brachypodium distachyon; Ta: Triticum aestivum; Hv: Hordeum vulgare; Sb: Sorghum bicolor; Zm: Zea mays; Si: Setaria italica; Sv: Se. viridis; Pv: Panicum virgatum; Pm: P. miliaceum; Ph: P. hallii
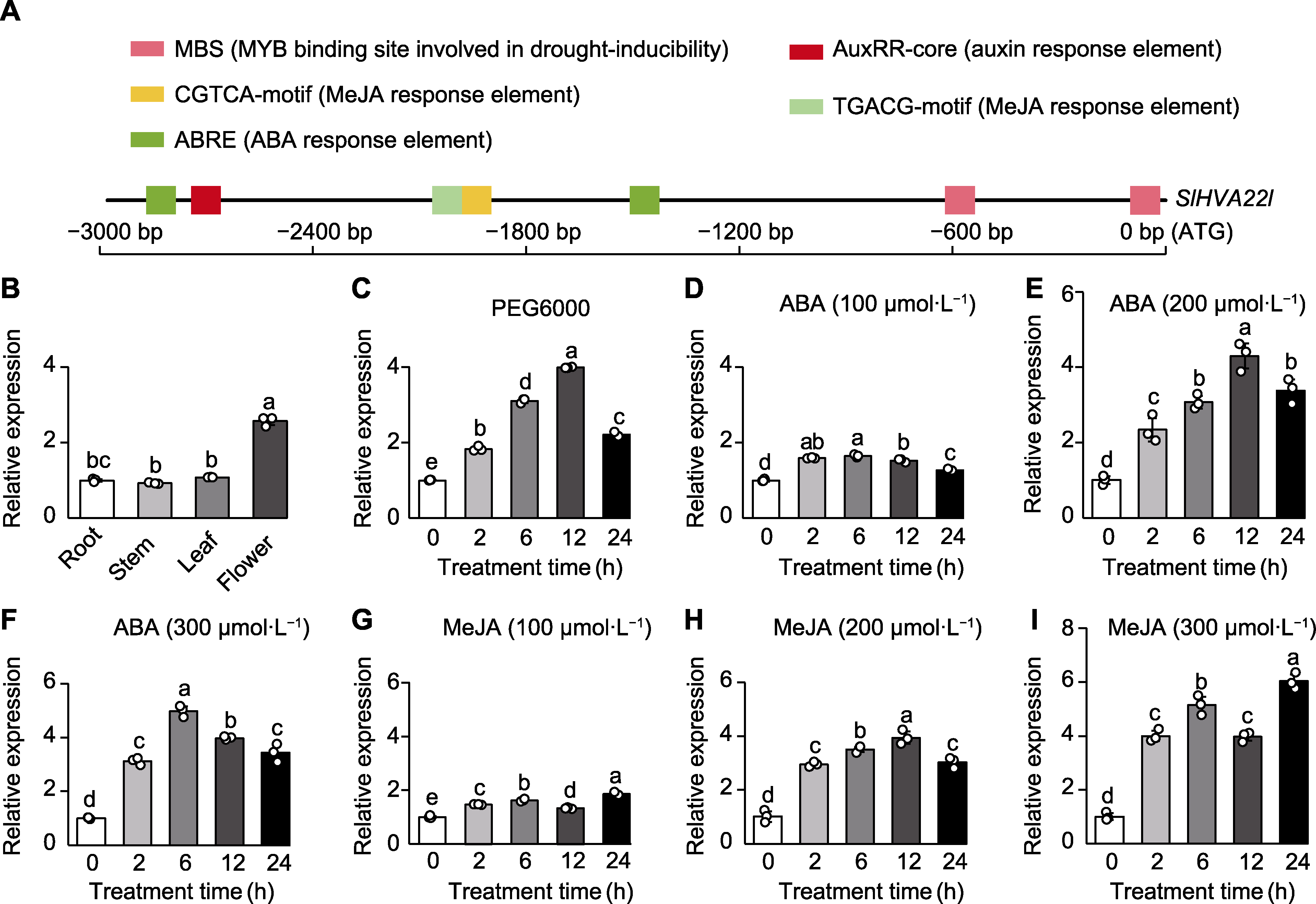
Figure 2 Cis-acting elements and expression pattern analysis of the SlHVA22l gene (A) Cis-acting elements within the 3 000 bp upstream promoter region of the SlHVA22l gene; (B) Tissue-specific expression of the SlHVA22l gene; (C)-(I) SlHVA22l expression pattern under PEG6000, ABA (100, 200, and 300 µmol∙L-1), and MeJA (100, 200, and 300 µmol∙L-1) treatments at 0-24 h. Slactin was used as a reference gene. Different lowercase letters indicate significant differences among different treatments (P<0.05).
