

Chinese Bulletin of Botany ›› 2019, Vol. 54 ›› Issue (4): 474-485.DOI: 10.11983/CBB18200 cstr: 32102.14.CBB18200
• EXPERIMENTAL COMMUNICATIONS • Previous Articles Next Articles
Xiaolong Wang,Fengzhi Liu,Xiangbin Shi,Xiaodi Wang,Xiaohao Ji,Zhiqiang Wang,Baoliang Wang,Xiaocui Zheng,Haibo Wang( )
)
Received:2018-09-19
Accepted:2019-01-15
Online:2019-07-01
Published:2020-01-08
Contact:
Haibo Wang
Xiaolong Wang,Fengzhi Liu,Xiangbin Shi,Xiaodi Wang,Xiaohao Ji,Zhiqiang Wang,Baoliang Wang,Xiaocui Zheng,Haibo Wang. Evolution and Expression of NCED Family Genes in Vitis vinifera[J]. Chinese Bulletin of Botany, 2019, 54(4): 474-485.
| Gene name | Forward primer (5'-3') | Reverse primer (5'-3') |
|---|---|---|
| VvNCED1 | GCTGGAGAAGCTGATAGTGAAG | GAAGATACCCAATGACCGGAAG |
| VvNCED2 | GGCACTTTCGGAGGTTGATAA | TGGATGAGCAGTGAAGGAATG |
| VvNCED3 | CGGTGGAGATGGTGAGAATAGA | CACTGCTGCGTACACGTATTT |
| VvNCED4 | CTCAGCAGTAGGTGATCCTTTG | CAGGCTCGTACATTCTCTTAGC |
| VvNCED6 | CTCGTGATTTGGGCTCTTTCT | GCTTGATGATGTGTGCTTTGG |
| VvNCED7 | CGCTCTTCTTCTTCCTCACTAC | GGCGTTCCCTCTTCTACTATTG |
| VvNCED9 | CCATGGACTTCCCGATGATAAA | ATCCCACAACTAGAGCTTGC |
| VvNCED10 | CAGGGAGGTGTTGAAGAAGATG | CCCTTTGAGGCAGTGTGATT |
| VvActin | TACAATTCCATCATGAAGTGTGATG | TTAGAAGCACTTCCTGTGAACAATG |
Table 1 The primer sequences used for quantitative RT-PCR
| Gene name | Forward primer (5'-3') | Reverse primer (5'-3') |
|---|---|---|
| VvNCED1 | GCTGGAGAAGCTGATAGTGAAG | GAAGATACCCAATGACCGGAAG |
| VvNCED2 | GGCACTTTCGGAGGTTGATAA | TGGATGAGCAGTGAAGGAATG |
| VvNCED3 | CGGTGGAGATGGTGAGAATAGA | CACTGCTGCGTACACGTATTT |
