

Chinese Bulletin of Botany ›› 2024, Vol. 59 ›› Issue (1): 22-33.DOI: 10.11983/CBB23025 cstr: 32102.14.CBB23025
• EXPERIMENTAL COMMUNICATIONS • Previous Articles Next Articles
Zhaoxuan Zhong1, Dongrui Zhang1, Lu Li1, Ying Su2, Daining Wang1, Zeran Wang1, Yang Liu1, Ying Chang1,*( )
)
Received:2023-02-27
Accepted:2023-08-29
Online:2024-01-10
Published:2024-01-10
Contact:
*E-mail: Zhaoxuan Zhong, Dongrui Zhang, Lu Li, Ying Su, Daining Wang, Zeran Wang, Yang Liu, Ying Chang. Bioinformatic and Expression Pattern Analysis of dfr-miR160a and Target Gene DfARF10 in Dryopteris fragrans[J]. Chinese Bulletin of Botany, 2024, 59(1): 22-33.
| Primer name | Sequence (5′-3′) | Purpose |
|---|---|---|
| DfARF10F | ATGCCCGGCCCTTTATCAAC | Gene clone |
| DfARF10R | TCACCTTGTAATGTTTTCACCG | Gene clone |
| dfr-pri-mir160aF | AAAATCACTCTGCCTGGCTC | Gene clone |
| dfr-pri-mir160aR | AGCGAGAAACTCTGCGTGG | Gene clone |
| DfARF10F-EGFP | CTCGGTACCCGGGGATCCATGCCCGGCCCTTTATCAAC | Gene clone |
| DfARF10R-EGFP | GGTGTCGACTCTAGAGGATCCCCTTGTAATGTTTTCACCG | Gene clone |
| pCAMBIA2301-dfr-pri-mir160aF | GGGCATCGATACGGGATCCATAAAATCACTCTGCCTGGCTC | Gene clone |
| pCAMBIA2301-dfr-pri-mir160aR | TCGAGCTCGATGGATCCCGTAAGCGAGAAACTCTGCGTGG | Gene clone |
| pCAMBIA2301-DfARF10F-GUS | GGGCATCGATACGGGATCCATATGCCCGGCCCTTTATCAAC | Gene clone |
| pCAMBIA2301-DfARF10R-GUS | TCGAGCTCGATGGATCCCGTACCTTGTAATGTTTTCACCG | Gene clone |