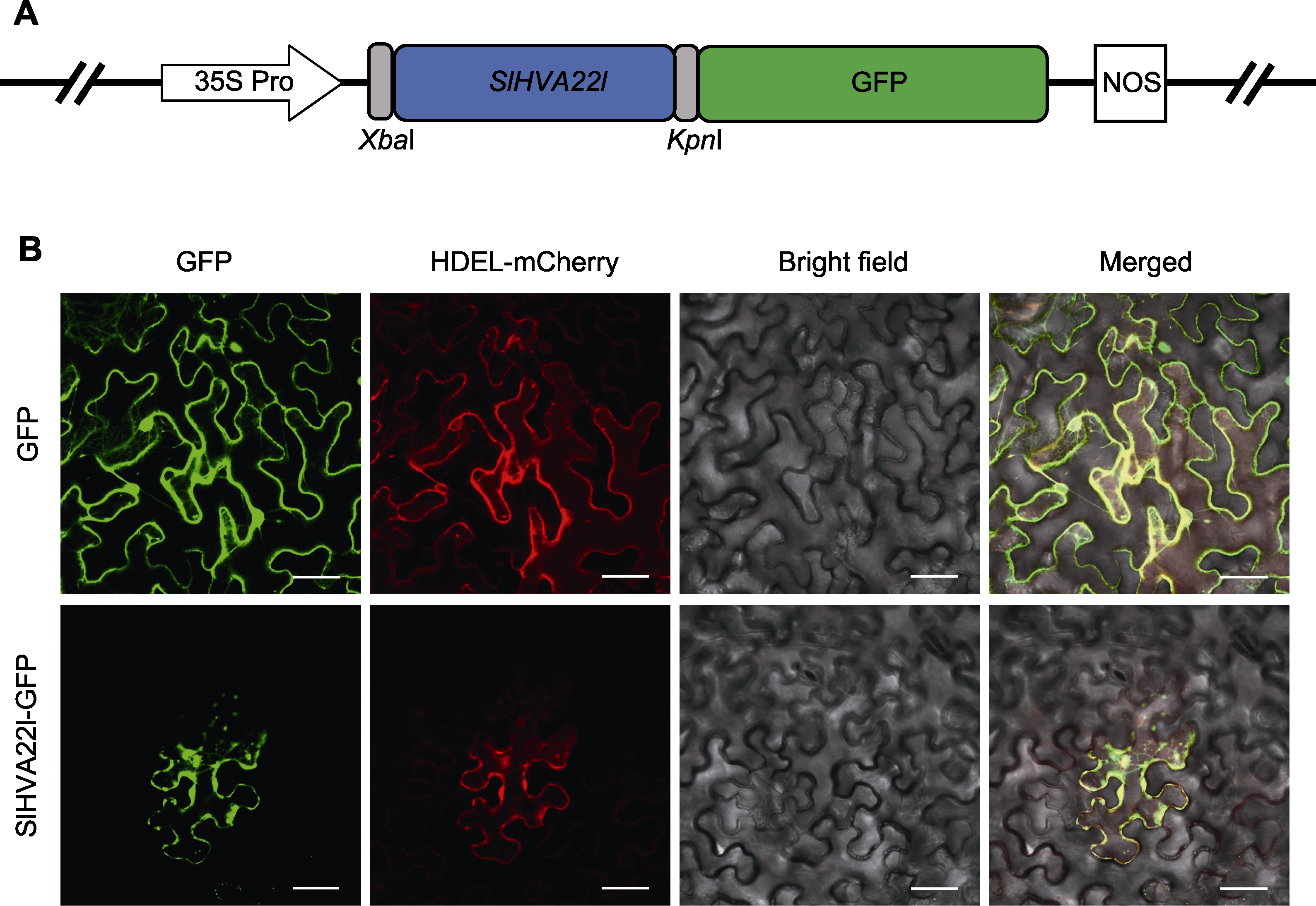
Figure 3 Subcellular localization of SlHVA22l (A) Schematic representation of SlHVA22l in the pCAMBIA1300-GFP vector; (B) Localization of the SlHVA22l protein in Ben’s tobacco cells. HDEL is an endoplasmic reticulum localization marker. Bars=50 μm
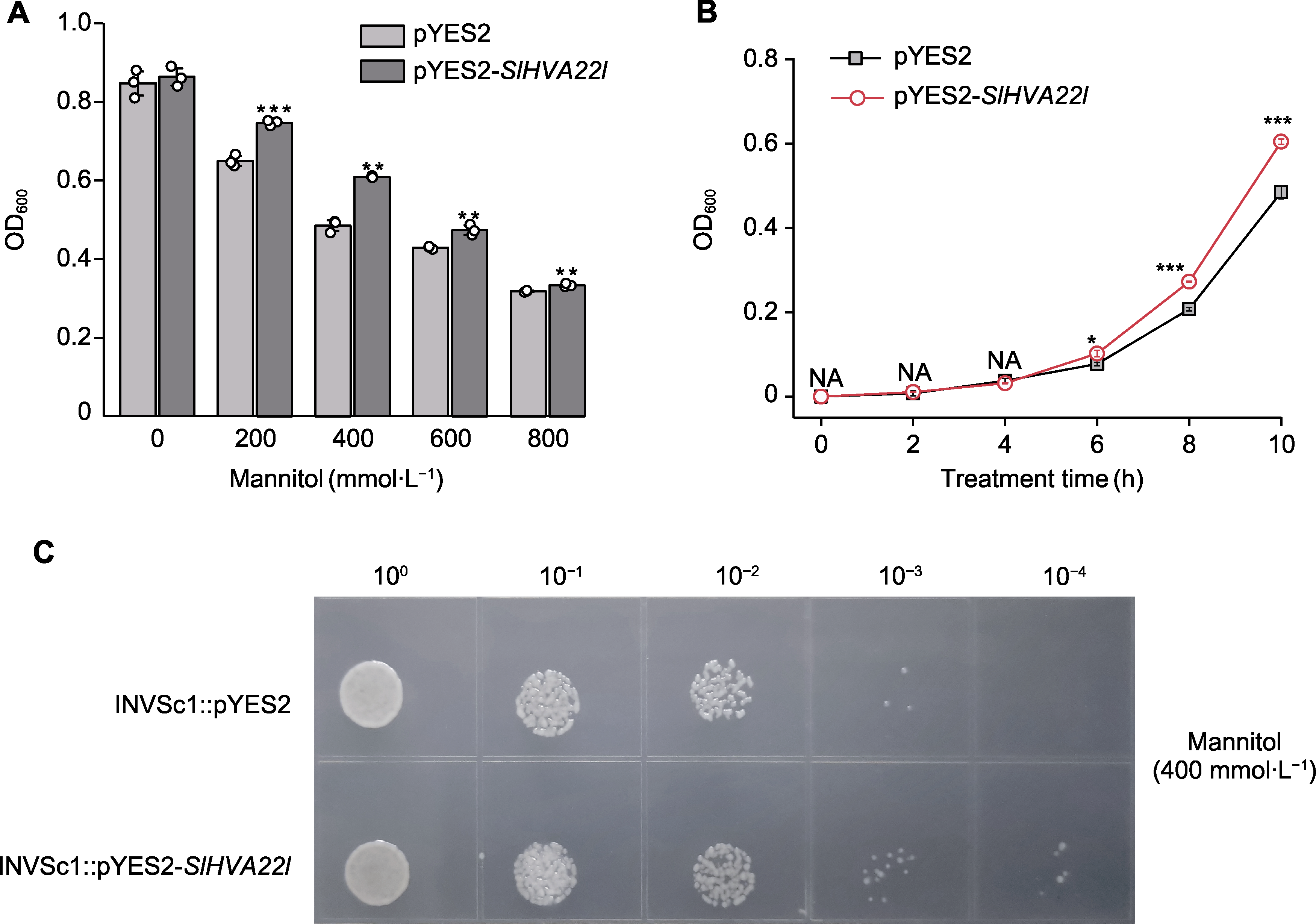
Figure 4 Expression analysis of SlHVA22l in Saccharomyces cerevisiae (A) Responses of recombinant pYES2-SlHVA22l and control pYES2 under simulated drought conditions; (B) Growth kinetics of recombinant pYES2-SlHVA22l and control pYES2 under drought stress; (C) Drip plate experiment of recombinant pYES2- SlHVA22l and control pYES2 under drought stress. * P<0.05; ** P<0.01; *** P<0.001
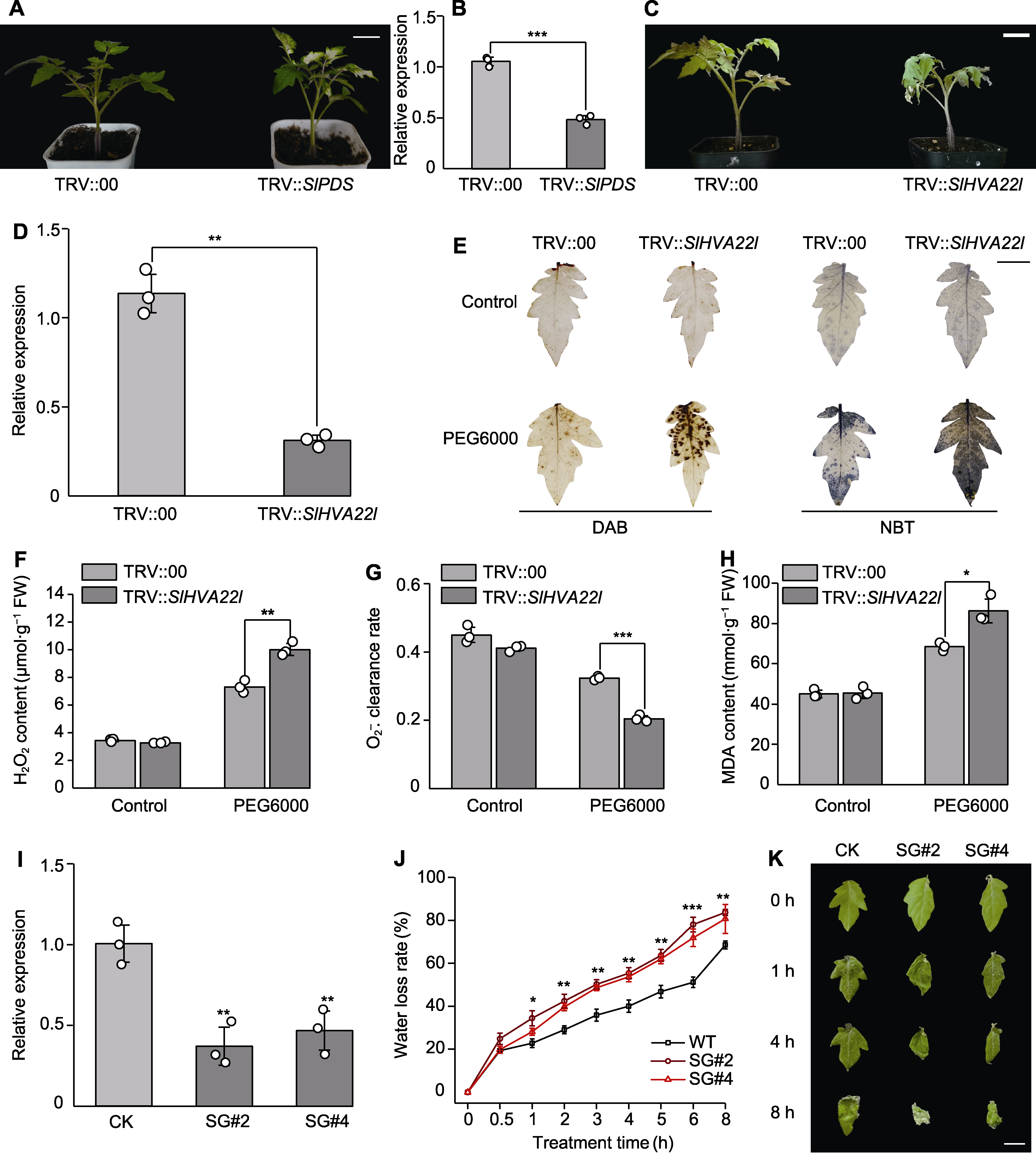
Figure 5 The effect of SlHVA22l gene silencing on drought tolerance in tomato (A), (B) TRV::SlPDS-carrying Agrobacterium was introduced into 2-week-old tomato plants, after two weeks, phenotypic changes (A) and the efficiency of SlPDS silencing was quantified using qRT-PCR (B); (C) The phenotypes of the control and silenced plants were visually documented and recorded 48 hours after treatment with a 20% PEG6000 solution; (D) The silencing efficiency of SlHVA22l