| VvNCED4 | CTCAGCAGTAGGTGATCCTTTG | CAGGCTCGTACATTCTCTTAGC |
| VvNCED6 | CTCGTGATTTGGGCTCTTTCT | GCTTGATGATGTGTGCTTTGG |
| VvNCED7 | CGCTCTTCTTCTTCCTCACTAC | GGCGTTCCCTCTTCTACTATTG |
| VvNCED9 | CCATGGACTTCCCGATGATAAA | ATCCCACAACTAGAGCTTGC |
| VvNCED10 | CAGGGAGGTGTTGAAGAAGATG | CCCTTTGAGGCAGTGTGATT |
| VvActin | TACAATTCCATCATGAAGTGTGATG | TTAGAAGCACTTCCTGTGAACAATG |
| Gene name | Accession No. | Chromosome location (start, end) | Length of amino acids (aa) | Molecular weight (kDa) | Theoretical pI | GRAVY |
|---|---|---|---|---|---|---|
| VvNCED1 | VIT_213s0064g00840.1 | Chr.13 (22672994, 22681910) | 546 | 61.63 | 6.13 | -0.271 |
| VvNCED2 | VIT_213s0064g00810.1 | Chr.13 (22587965, 22596719) | 510 | 57.85 | 6.23 | -0.250 |
| VvNCED3 | VIT_202s0087g00910.1 | Chr.2 (18560696, 18562591) | 599 | 65.90 | 6.88 | -0.236 |
| VvNCED4 | VIT_202s0087g00930.1 | Chr.2 (18588853, 18590786) | 589 | 65.61 | 6.63 | -0.187 |
| VvNCED5 | VIT_216s0039g01370.1 | Chr.16 (789473, 791221) | 558 | 62.09 | 5.57 | -0.197 |
| VvNCED6 | VIT_219s0093g00550.1 | Chr.19 (17645348, 17647649) | 609 | 67.13 | 6.38 | -0.317 |
| VvNCED7 | VIT_210s0003g03750.1 | Chr.10 (6374432, 6376728) | 605 | 67.34 | 6.36 | -0.365 |
| VvNCED8 | VIT_205s0051g00670.1 | Chr.5 (11589343, 11591102) | 575 | 63.14 | 8.24 | -0.202 |
| VvNCED9 | VIT_204s0008g03510.1 | Chr.4 (2883265, 2886523) | 567 | 63.72 | 5.73 | -0.335 |
| VvNCED10 | VIT_204s0008g03480.1 | Chr.4 (2873553, 2878309) | 625 | 70.53 | 6.43 | -0.305 |
| VvNCED11 | VIT_204s0008g03380.1 | Chr.4 (2784465, 2788790) | 563 | 62.34 | 7.27 | -0.339 |