| pGreenII-dfr-pri-mir160aF | CAGTGGTCTCACACC AAAATCACTCTGCCTGGCTC | Gene clone |
| pGreenII-dfr-pri-mir160aR | CAGTGGTCTCAAGCGAGCGAGAAACTCTGCGTGG | Gene clone |
| pGreenII-LUC-DfARF10F | CAGTGGTCTCAGATCTCCATGGCAAGTGGAGCTA | Gene clone |
| pGreenII-LUC-DfARF10R | CAGTGGTCTCAAATTGTTCACAAGGCTACCCATGTTA | Gene clone |
| Df18sRNAF | GCTTTCGCAGTAGTTCGTCTTTC | qRT-PCR |
| Df18sRNAR | TGGTCCTATTATGTTGGTCTTCGG | qRT-PCR |
| DfARF10F | GCAATGCGGCGGGAGATCTT | qRT-PCR |
| DfARF10R | CAGAGCTCGAGCGCAAAGCC | qRT-PCR |
| dfr-miR160aF | TGCCTGGCTCCCTGTATGCCA | qRT-PCR |
| dfr-miR160aR | mRQ 3′ Primer | qRT-PCR |
Table1 Primer sequences
| Primer name | Sequence (5′-3′) | Purpose |
|---|---|---|
| DfARF10F | ATGCCCGGCCCTTTATCAAC | Gene clone |
| DfARF10R | TCACCTTGTAATGTTTTCACCG | Gene clone |
| dfr-pri-mir160aF | AAAATCACTCTGCCTGGCTC | Gene clone |
| dfr-pri-mir160aR | AGCGAGAAACTCTGCGTGG | Gene clone |
| DfARF10F-EGFP | CTCGGTACCCGGGGATCCATGCCCGGCCCTTTATCAAC | Gene clone |
| DfARF10R-EGFP | GGTGTCGACTCTAGAGGATCCCCTTGTAATGTTTTCACCG | Gene clone |
| pCAMBIA2301-dfr-pri-mir160aF | GGGCATCGATACGGGATCCATAAAATCACTCTGCCTGGCTC | Gene clone |
| pCAMBIA2301-dfr-pri-mir160aR | TCGAGCTCGATGGATCCCGTAAGCGAGAAACTCTGCGTGG | Gene clone |
| pCAMBIA2301-DfARF10F-GUS | GGGCATCGATACGGGATCCATATGCCCGGCCCTTTATCAAC | Gene clone |