was assessed via qRT-PCR analysis; (E) DAB and NBT staining of leaves from drought-stressed control and silenced plants, respectively, were performed before and after the stress treatment; (F) H2O2 contents; (G) O2-. clearance rate; (H) Malondialdehyde (MDA) contents; (I) Identification of silenced plants; (J) Water loss rate; (K) Dehydration phenotype of detached leaves. CK: TRV::00; SG: TRV::SlHVA22l. * P<0.05; ** P<0.01; *** P<0.001. (A), (C) Bars=3 cm; (E), (K) Bars=1 cm
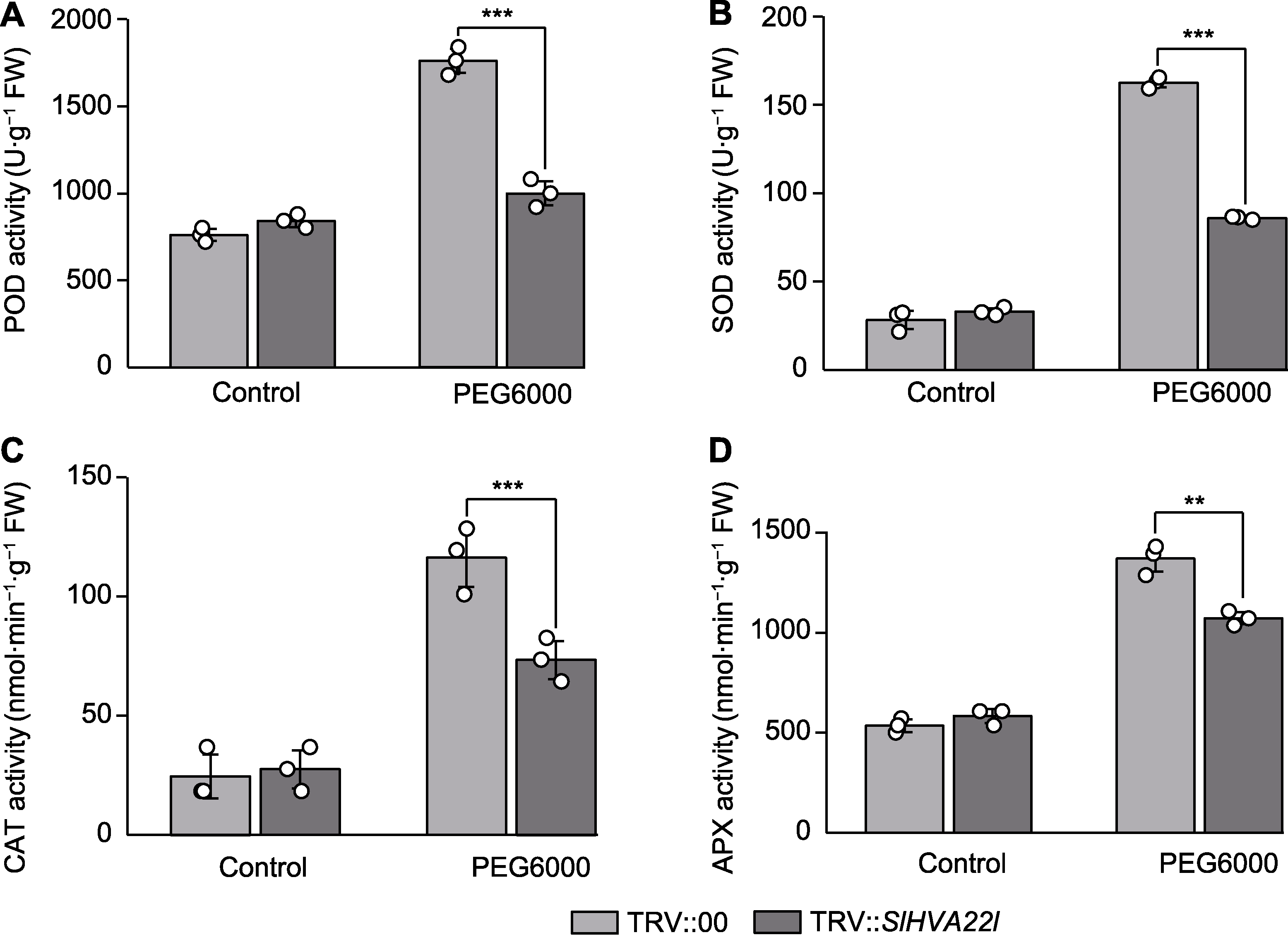
Figure 6 The antioxidant enzyme activities in SlHVA22l silenced tomato leaf under drought stress (A) Peroxidase (POD) activity; (B) Superoxide dismutase (SOD) activity; (C) Catalase (CAT) activity; (D) Ascorbate peroxidase (APX) activity. ** P<0.01; *** P<0.001
| [1] |
Argasinska J, Rana AA, Gilchrist MJ, Lachani K, Young A, Smith JC (2009). Loss of REEP4 causes paralysis of the Xenopus embryo. Int J Dev Biol 53, 37-43.
DOI PMID |
| [2] |
Casaretto JA, Ho THD (2005). Transcriptional regulation by abscisic acid in barley (Hordeum vulgare L.) seeds involves autoregulation of the transcription factor HvABI5. Plant Mol Biol 57, 21-34.
DOI PMID |