| VvNCED12 | VIT_215s0021g02190.1 | Chr.15 (13131078, 13135539) | 610 | 68.46 | 7.31 | -0.313 |
Table 2 NCED genes identified in grapevine and their detailed information
| Gene name | Accession No. | Chromosome location (start, end) | Length of amino acids (aa) | Molecular weight (kDa) | Theoretical pI | GRAVY |
|---|---|---|---|---|---|---|
| VvNCED1 | VIT_213s0064g00840.1 | Chr.13 (22672994, 22681910) | 546 | 61.63 | 6.13 | -0.271 |
| VvNCED2 | VIT_213s0064g00810.1 | Chr.13 (22587965, 22596719) | 510 | 57.85 | 6.23 | -0.250 |
| VvNCED3 | VIT_202s0087g00910.1 | Chr.2 (18560696, 18562591) | 599 | 65.90 | 6.88 | -0.236 |
| VvNCED4 | VIT_202s0087g00930.1 | Chr.2 (18588853, 18590786) | 589 | 65.61 | 6.63 | -0.187 |
| VvNCED5 | VIT_216s0039g01370.1 | Chr.16 (789473, 791221) | 558 | 62.09 | 5.57 | -0.197 |
| VvNCED6 | VIT_219s0093g00550.1 | Chr.19 (17645348, 17647649) | 609 | 67.13 | 6.38 | -0.317 |
| VvNCED7 | VIT_210s0003g03750.1 | Chr.10 (6374432, 6376728) | 605 | 67.34 | 6.36 | -0.365 |
| VvNCED8 | VIT_205s0051g00670.1 | Chr.5 (11589343, 11591102) | 575 | 63.14 | 8.24 | -0.202 |
| VvNCED9 | VIT_204s0008g03510.1 | Chr.4 (2883265, 2886523) | 567 | 63.72 | 5.73 | -0.335 |
| VvNCED10 | VIT_204s0008g03480.1 | Chr.4 (2873553, 2878309) | 625 | 70.53 | 6.43 | -0.305 |
| VvNCED11 | VIT_204s0008g03380.1 | Chr.4 (2784465, 2788790) | 563 | 62.34 | 7.27 | -0.339 |
| VvNCED12 | VIT_215s0021g02190.1 | Chr.15 (13131078, 13135539) | 610 | 68.46 | 7.31 | -0.313 |
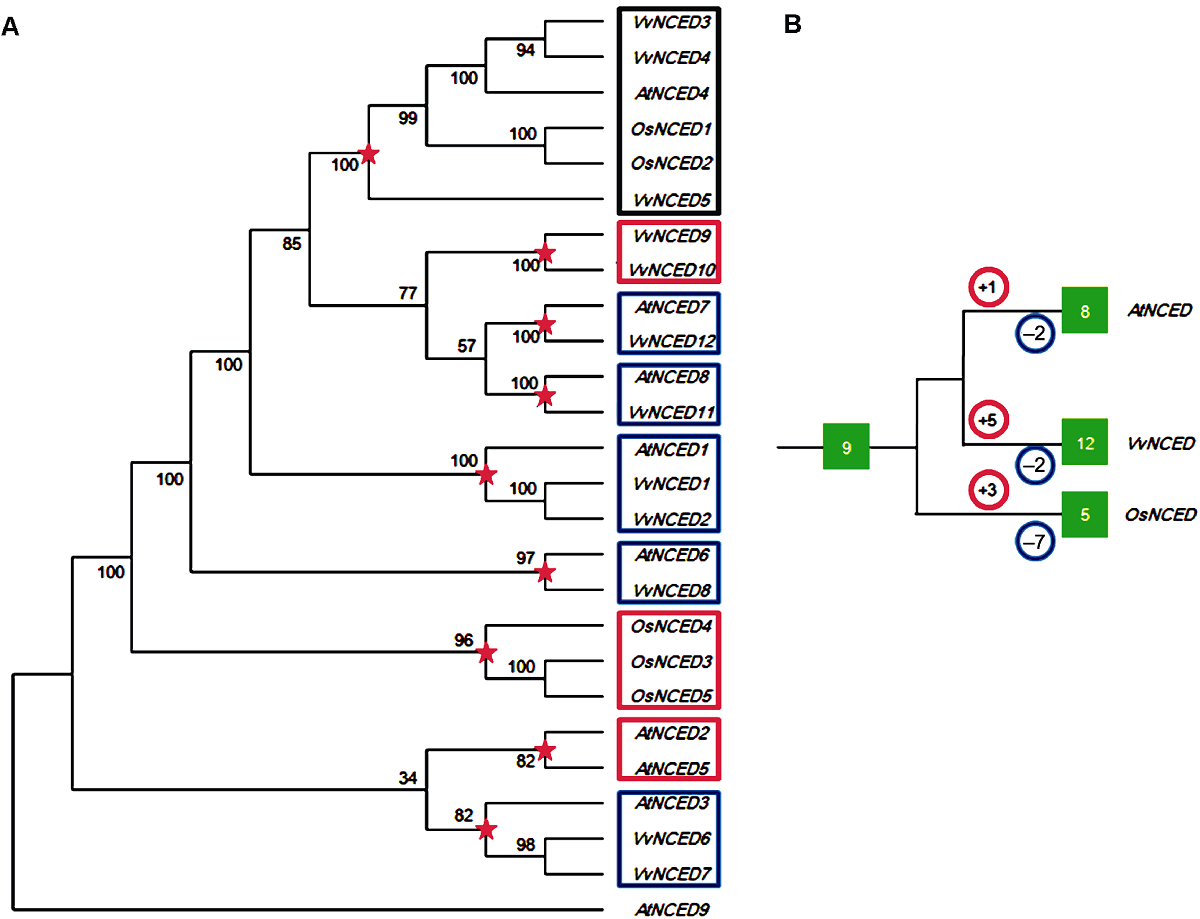