| pCAMBIA2301-DfARF10R-GUS | TCGAGCTCGATGGATCCCGTACCTTGTAATGTTTTCACCG | Gene clone |
| pGreenII-dfr-pri-mir160aF | CAGTGGTCTCACACC AAAATCACTCTGCCTGGCTC | Gene clone |
| pGreenII-dfr-pri-mir160aR | CAGTGGTCTCAAGCGAGCGAGAAACTCTGCGTGG | Gene clone |
| pGreenII-LUC-DfARF10F | CAGTGGTCTCAGATCTCCATGGCAAGTGGAGCTA | Gene clone |
| pGreenII-LUC-DfARF10R | CAGTGGTCTCAAATTGTTCACAAGGCTACCCATGTTA | Gene clone |
| Df18sRNAF | GCTTTCGCAGTAGTTCGTCTTTC | qRT-PCR |
| Df18sRNAR | TGGTCCTATTATGTTGGTCTTCGG | qRT-PCR |
| DfARF10F | GCAATGCGGCGGGAGATCTT | qRT-PCR |
| DfARF10R | CAGAGCTCGAGCGCAAAGCC | qRT-PCR |
| dfr-miR160aF | TGCCTGGCTCCCTGTATGCCA | qRT-PCR |
| dfr-miR160aR | mRQ 3′ Primer | qRT-PCR |
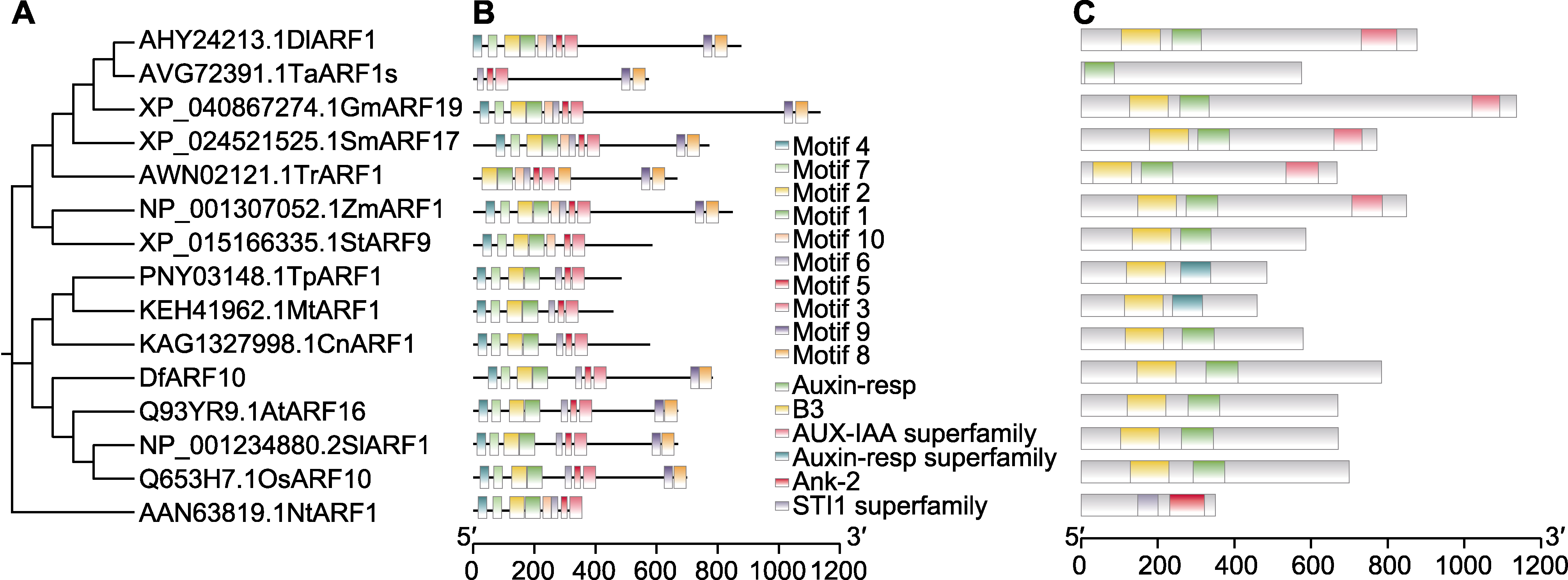
Figure 3 Conservative motif analysis of DfARF10 protein in Dryopteris fragrans (A) Phylogenetic tree of DfARF10 protein; (B) Motif analysis of DfARF10; (C) Conservative domains analysis of DfARF10
| Motif | Sequence | PFAM analysis |
|---|---|---|
| Motif1 | WKFRHIYRGQPRRHLLTTGWSVFVNQKKLVAGDSVVFLRNENGELRVGIR | B3 |
| Motif2 | SFCKTLTASDTNNGGGFSVPRRCAETIFPPLDYSQDPPVQELVAKDVHG | Auxin-resp |
| Motif3 | DPVRWPNSKWRMLQVGWDEPEALZRPKRVSPWZIEPVSAPP | - |
| Motif4 | LWHACAGPLVSIPPVGSKVYYFPQGHAEQ | - |
| Motif5 | GMRFKMAFETEESSRRRYFG | - |
| Motif6 | FEVVYYPRASPSEFVVPAKKV | - |
| Motif7 | PKILCRVLNVKLLADPETDEVYAKITLQP | - |