| [3] | Chaudhary J, Khatri P, Singla P, Kumawat S, Kumari A, Vikram A, Jindal SK, Kardile H, Kumar R, Sonah H (2019). Advances in omics approaches for abiotic stress tolerance in tomato. Biology 8, 90. |
| [4] | Chen CN, Chu CC, Zentella R, Pan SM, Ho THD (2002). AtHVA22 gene family in Arabidopsis: phylogenetic relationship, ABA and stress regulation, and tissue-specific expression. Plant Mol Biol 49, 631-642. |
| [5] | Choudhury FK, Rivero RM, Blumwald E, Mittler R (2017). Reactive oxygen species, abiotic stress and stress combination. Plant J 90, 856-867. |
| [6] | Danquah A, de Zélicourt A, Boudsocq M, Neubauer J, Frei dit Frey N, Leonhardt N, Pateyron S, Gwinner F, Tamby JP, Ortiz-Masia D, Marcote MJ, Hirt H, Colcombet J (2015). Identification and characterization of an ABA-activated MAP kinase cascade in Arabidopsis thaliana. Plant J 82, 232-244. |
| [7] | Eggert E, Obata T, Gerstenberger A, Gier K, Brandt T, Fernie AR, Schulze W, Kühn C (2016). A sucrose transporter-interacting protein disulphide isomerase affects redox homeostasis and links sucrose partitioning with abiotic stress tolerance. Plant Cell Environ 39, 1366- 1380. |
| [8] | Fukuda T, Saigusa T, Furukawa K, Inoue K, Yamashita SI, Kanki T (2023). Hva22, a REEP family protein in fission yeast, promotes reticulophagy in collaboration with a receptor protein. Autophagy 19, 2657-2667. |