Figure 1 Joint phylogenetic tree (A) and amplification model (B) of NCED gene from grape, Arabidopsis and rice The red five-pointed stars in the figure represent the most recent common ancestral differentiation node before species differentiation. The numbers in the red and blue circles represent the number of duplication and loss events that occur after the species differentiated nodes, respectively. The numbers in the green solid box represent the number of NCED genes.
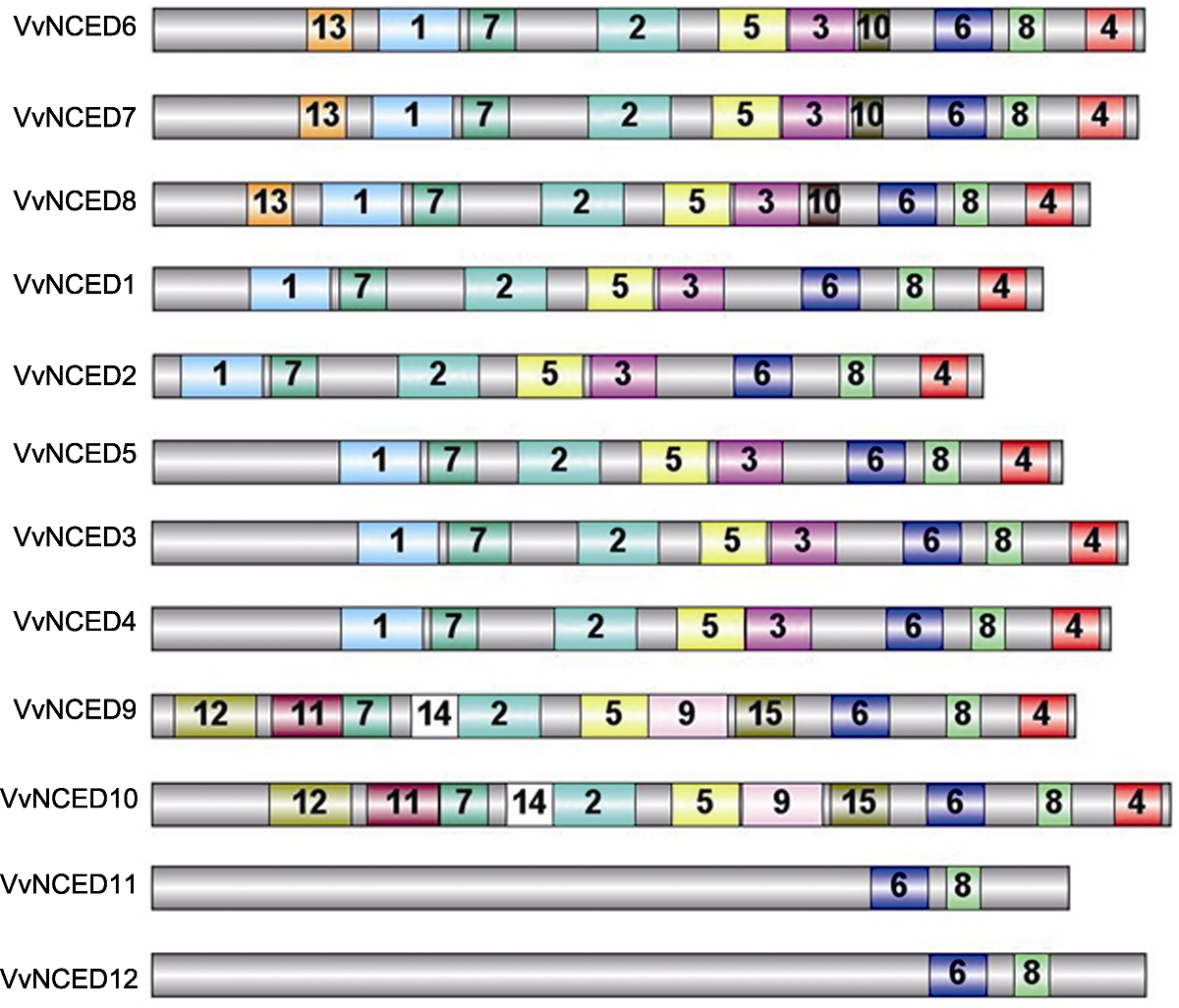
Figure 3 Schematic structure of conserved motifs identified in grapevine NCED proteins The grey bars represent the full length of NCED proteins, and the numbers in the other colored boxes are random numbers for the conserved domains located at NCED protein. The same number represents the same conserved domain.
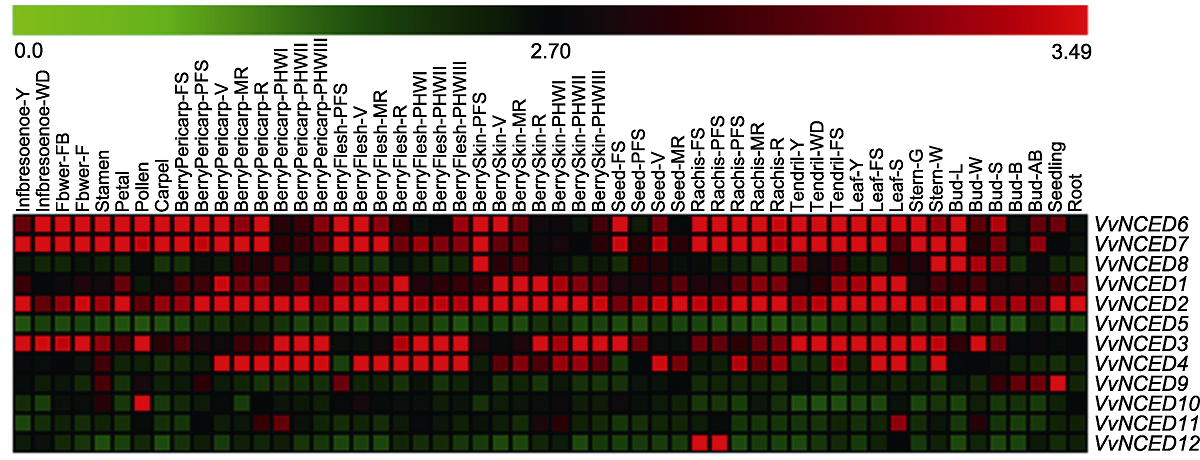
Figure 4 Expression pattern of NCED genes at different developmental stages and in some specialized tissues of grapevine The 54 tissue names and gene names are located above and to the right of the heat map, respectively. The scale bar above the heat map indicates the geng expression level from 0.0 to 3.49.

Figure 5 Expression pattern of grapevine NCED genes under drought treatments The drought treatments (DL1-DL5 indicate drought treatment for 0, 7, 14, 21 and 28 days of leaf tissue, respectively; DR1-DR5 indicate drought treatment for 0, 7, 14, 21 and 28 days of root tissue, respectively) and gene names are located above and to the right of the heat map, respectively. The scale bar above the heat map indicates the gene expression level from 0.0 to 77.06. Solid dots of the same color represent duplicated gene pairs.