| Motif8 | WQVVYVDAEGDILLVGDDPWSEFVKTVRRIKILSPEEVQKM | AUX-IAA |
| Motif9 | VGRSLDLSKFSSYEELREELARMFGIEG | - |
| Motif10 | MPSSVISSHSMHIGVLAAAAHAVATNTM | - |
Table 2 Motif sequences of DfARF10
| Motif | Sequence | PFAM analysis |
|---|---|---|
| Motif1 | WKFRHIYRGQPRRHLLTTGWSVFVNQKKLVAGDSVVFLRNENGELRVGIR | B3 |
| Motif2 | SFCKTLTASDTNNGGGFSVPRRCAETIFPPLDYSQDPPVQELVAKDVHG | Auxin-resp |
| Motif3 | DPVRWPNSKWRMLQVGWDEPEALZRPKRVSPWZIEPVSAPP | - |
| Motif4 | LWHACAGPLVSIPPVGSKVYYFPQGHAEQ | - |
| Motif5 | GMRFKMAFETEESSRRRYFG | - |
| Motif6 | FEVVYYPRASPSEFVVPAKKV | - |
| Motif7 | PKILCRVLNVKLLADPETDEVYAKITLQP | - |
| Motif8 | WQVVYVDAEGDILLVGDDPWSEFVKTVRRIKILSPEEVQKM | AUX-IAA |
| Motif9 | VGRSLDLSKFSSYEELREELARMFGIEG | - |
| Motif10 | MPSSVISSHSMHIGVLAAAAHAVATNTM | - |
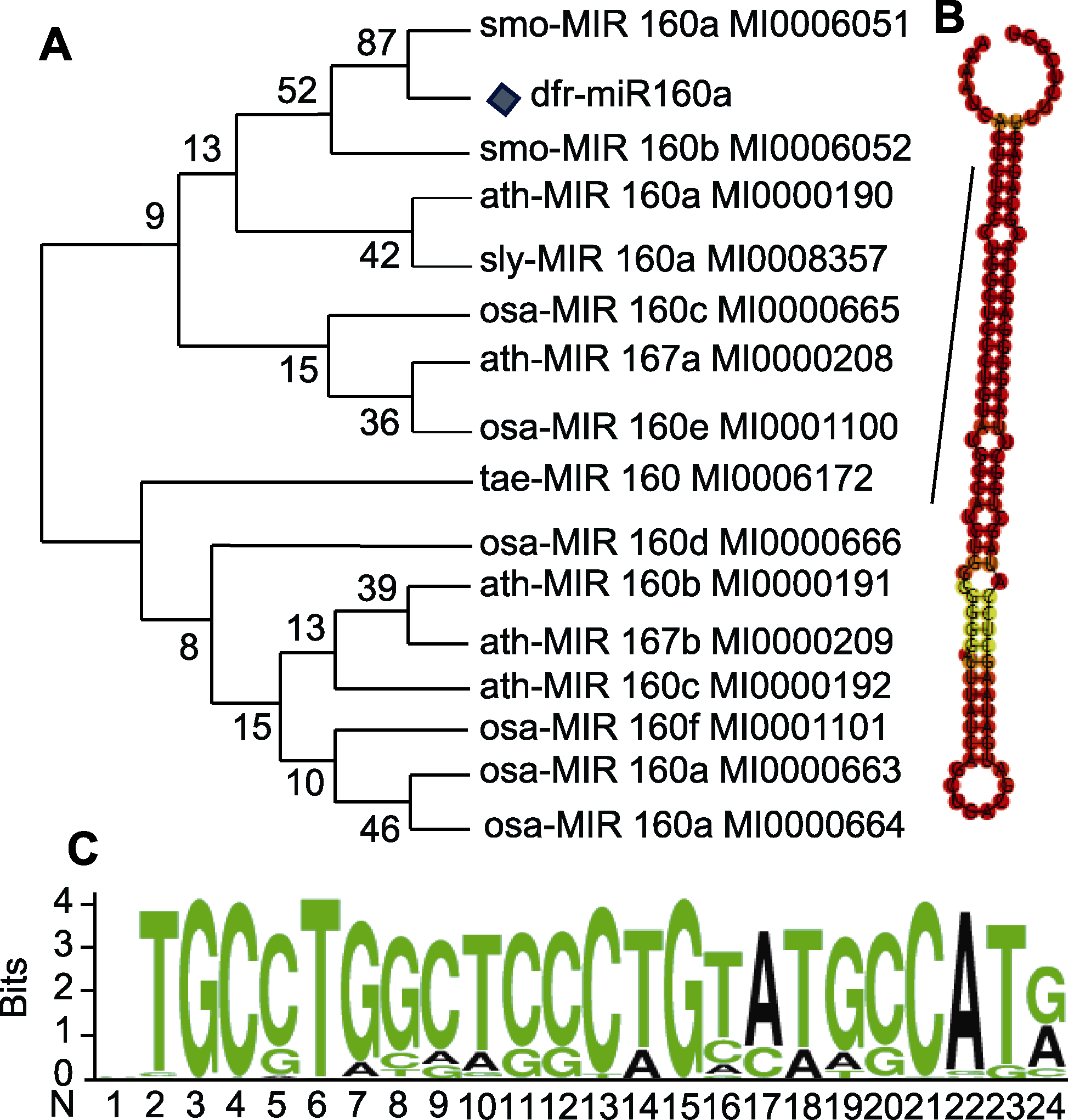
Figure 4 Conservative motif analysis of dfr-miR160a in Dryopteris fragrans (A) Phylogenetic tree of dfr-miR160a; (B) Stem-loop structure of dfr-miR160a (black line represents the mature miR160a sequence); (C) Conservative base analysis of dfr-miR160a mature sequence
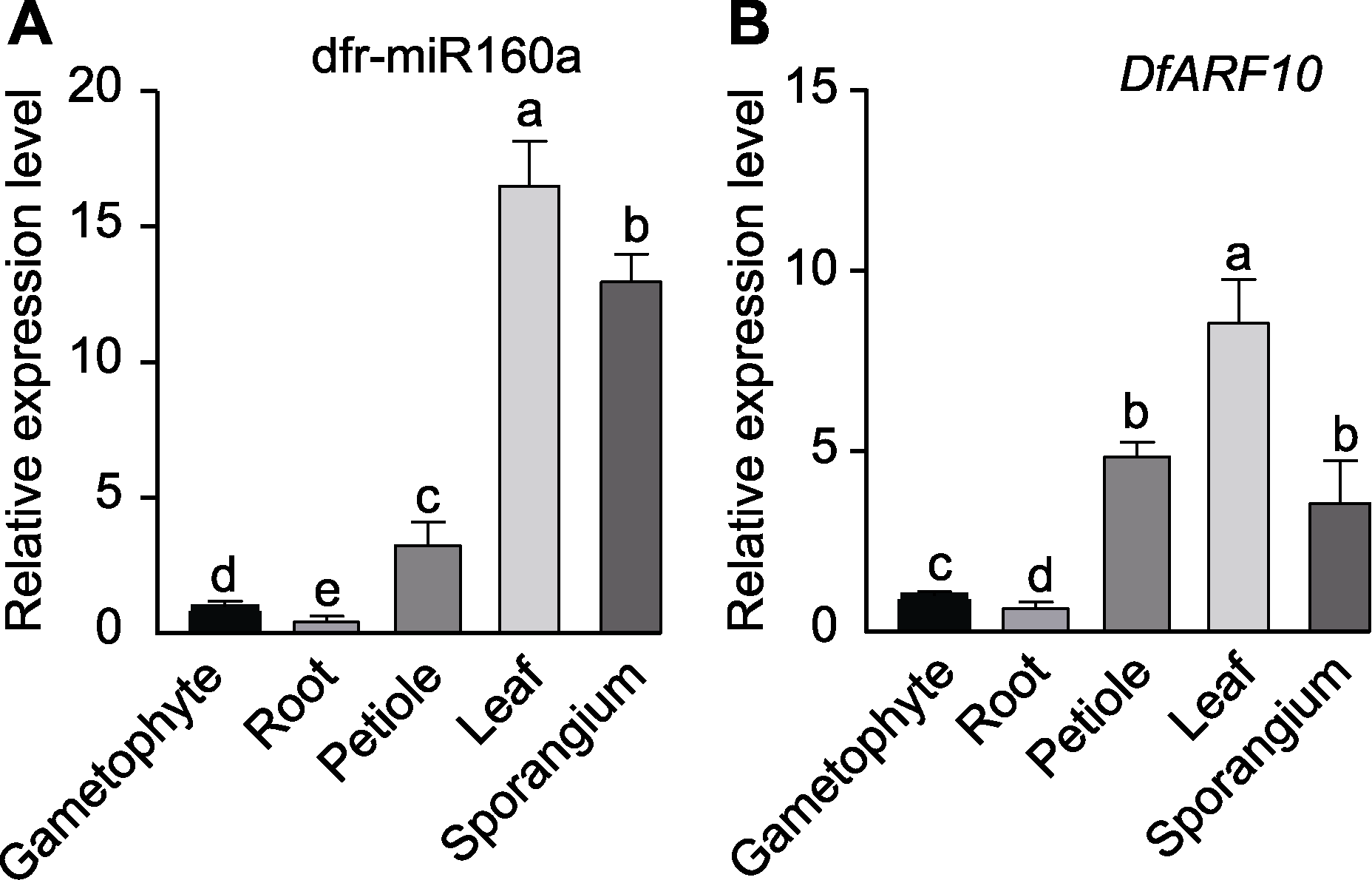
Figure 5 Relative expression of dfr-miR160a (A) and target gene DfARF10 (B) in different tissues of Dryopteris fragrans P value is calculated with One-way ANOVA, different lowercase letters indicate significant differences among different tissues (P<0.05).