| [9] | Gomes Ferreira MD, Araújo Castro J, Santana Silva RJ, Micheli F (2019). HVA22 from citrus: a small gene family whose some members are involved in plant response to abiotic stress. Plant Physiol Biochem 142, 395-404. |
| [10] | Grzesiak MT, Hordyńska N, Maksymowicz A, Grzesiak S, Szechyńska-Hebda M (2019). Variation among spring wheat (Triticum aestivum L.) genotypes in response to the drought stress. II-root system structure. Plants 8, 584. |
| [11] | Guo WJ, Ho THD (2008). An abscisic acid-induced protein, HVA22, inhibits gibberellin-mediated programmed cell death in cereal aleurone cells. Plant Physiol 147, 1710- 1722. |
| [12] |
Hata T, Nan HT, Koh K, Ishiura H, Tsuji S, Takiyama Y (2022). A clinical and genetic study of SPG31 in Japan. J Hum Genet 67, 421-425.
DOI PMID |
| [13] |
Iovieno P, Punzo P, Guida G, Mistretta C, Van Oosten MJ, Nurcato R, Bostan H, Colantuono C, Costa A, Bagnaresi P, Chiusano ML, Albrizio R, Giorio P, Batelli G, Grillo S (2016). Transcriptomic changes drive physiological responses to progressive drought stress and rehydration in tomato. Front Plant Sci 7, 371.