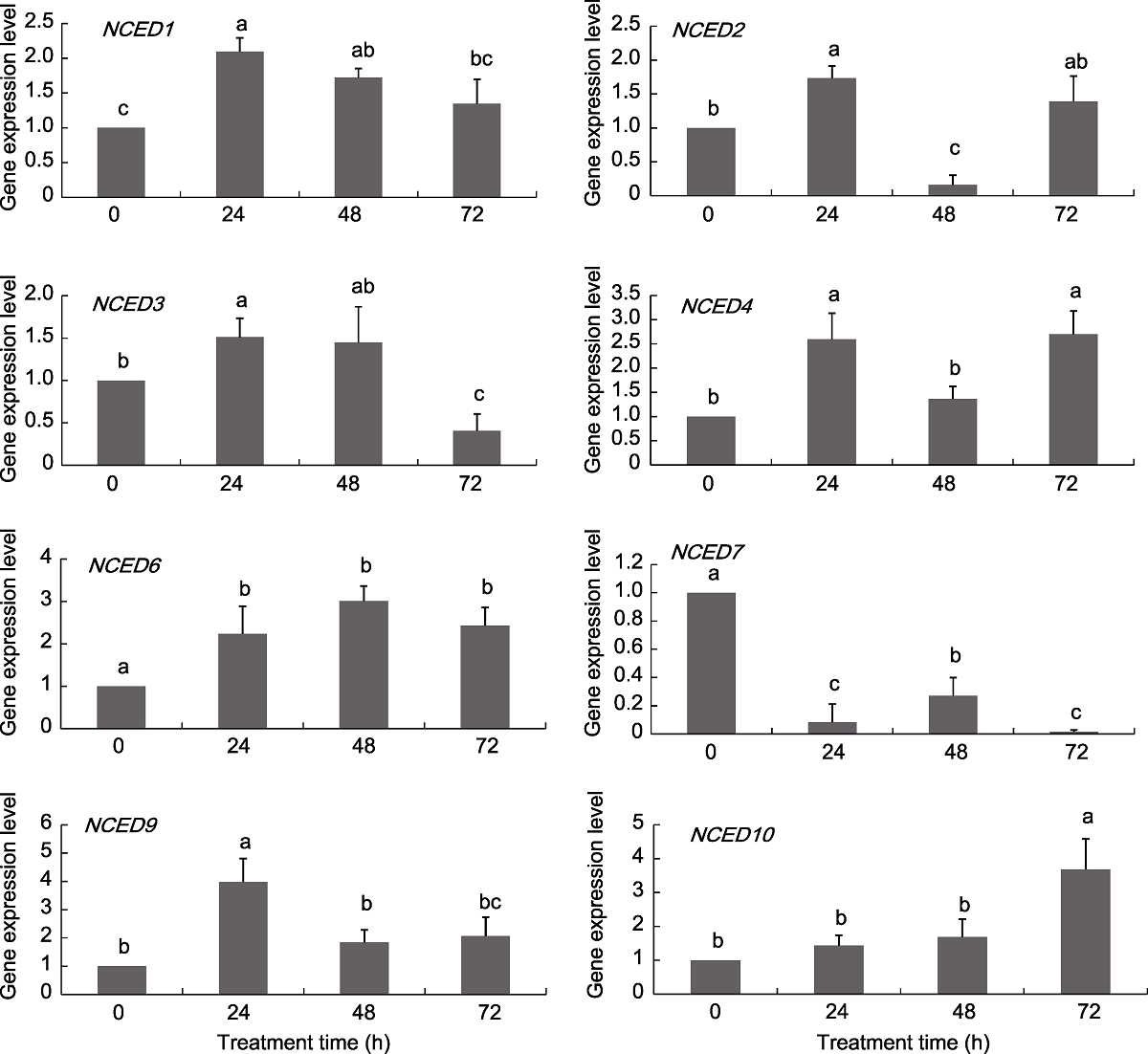
Figure 6 Expression pattern of grapevine NCED genes under ABA treatments Different lowercase letters above the histogram indicate significant differences among the different time points of the treatment (P<0.05).