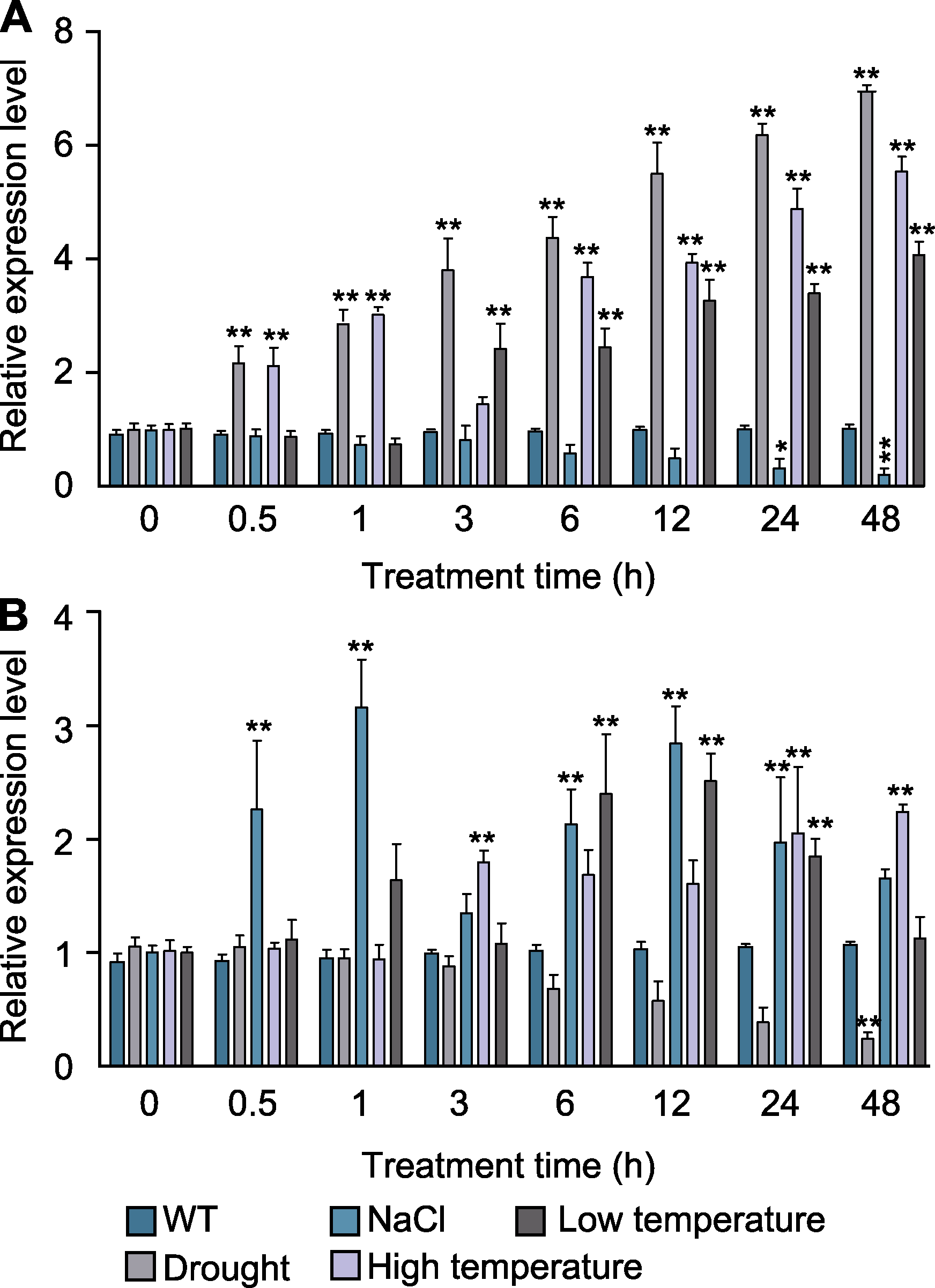
Figure 6 Effects of abiotic stress on the relative expression level of dfr-miR160a (A) and target gene DfARF10 (B) in the leaves of Dryopteris fragrans under different treatment times The expression level under four kinds of stresses were compared with control (WT) at different treatment times. P value (wild type as control) is calculated with One-way ANOVA; * P<0.05; ** P<0.01
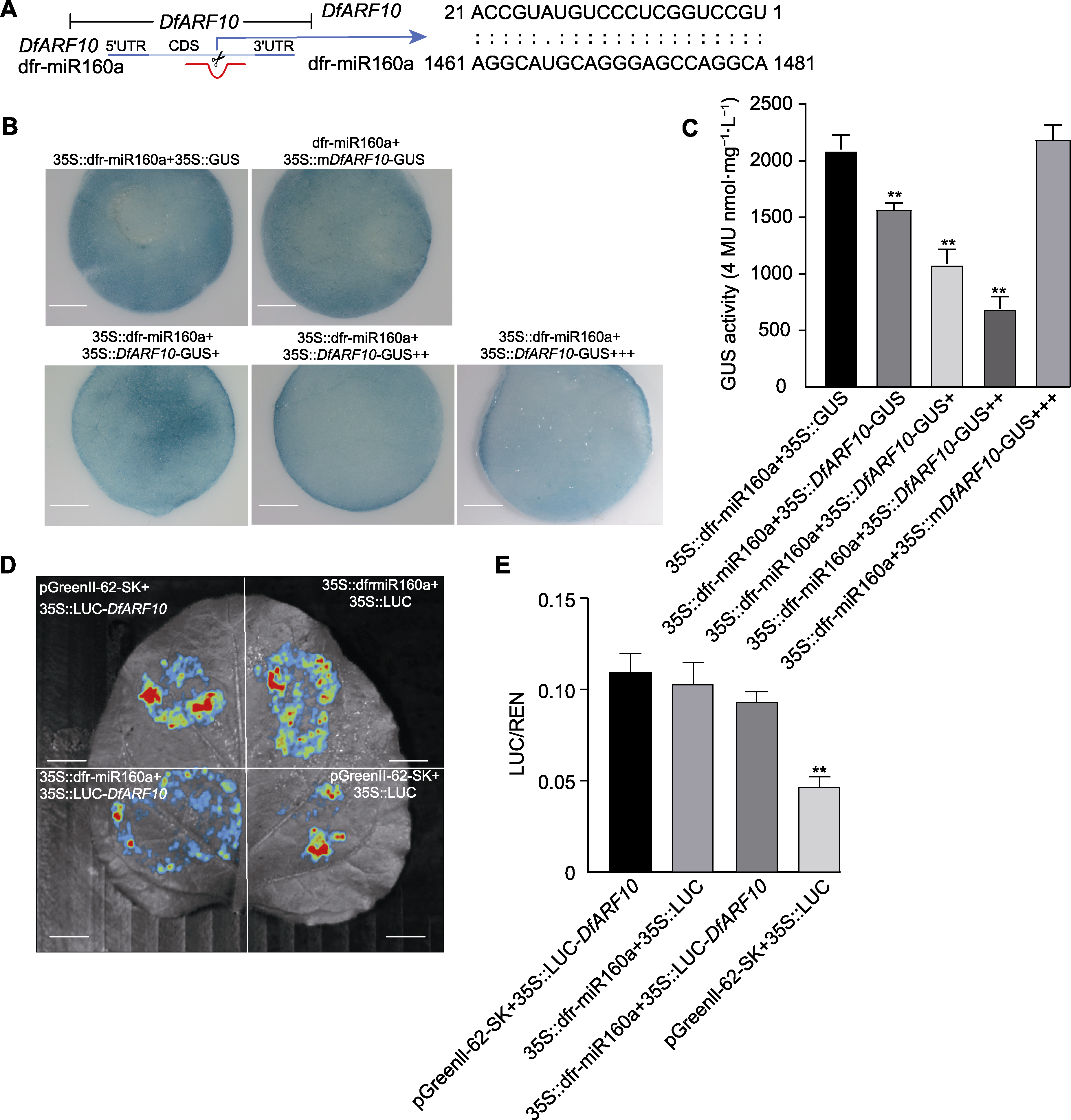
Figure 7 Agrobacterium mediated transient co-transformation of tobacco and double luciferase (LUC) activity to verify that dfr-miR160a targeted cleavage of DfARF10 (A) Cutting site of dfr-miR160a in DfARF10 gene structure; (B) β-glucuronidase (GUS) phenotype observed by histochemical staining analysis of 35S::dfr-pri-mir160a and 35S::DfARF10-GUS (+, ++, and +++ represent different concentrations of 35S::DfARF10-GUS, respectively (bars=5 mm)); (C) Co-expression of the constructs containing 35S::DfARF10-GUS and 35S::dfr-pri-mir160a in tobacco leaves (GUS activity were normalized to the expression levels of tobacco, +, ++, and +++ represent different concentrations of 35S::DfARF10-GUS, respectively); (D) The dual luciferase validation system visualizes the targeted degradation ability of dfr-miR160a through LUC activity (bars=3 cm); (E) Quantitative calculate intensity of fluorescence by LUC method.