DOI PMID |
| [14] | Jiang LL, Wang YB, Zhang SH, He R, Li W, Han J, Cheng XG (2017). Tomato SlDREB1 gene conferred the transcriptional activation of drought-induced gene and an enhanced tolerance of the transgenic Arabidopsis to drought stress. Plant Growth Regul 81, 131-145. |
| [15] | Jiao P, Liu TY, Zhao CL, Fei JB, Guan SY, Ma YY (2023). ZmTCP14, a TCP transcription factor, modulates drought stress response in Zea mays L. Environ Exp Bot 208, 105232. |
| [16] | Kumar MN, Jane WN, Verslues PE (2013). Role of the putative osmosensor Arabidopsis Histidine kinase1 in dehydration avoidance and low-water-potential response. Plant Physiol 161, 942-953. |
| [17] | Letunic I, Bork P (2021). Interactive tree of life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res 49, W293-W296. |
| [18] | Li Q, Shen H, Yuan SJ, Dai XG, Yang CX (2023). miRNAs and lncRNAs in tomato: roles in biotic and abiotic stress responses. Front Plant Sci 13, 1094459. |
| [19] | Liang MW, Li HJ, Zhou F, Li HY, Liu J, Hao Y, Wang YD, Zhao HP, Han SC (2015). Subcellular distribution of NTL transcription factors in Arabidopsis thaliana. Traffic 16, 1062- 1074. |
| [20] |
Liu MM, Yu HY, Zhao GJ, Huang QF, Lu YE, Ouyang B (2017). Profiling of drought-responsive microRNA and mRNA in tomato using high-throughput sequencing. BMC Genomics 18, 481.
DOI PMID |
| [21] |
Livak KJ, Schmittgen TD (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-∆∆CT method. Methods 25, 402-408.
DOI PMID |
| [22] | Lu P (2013). Physiological functional analysis of a stress- induced protein, HVA22, in Escherichia coli. Acce Inter J 1, 14-23. |
| [23] | Massimi M (2021). Tomato (Lycopersicon esculentum Mill.) anatomical, physiological, biochemical and production responses to drought stress-a mini-review essay. Int J Hortic Sci 27, 40-45. |
| [24] | Meng FW, Zhao QQ, Zhao X, Yang C, Liu R, Pang JH, Zhao WS, Wang Q, Liu MX, Zhang ZG, Kong ZS, Liu J (2022). A rice protein modulates endoplasmic reticulum homeostasis and coordinates with a transcription factor to initiate blast disease resistance. Cell Rep 39, 110941. |
| [25] | Nemeskéri E, Helyes L (2019). Physiological responses of selected vegetable crop species to water stress. Agronomy 9, 447. |
| [26] | Nemeskéri E, Neményi A, Bőcs A, Pék Z, Helyes L (2019). Physiological factors and their relationship with the productivity of processing tomato under different water supplies. Water 11, 586. |
| [27] |
Nicolas P, Shinozaki Y, Powell A, Philippe G, Snyder SI, Bao K, Zheng Y, Xu YM, Courtney L, Vrebalov J, Casteel CL, Mueller LA, Fei ZJ, Giovannoni JJ, Rose JKC, Catalá C (2022). Spatiotemporal dynamics of the tomato fruit transcriptome under prolonged water stress. Plant Physiol 190, 2557-2578.
DOI PMID |
| [28] | Orellana S, Yañez M, Espinoza A, Verdugo I, González E, Ruiz-Lara S, Casaretto JA (2010). The transcription factor SlAREB1 confers drought, salt stress tolerance and regulates biotic and abiotic stress-related genes in tomato. Plant Cell Environ 33, 2191-2208. |
| [29] |
Park S, Li JS, Pittman JK, Berkowitz GA, Yang HB, Undurraga S, Morris J, Hirschi KD, Gaxiola RA (2005). Up-regulation of a H+-pyrophosphatase (H+-PPase) as a strategy to engineer drought-resistant crop plants. Proc Natl Acad Sci USA 102, 18830-18835.
DOI PMID |
| [30] |
Reichardt S, Piepho HP, Stintzi A, Schaller A (2020). Peptide signaling for drought-induced tomato flower drop. Science 367, 1482-1485.
DOI PMID |
| [31] | Ripoll J, Urban L, Brunel B, Bertin N (2016). Water deficit effects on tomato quality depend on fruit developmental stage and genotype. J Plant Physiol 190, 26-35. |
| [32] | Rombauts S, Déhais P, Van Montagu M, Rouzé P (1999). PlantCARE, a plant cis-acting regulatory element database. Nucleic Acids Res 27, 295-296. |
| [33] | Sharon K, Suvarna S (2017). Cloning of HVA22 homolog from aloe vera and preliminary study of transgenic plant development. Int J Pure App Biosci 5, 1113-1121. |
| [34] | Shen QX, Chen CN, Brands A, Pan SM, Tuan-Hua DH (2001). The stress- and abscisic acid-induced barley gene HVA22: developmental regulation and homologues in diverse organisms. Plant Mol Biol 45, 327-340. |
| [35] |
Shen QX, Uknes S, Ho THD (1993). Hormone respon-se complex in a novel abscisic acid and cycloheximide-inducible barley gene. J Biol Chem 268, 23652-23660.