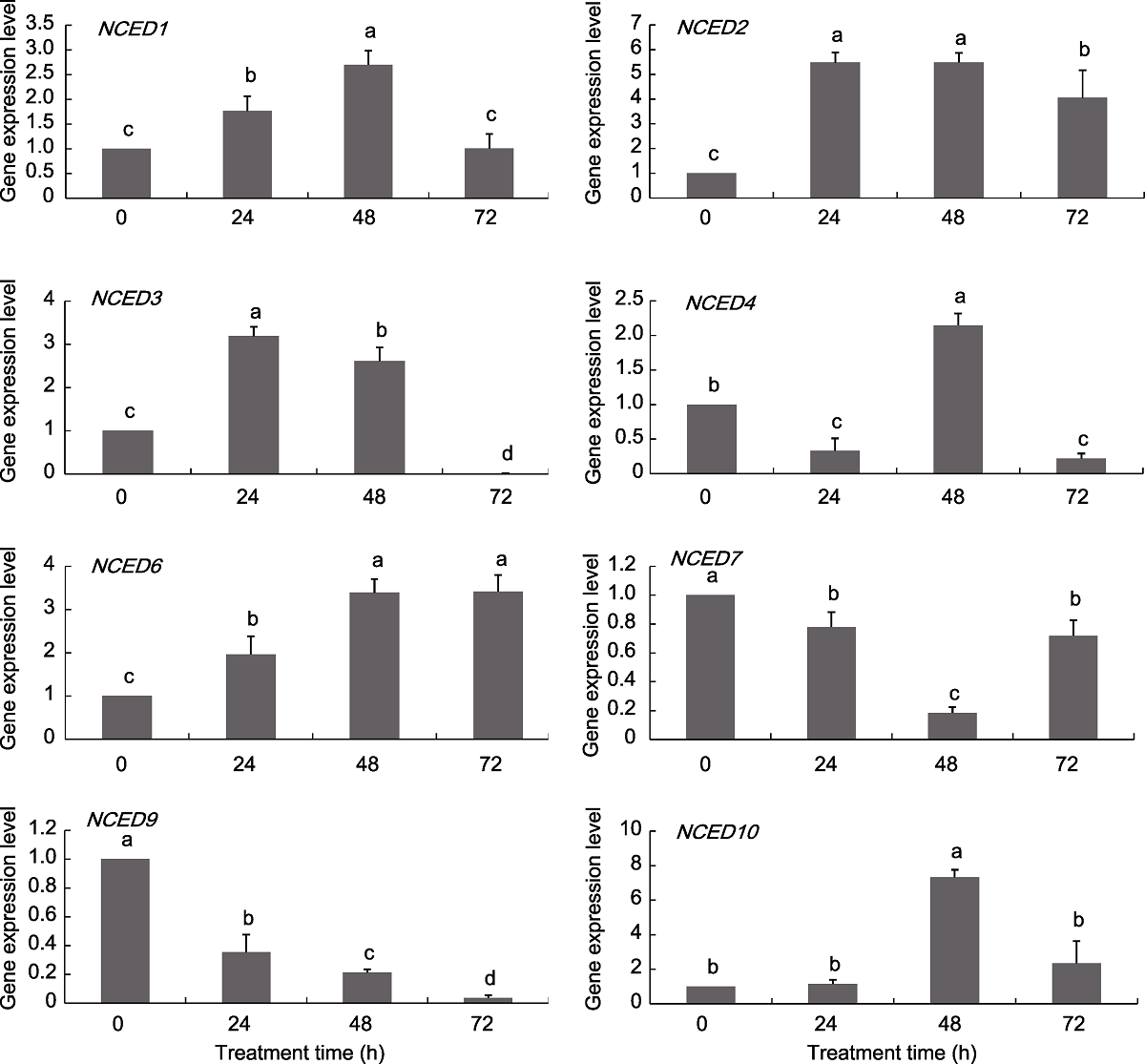
Figure 7 Expression pattern of grapevine NCED genes under NaCl treatments Different lowercase letters above the histogram indicate significant differences among the different time points of the treatment (P<0.05).
| [1] | 白戈, 杨大海, 姚恒, 谢贺 ( 2017). 烟草NtNCED基因的鉴定分析. 分子植物育种 15, 3907-3912. |
| [2] | 徐学中, 汪婷, 万旺, 李思慧, 朱国辉 ( 2018). 水稻ABA生物合成基因OsNCED3响应干旱胁迫. 作物学报 44, 24-31. |
| [3] | Adams KL, Cronn R, Percifield R, Wendel JF ( 2003). Genes duplicated by polyploidy show unequal contributions to the transcriptome and organ-specific reciprocal silencing. Proc Natl Acad Sci USA 100, 4649-4654. |
| [4] | Ahrazem O, Rubio-Moraga A, Trapero A, Gómez-Gómez L ( 2012). Developmental and stress regulation of gene expression for a 9-cis-epoxycarotenoid dioxygenase, Cst NCED, isolated from Crocus sativus stigmas. J Exp Bot 63, 681-694. |
| [5] | Chaw SM, Chang CC, Chen HL, Li WH ( 2004). Dating the monocot-dicot divergence and the origin of core eudicots using whole chloroplast genomes. J Mol Evol 58, 424-441. |
| [6] | Cohen-Gihon I, Sharan R, Nussinov R ( 2011). Processes of fungal proteome evolution and gain of function: gene duplication and domain rearrangement. Phys Biol 8, 035009. |
| [7] | Fasoli M, Dal Santo S, Zenoni S, Tornielli GB, Farina L, Zamboni A, Porceddu A, Venturini L, Bicego M, Murino V, Ferrarini A, Delledonne M, Pezzotti M ( 2012). The grapevine expression atlas reveals a deep transcriptome shift driving the entire plant into a maturation program. Plant Cell 24, 3489-3505. |
| [8] | Gaut BS, Morton BR, McCaig BC, Clegg MT ( 1996). Substitution rate comparisons between grasses and palms: synonymous rate differences at the nuclear gene Adh parallel rate differences at the plastid gene rbcL. Proc Natl Acad Sci USA 93, 10274-10279. |
| [9] | Gu ZL, Nicolae D, Lu HHS, Li WH ( 2002). Rapid divergence in expression between duplicate genes inferred from microarray data. Trends Genet 18, 609-613. |
| [10] | Gu ZL, Rifkin SA, White KP, Li WH ( 2004). Duplicate genes increase gene expression diversity within and between species. Nat Genet 36, 577-579. |
| [11] | Guo CL, Guo RR, Xu XZ, Gao M, Li XQ, Song JY, Zheng Y, Wang XP ( 2014). Evolution and expression analysis of the grape (Vitis vinifera L.) WRKY gene family. J Exp Bot 65, 1513-1528. |
| [12] | Jaillon O, Aury JM, Noel B, Policriti A, Clepet C, Casagrande A, Choisne N, Aubourg S, Vitulo N, Jubin C, Vezzi A, Legeai F, Hugueney P, Dasilva C, Horner D, Mica E, Jublot D, Poulain J, Bruyère C, Billault A, Segurens B, Gouyvenoux M, Ugarte E, Cattonaro F, Anthouard V, Vico V, Del Fabbro C, Alaux M, Di Gaspero G, Dumas V, Felice N, Paillard S, Juman I, Moroldo M, Scalabrin S, Canaguier A, Le Clainche I, Malacrida G, Durand E, Pesole G, Laucou V, Chatelet P, Merdinoglu D, Delledonne M, Pezzotti M, Lecharny A, Scarpelli C, Artiguenave F, Pè ME, Valle G, Morgante M, Caboche M, Adam-Blondon AF, Weissenbach J, Quétier F, Wincker P ( 2007). The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449, 463-467. |