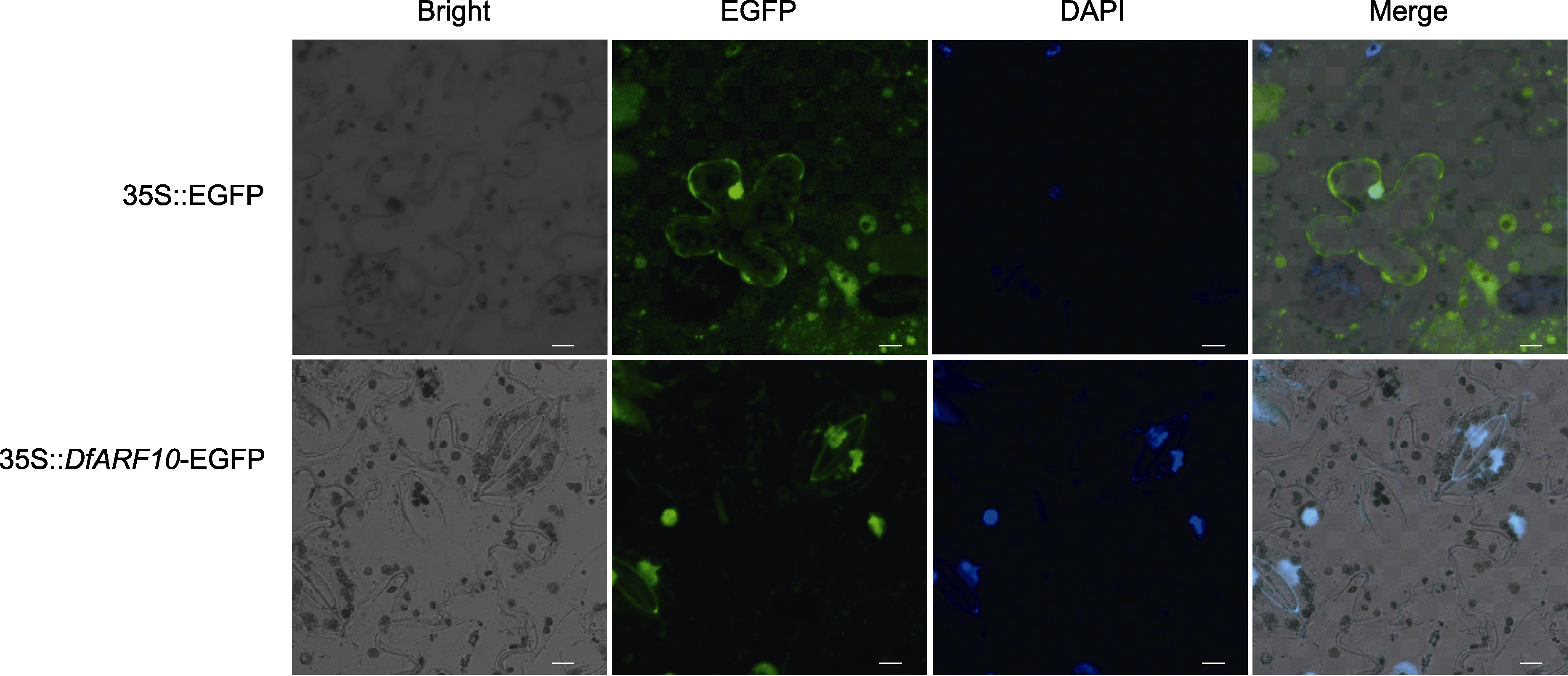
Figure 8 Subcellular localization of DfARF10 proteins of Dryopteris fragrans in tobacco leaf Blue fluorescence of DAPI indicate cell nucleus (bars=10 µm).
| [1] |
曹丽茹, 张前进, 郭子宁, 鲁晓民, 张新, 魏昕, 皇甫柏树, 王振华 (2021). 玉米生长素响应因子基因家族全基因组鉴定及表达分析. 核农学报 35, 2016-2026.