PMID |
| [36] | Sun HM, Li JT, Li X, Lv Q, Chen LP, Wang BX, Li LQ (2022a). RING E3 ubiquitin ligase TaSADR1 negatively regulates drought resistance in transgenic Arabidopsis. Plant Physiol Biochem 170, 255-265. |
| [37] | Sun ZH, Feng ZK, Ding YL, Qi YP, Jiang S, Li Z, Wang Y, Qi JS, Song CP, Yang SH, Gong ZZ (2022b). RAF22, ABI1 and OST1 form a dynamic interactive network that optimizes plant growth and responses to drought stress in Arabidopsis. Mol Plant 15, 1192-1210. |
| [38] |
Tamura K, Stecher G, Kumar S (2021). MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol 38, 3022-3027.
DOI PMID |
| [39] |
Wagih O (2017). ggseqlogo: a versatile R package for drawing sequence logos. Bioinformatics 33, 3645-3647.
DOI PMID |
| [40] | Wai AH, Waseem M, Cho LH, Kim ST, Lee DJ, Kim CK, Chung MY (2022). Comprehensive genome-wide analysis and expression pattern profiling of the SlHVA22 gene family unravels their likely involvement in the abiotic stress adaptation of tomato. Int J Mol Sci 23, 12222. |
| [41] | Wang L, Zheng B, Yuan Y, Xu QL, Chen P (2020a). Transcriptome profiling of Fagopyrum tataricum leaves in response to lead stress. BMC Plant Biol 20, 54. |
| [42] | Wang M, Yuan JR, Qin LM, Shi WM, Xia GM, Liu SW (2020b). TaCYP81D5, one member in a wheat cytochrome P450 gene cluster, confers salinity tolerance via reactive oxygen species scavenging. Plant Biotechnol J 18, 791-804. |
| [43] | Wang YJ, Zhang XY, Huang GR, Liu XY, Guo R, Gu FX, Zhong XL, Mei XR (2019). Characteristics of phosphatidic acid and the underlying mechanisms of ABA-induced stomatal movement in plants. Chin Bull Bot 54, 245-254. (in Chinese) |
|
王雅静, 张欣莹, 黄桂荣, 刘晓英, 郭瑞, 顾峰雪, 钟秀丽, 梅旭荣 (2019). 植物磷脂酸的特性及其在ABA诱导气孔运动中的作用. 植物学报 54, 245-254.
DOI |
|
| [44] | Wang YQ, Cao XY, Zhang DK, Li YQ, Wang QQ, Ma F, Xu X, Zhan XQ, Hu TX (2023). SlGATA17, a tomato GATA protein, interacts with SlHY5 to modulate salinity tolerance and germination. Environ Exp Bot 206, 105191. |
| [45] | Wei XT, Fan XH, Zhang HL, Jiao P, Jiang ZZ, Lu X, Liu SY, Guan SY, Ma YY (2022). Overexpression of ZmSRG7 improves drought and salt tolerance in maize (Zea mays L.). Int J Mol Sci 23, 13349. |
| [46] | Xiong HY, Yu JP, Miao JL, Li JJ, Zhang HL, Wang X, Liu PL, Zhao Y, Jiang CH, Yin ZG, Li Y, Guo Y, Fu BY, Wang WS, Li ZK, Ali J, Li ZC (2018). Natural variation in OsLG3 increases drought tolerance in rice by inducing ROS scavenging. Plant Physiol 178, 451-467. |
| [47] |
Yamaguchi-Shinozaki K, Shinozaki K (2006). Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu Rev Plant Biol 57, 781-803.
PMID |
| [48] |
Yang YG, Lv WT, Li MJ, Wang B, Sun DM, Deng X (2013). Maize membrane-bound transcription factor Zmbzip17 is a key regulator in the cross-talk of ER quality control and ABA signaling. Plant Cell Physiol 54, 2020-2033.