| [13] | Malacarne G, Perazzolli M, Cestaro A, Sterck L, Fontana P, Van de Peer Y, Viola R, Velasco R, Salamini F ( 2012). Deconstruction of the (Paleo) polyploid grapevine genome based on the analysis of transposition events involving NBS resistance genes. PLoS One 7, e29762. |
| [14] | McAdam SAM, Brodribb TJ ( 2015). The evolution of mechanisms driving the stomatal response to vapor pressure deficit. Plant Physiol 167, 833-843. |
| [15] | McAdam SAM, Brodribb TJ, Banks JA, Hedrich R, Atallah NM, Cai C, Geringer MA, Lind C, Nichols DS, Stachowski K, Geiger D, Sussmilch FC ( 2016). Abscisic acid controlled sex before transpiration in vascular plants. Proc Natl Acad Sci USA 113, 12862-12867. |
| [16] | Raghavendra AS, Gonugunta VK, Christmann A, Grill E ( 2010). ABA perception and signaling. Trends Plant Sci 15, 395-401. |
| [17] | Roychoudhury A, Paul S, Basu S ( 2013). Cross-talk between abscisic acid-dependent and abscisic acid-independent pathways during abiotic stress. Plant Cell Rep 32, 985-1006. |
| [18] | Saeed AI, Bhagabati NK, Braisted JC, Liang W, Sharov V, Howe EA, Li JW, Thiagarajan M, White JA, Quackenbush J ( 2006). TM4 microarray software suite. Methods Enzymol 411, 134-193. |
| [19] | Sussmilch FC, Brodribb TJ, McAdam SAM ( 2017). What are the evolutionary origins of stomatal responses to abscisic acid in land plants? J Integr Plant Biol 59, 240-260. |
| [20] | Tan BC, Joseph LM, Deng WT, Liu LJ, Li QB, Cline K, McCarty DR ( 2010). Molecular characterization of the Arabidopsis 9-cis epoxycarotenoid dioxygenase gene family. Plant J 35, 44-56. |
| [21] | Wang RY, Yang Y, Wang HG, Chen L, Wang L, Lu P, Liu MX, Qiao ZJ ( 2018). Cloning of gene PmNCED1 and its response to PEG stress in common millet. J Nuclear Agric Sci 32, 244-256. |
| [22] | Yang SH, Zhang XH, Yue JX, Tian DC, Chen JQ ( 2008). Recent duplications dominate NBS-encoding gene expan- sion in two woody species. Mol Genet Genom 280, 187-198. |
| [23] | Ye X, Kang BG, Osburn LD, Li Y, Cheng ZM ( 2009). Identification of the flavin-dependent monooxygenase- encoding YUCCA gene family in Populus trichocarpa and their expression in vegetative tissues and in response to hormone and environmental stresses. Plant Cell Tissue Organ Cult 97, 271-283. |
| [24] | Zhang JZ ( 2003). Evolution by gene duplication: an update. Trends Ecol Evol 18, 292-298. |
| [1] | Ziyun Wang, Yanwen Lv, Yu Xiao, Chao Wu, Xinsheng Hu. Advances in Regulation and Evolutionary Mechanisms of Plant Gene Expression [J]. Chinese Bulletin of Botany, 2025, 60(4): 1-0. |
| [2] | Xiong Lianglin, Liang Guolu, Guo Qigao, Jing Danlong. Advances in the Regulation of Alternative Splicing of Genes in Plants in Response to Abiotic Stress [J]. Chinese Bulletin of Botany, 2025, 60(3): 435-448. |
| [3] | Qingguo Du, Wenxue Li. Research Progress in the Regulation of Development and Stress Responses by Long Non-coding RNAs in Maize [J]. Chinese Bulletin of Botany, 2024, 59(6): 950-962. |