DOI |
| [2] | 陈丽 (2018). 甘蓝型油菜株型及角果长度相关miRNA和靶基因的挖掘. 博士论文. 武汉: 华中农业大学. pp. 1-74. |
| [3] | 陈文浩, 宋国强, 贾小舟, 唐春萍, 冯淡开, 沈志滨 (2017). 香鳞毛蕨中1对间苯三酚类同分异构体的分离与抗真菌活性研究. 中草药 48, 433-436. |
| [4] |
官亚琳, 汤珣, 张冬瑞, 夏德鑫, 李杰, 刘守银, 宋春华, 常缨 (2020). 香鳞毛蕨DfTCP的生物信息学及其表达模式分析. 华北农学报 35(5), 39-46.
DOI |
| [5] | 邱晓杰, 汤珣, 官亚琳, 张冬瑞, 苏颖, 常缨 (2021). 香鳞毛蕨DfGNOM基因的克隆及其表达分析. 西北植物学报 41, 1279-1286. |
| [6] | 苏佳萌, 袁强, 张冬瑞, 汤珣, 常缨 (2022). 香鳞毛蕨1-脱氧- D-木酮糖-5-磷酸合酶基因的克隆及表达分析. 西北植物学报 42, 1441-1449. |
| [7] | 孙仙泽, 王政委, 娄贵诚, 王多佳, 齐越, 于晶, 苍晶 (2022). 外源脱落酸和茉莉酸甲酯调控低温胁迫下冬小麦miR444a及其靶基因TaMADS57表达. 植物生理学报 58, 708-722. |
| [8] | 汤珣, 官亚琳, 陈玲玲, 夏德鑫, 宋春华, 刘丽艳, 常缨 (2020). 香鳞毛蕨DfDREB基因克隆与表达模式分析. 西北植物学报 40, 1105-1113. |
| [9] | 吴书昌 (2016). MiR160在棉花胚珠发育中功能分析. 硕士论文. 武汉: 华中农业大学. pp. 1-39. |
| [10] | 张冬瑞, 卜志刚, 陈玲玲, 常缨 (2020). 香鳞毛蕨的组织培养和快速繁殖体系构建. 植物学报 55, 760-767. |
| [11] | 朱冲冲, 彭冰, 曾祖平, 韩旭阳, 王宏, 王天园, 何薇 (2017). 香鳞毛蕨的化学成分及药理作用研究进展. 中国药房 28, 1418-1423. |
| [12] |
Bustos-Sanmamed P, Mao GH, Deng Y, Elouet M, Khan GA, Bazin JRM, Turner M, Subramanian S, Yu O, Crespi M, Lelandais-Brière C (2013). Overexpression of miR160 affects root growth and nitrogen-fixing nodule number in Medicago truncatula. Funct Plant Biol 40, 1208-1220.
DOI PMID |
| [13] |
Cui J, Li XY, Li JL, Wang CY, Cheng DY, Dai CH (2020). Genome-wide sequence identification and expression analysis of ARF family in sugar beet (Beta vulgaris L.) under salinity stresses. PeerJ 8, e9131.
DOI URL |
| [14] |
Damodharan S, Zhao DZ, Arazi T (2016). A common miRNA160-based mechanism regulates ovary patterning, floral organ abscission and lamina outgrowth in tomato. Plant J 86, 458-471.
DOI URL |
| [15] |
Ferdous J, Hussain SS, Shi BJ (2015). Role of microRNAs in plant drought tolerance. Plant Biotechnol J 13, 293-305.
DOI PMID |
| [16] |
Guo ZY, Hao K, Lv ZY, Yu LY, Bu QT, Ren JZ, Zhang HN, Chen RB, Zhang L (2023). Profiling of phytohormone- specific microRNAs and characterization of the miR160- ARF1 module involved in glandular trichome development and artemisinin biosynthesis in Artemisia annua. Plant Biotechnol J 21, 591-605.
DOI URL |
| [17] |
Gutierrez L, Bussell JD, Pacurar DI, Schwambach J, Pacurar M, Bellini C (2009). Phenotypic plasticity of adventitious rooting in Arabidopsis is controlled by complex regulation of AUXIN RESPONSE FACTOR transcripts and microRNA abundance. Plant Cell 21, 3119-3132.
DOI URL |
| [18] |
Hao K, Wang Y, Zhu ZP, Wu Y, Chen RB, Zhang L (2022). miR160: an indispensable regulator in plant. Front Plant Sci 13, 833322.
DOI URL |
| [19] |
Huang J, Zhao L, Malik S, Gentile BR, Xiong V, Arazi T, Owen HA, Friml J, Zhao DZ (2022). Specification of female germline by microRNA orchestrated auxin signaling in Arabidopsis. Nat Commun 13, 6960.
DOI PMID |
| [20] |
Kumar R, Dhanda SK (2020). Bird eye view of protein subcellular localization prediction. Life (Basel) 10, 347.
DOI URL |
| [21] |
Liu XD, Zhang H, Zhao Y, Feng ZY, Li Q, Yang HQ, Luan S, Li JM, He ZH (2013). Auxin controls seed dormancy through stimulation of abscisic acid signaling by inducing ARF-mediated ABI3 activation in Arabidopsis. Proc Natl Acad Sci USA 110, 15485-15490.
DOI URL |
| [22] |
Luo J, Zhou JJ, Zhang JZ (2018). Aux/IAA gene family in plants: molecular structure, regulation, and function. Int J Mol Sci 19, 259.
DOI URL |
| [23] |
Peng Y, Fang T, Zhang YY, Zhang MY, Zeng LH (2020). Genome-wide identification and expression analysis of auxin response factor (ARF) gene family in Longan (Dimocarpus longan L.). Plants (Basel) 9, 221.