DOI PMID |
| [49] | Zhang HJ, Yuan YC, Xing HX, Xin M, Saeed M, Wu Q, Wu J, Zhuang T, Zhang XP, Mao LL, Sun XZ, Song XL, Wang ZW (2023). Genome-wide identification and expression analysis of the HVA22 gene family in cotton and functional analysis of GhHVA22E1D in drought and salt tolerance. Front Plant Sci 14, 1139526. |
| [50] | Zhang YG, Zhang Y, Ayibaiheremu Mutailifu, Zhang DY (2021). Heterologous overexpression of desiccation-tole- rance moss ScABI3gene changes stomatal phenotype and improves drought resistance in transgenic Arabidopsis. Chin Bull Bot 56, 414-421. (in Chinese) |
| 张一弓, 张怡, 阿依白合热木·木台力甫, 张道远 (2021). 异源过表达齿肋赤藓ScABI3基因改变拟南芥气孔表型并提高抗旱性. 植物学报 56, 414-421. | |
| [51] | Zhao LP, Wang BK, Yang T, Yan HZ, Yu QH, Wang J (2023). Genome-wide identification and analysis of the evolution and expression pattern of the HVA22 gene family in three wild species of tomatoes. PeerJ 11, e14844. |
| [52] | Zhou P, An Y, Wang ZL, Du HM, Huang BR (2014). Characterization of gene expression associated with drought avoidance and tolerance traits in a perennial grass species. PLoS One 9, e103611. |
| [1] | Ziyun Wang, Yanwen Lv, Yu Xiao, Chao Wu, Xinsheng Hu. Advances in Regulation and Evolutionary Mechanisms of Plant Gene Expression [J]. Chinese Bulletin of Botany, 2025, 60(4): 1-0. |
| [2] | Bei Fan, Min Ren, Yanfeng Wang, Fengfeng Dang, Guoliang Chen, Guoting Cheng, Jinyu Yang, Huiru Sun. Functions of SlWRKY45 in Response to Low-temperature and Drought Stress in Tomato [J]. Chinese Bulletin of Botany, 2025, 60(2): 186-203. |
| [3] | LONG Ji-Lan, JIANG Zheng, LIU Ding-Qin, MIAO Yu-Xuan, ZHOU Ling-Yan, FENG Ying, PEI Jia-Ning, LIU Rui-Qiang, ZHOU Xu-Hui, FU Yu-Ling. Effects of drought on plant root exudates and associated rhizosphere priming effect: review and prospect [J]. Chin J Plant Ecol, 2024, 48(7): 817-827. |
| [4] | Ren-Yu Liao, Jia-Wei Wang. From Wound to Rebirth: How does REF1 Peptide Activate Intrinsic Regenerative Potential of Plants? [J]. Chinese Bulletin of Botany, 2024, 59(3): 347-350. |
| [5] | Hanqian Zhao, Jiayi Song, Jie Yang, Yongjing Zhao, Wennian Xia, Weizhuo Gu, Zhongyi Wang, Nan Yang, Huizhen Hu. Identification of XTH Family Genes in Antirrhinum majus and Screening of Genes Involoved in Sclerotinia sclerotiorum Resistance and Stamen Petalization [J]. Chinese Bulletin of Botany, 2024, 59(2): 188-203. |
| [6] | Zhengyong Duan, Min Ding, Yuzhuo Wang, Yibing Ding, Ling Chen, Ruiyun Wang, Zhijun Qiao. Genome-wide Identification and Expression Analysis of SBP Genes in Panicum miliaceum [J]. Chinese Bulletin of Botany, 2024, 59(2): 231-244. |
| [7] | Zhang Yingchuan, Wu Xiaomingyu, Tao Baolong, Chen Li, Lu Haiqin, Zhao Lun, Wen Jing, Yi Bin, Tu Jinxing, Fu Tingdong, Shen Jinxiong. Bna-miR43 Mediates the Response of Drought Tolerance in Brassica napus [J]. Chinese Bulletin of Botany, 2023, 58(5): 701-711. |
| [8] | Cai Shuyu, Liu Jianxin, Wang Guofu, Wu Liyuan, Song Jiangping. Regulatory Mechanism of Melatonin on Tomato Seed Germination Under Cd2+ Stress [J]. Chinese Bulletin of Botany, 2023, 58(5): 720-732. |
| [9] | Nan Wu, Lei Qin, Kan Cui, Haiou Li, Zhongsong Liu, Shitou Xia. Cloning of Brassica napus EXA1 Gene and Its Regulation on Plant Disease Resistance [J]. Chinese Bulletin of Botany, 2023, 58(3): 385-393. |
| [10] | CHEN Tu-Qiang, XU Gui-Qing, LIU Shen-Si, LI Yan. Hydraulic traits adjustments and nonstructural carbohydrate dynamics of Haloxylon ammodendron under drought stress [J]. Chin J Plant Ecol, 2023, 47(10): 1407-1421. |
| [11] | Feifei Wang, Zhenxiang Zhou, Yi Hong, Yangyang Gu, Chao Lü, Baojian Guo, Juan Zhu, Rugen Xu. Identification of the NF-YC Genes in Hordeum vulgare and Expression Analysis Under Salt Stress [J]. Chinese Bulletin of Botany, 2023, 58(1): 140-149. |
| [12] | ZHOU Jie, YANG Xiao-Dong, WANG Ya-Yun, LONG Yan-Xin, WANG Yan, LI Bo-Rui, SUN Qi-Xing, SUN Nan. Difference in adaptation strategy between Haloxylon ammodendron and Alhagi sparsifolia to drought [J]. Chin J Plant Ecol, 2022, 46(9): 1064-1076. |
| [13] | Xiaomin Wang, Honglei Li, Lin Wang, Pengze Zhou, Shengyi Bai, Guohua Li, Fushun Zheng, Xiaorong Tao, Guoxin Cheng, Yanming Gao, Jianshe Li. Molecular Identification of Tomato Spotted Wilt Virus on Tomato in Yinchuan [J]. Chinese Bulletin of Botany, 2021, 56(6): 715-721. |
| [14] | Yongmei Che, Yanjun Sun, Songchong Lu, Lixia Hou, Xinxin Fan, Xin Liu. AtMYB77 Involves in Lateral Root Development via Regulating Nitric Oxide Biosynthesis under Drought Stress in Arabidopsis thaliana [J]. Chinese Bulletin of Botany, 2021, 56(4): 404-413. |
| [15] | Jiaxin Li, Xia Li, Yinfeng Xie. Mechanism on Drought Tolerance Enhanced by Exogenous Trehalose in C4-PEPC Rice [J]. Chinese Bulletin of Botany, 2021, 56(3): 296-314. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||