| [4] | Wenjie Zhou, Wenhan Zhang, Wei Jia, Zicheng Xu, Wuxing Huang. Advances in Plant miRNAs Responses to Abiotic Stresses [J]. Chinese Bulletin of Botany, 2024, 59(5): 810-833. |
| [5] | Laipeng Zhao, Baike Wang, Tao Yang, Ning Li, Haitao Yang, Juan Wang, Huizhuan Yan. Investigation of the Regulation of Drought Tolerance by the SlHVA22l Gene in Tomato [J]. Chinese Bulletin of Botany, 2024, 59(4): 558-573. |
| [6] | Zhengyong Duan, Min Ding, Yuzhuo Wang, Yibing Ding, Ling Chen, Ruiyun Wang, Zhijun Qiao. Genome-wide Identification and Expression Analysis of SBP Genes in Panicum miliaceum [J]. Chinese Bulletin of Botany, 2024, 59(2): 231-244. |
| [7] | Hanqian Zhao, Jiayi Song, Jie Yang, Yongjing Zhao, Wennian Xia, Weizhuo Gu, Zhongyi Wang, Nan Yang, Huizhen Hu. Identification of XTH Family Genes in Antirrhinum majus and Screening of Genes Involoved in Sclerotinia sclerotiorum Resistance and Stamen Petalization [J]. Chinese Bulletin of Botany, 2024, 59(2): 188-203. |
| [8] | Yuejing Zhang, Hetian Sang, Hanqi Wang, Zhenzhen Shi, Li Li, Xin Wang, Kun Sun, Ji Zhang, Hanqing Feng. Research Progress of Plant Signaling in Systemic Responses to Abiotic Stresses [J]. Chinese Bulletin of Botany, 2024, 59(1): 122-133. |
| [9] | Zhaoxuan Zhong, Dongrui Zhang, Lu Li, Ying Su, Daining Wang, Zeran Wang, Yang Liu, Ying Chang. Bioinformatic and Expression Pattern Analysis of dfr-miR160a and Target Gene DfARF10 in Dryopteris fragrans [J]. Chinese Bulletin of Botany, 2024, 59(1): 22-33. |
| [10] | Yanan Xu, Jiarong Yan, Xin Sun, Xiaomei Wang, Yufeng Liu, Zhouping Sun, Mingfang Qi, Tianlai Li, Feng Wang. Red and Far-red Light Regulation of Plant Growth, Development, and Abiotic Stress Responses [J]. Chinese Bulletin of Botany, 2023, 58(4): 622-637. |
| [11] | Jia Zhang, Qidong Li, Cui Li, Qinghai Wang, Xincun Hou, Chunqiao Zhao, Shuhe Li, Qiang Guo. Research Progress on MATE Transporters in Plants [J]. Chinese Bulletin of Botany, 2023, 58(3): 461-474. |
| [12] | Xiaotong Ren, Ranran Zhang, Shaowei Wei, Xiaofeng Luo, Jiahui Xu, Kai Shu. Research Progress of Spermosphere Microorganisms [J]. Chinese Bulletin of Botany, 2023, 58(3): 499-509. |
| [13] | Nan Wu, Lei Qin, Kan Cui, Haiou Li, Zhongsong Liu, Shitou Xia. Cloning of Brassica napus EXA1 Gene and Its Regulation on Plant Disease Resistance [J]. Chinese Bulletin of Botany, 2023, 58(3): 385-393. |
| [14] | Feifei Wang, Zhenxiang Zhou, Yi Hong, Yangyang Gu, Chao Lü, Baojian Guo, Juan Zhu, Rugen Xu. Identification of the NF-YC Genes in Hordeum vulgare and Expression Analysis Under Salt Stress [J]. Chinese Bulletin of Botany, 2023, 58(1): 140-149. |
| [15] | WU Lin-Sheng, ZHANG Yong-Guang, ZHANG Zhao-Ying, ZHANG Xiao-Kang, WU Yun-Fei. Remote sensing of solar-induced chlorophyll fluorescence and its applications in terrestrial ecosystem monitoring [J]. Chin J Plant Ecol, 2022, 46(10): 1167-1199. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||