DOI URL |
| [24] |
Shen XX, He JQ, Ping YK, Guo JX, Hou N, Cao FG, Li XW, Geng DL, Wang SC, Chen PX, Qin GG, Ma FW, Guan QM (2022). The positive feedback regulatory loop of miR160-auxin response factor 17-HYPONASTIC LEAVES 1 mediates drought tolerance in apple trees. Plant Physiol 188, 1686-1708.
DOI URL |
| [25] |
Song CH, Fan Q, Tang YQ, Sun YN, Wang L, Wei MC, Chang Y (2022a). Overexpression of DfRaf from fragrant woodfern (Dryopteris fragrans) enhances high-temperature tolerance in tobacco (Nicotiana tabacum). Genes (Basel) 13, 1212.
DOI URL |
| [26] |
Song CH, Guan YL, Zhang DR, Tang X, Chang Y (2022b). Integrated mRNA and miRNA transcriptome analysis suggests a regulatory network for UV-B-controlled terpenoid synthesis in fragrant woodfern (Dryopteris fragrans). Int J Mol Sci 23, 5708.
DOI URL |
| [27] |
Su LY, Xu M, Zhang JD, Wang YH, Lei YS, Li Q (2021). Genome-wide identification of auxin response factor (ARF) family in kiwifruit (Actinidia chinensis) and analysis of their inducible involvements in abiotic stresses. Physiol Mol Biol Plants 27, 1261-1276.
DOI |
| [28] |
Tang YY, Du GN, Xiang J, Hu CL, Li XT, Wang WH, Zhu H, Qiao LX, Zhao CM, Wang JS, Yu SL, Sui JM (2022). Genome-wide identification of auxin response factor (ARF) gene family and the miR160-ARF18-mediated response to salt stress in peanut (Arachis hypogaea L.). Genomics 114, 171-184.
DOI URL |
| [29] |
Wang M, Wu HJ, Fang J, Chu CC, Wang XJ (2017). A long noncoding RNA involved in rice reproductive development by negatively regulating osa-miR160. Sci Bull 62, 470-475.
DOI PMID |
| [30] |
Wang YJ, Deng DX, Shi YT, Miao N, Bian YL, Yin ZT (2012). Diversification, phylogeny and evolution of auxin response factor (ARF) family: insights gained from analyzing maize ARF genes. Mol Biol Rep 39, 2401-2415.
DOI URL |
| [31] |
Wójcik AM, Nodine MD, Gaj MD (2017). miR160 and miR166/165 contribute to the LEC2-mediated auxin response involved in the somatic embryogenesis induction in Arabidopsis. Front Plant Sci 8, 2024.
DOI PMID |
| [32] | Yang TX, Wang YY, Teotia S, Wang ZH, Shi CN, Sun HW, Gu YY, Zhang ZH, Tang GL (2019). The interaction between miR160 and miR165/166 in the control of leaf development and drought tolerance in Arabidopsis. Sci Rep 9, 2832. |
| [33] |
Zhang YQ, Zeng ZH, Chen CJ, Li CQ, Xia R, Li JG (2019). Genome-wide characterization of the auxin response factor (ARF) gene family of litchi (Litchi chinensis Sonn.): phylogenetic analysis, miRNA regulation and expression changes during fruit abscission. PeerJ 7, e6677.
DOI URL |
| [1] | Xiong Lianglin, Liang Guolu, Guo Qigao, Jing Danlong. Advances in the Regulation of Alternative Splicing of Genes in Plants in Response to Abiotic Stress [J]. Chinese Bulletin of Botany, 2025, 60(3): 435-448. |
| [2] | Qingguo Du, Wenxue Li. Research Progress in the Regulation of Development and Stress Responses by Long Non-coding RNAs in Maize [J]. Chinese Bulletin of Botany, 2024, 59(6): 950-962. |
| [3] | Wenjie Zhou, Wenhan Zhang, Wei Jia, Zicheng Xu, Wuxing Huang. Advances in Plant miRNAs Responses to Abiotic Stresses [J]. Chinese Bulletin of Botany, 2024, 59(5): 810-833. |
| [4] | Yuejing Zhang, Hetian Sang, Hanqi Wang, Zhenzhen Shi, Li Li, Xin Wang, Kun Sun, Ji Zhang, Hanqing Feng. Research Progress of Plant Signaling in Systemic Responses to Abiotic Stresses [J]. Chinese Bulletin of Botany, 2024, 59(1): 122-133. |
| [5] | Yanan Xu, Jiarong Yan, Xin Sun, Xiaomei Wang, Yufeng Liu, Zhouping Sun, Mingfang Qi, Tianlai Li, Feng Wang. Red and Far-red Light Regulation of Plant Growth, Development, and Abiotic Stress Responses [J]. Chinese Bulletin of Botany, 2023, 58(4): 622-637. |
| [6] | Jia Zhang, Qidong Li, Cui Li, Qinghai Wang, Xincun Hou, Chunqiao Zhao, Shuhe Li, Qiang Guo. Research Progress on MATE Transporters in Plants [J]. Chinese Bulletin of Botany, 2023, 58(3): 461-474. |
| [7] | Xiaotong Ren, Ranran Zhang, Shaowei Wei, Xiaofeng Luo, Jiahui Xu, Kai Shu. Research Progress of Spermosphere Microorganisms [J]. Chinese Bulletin of Botany, 2023, 58(3): 499-509. |
| [8] | WU Lin-Sheng, ZHANG Yong-Guang, ZHANG Zhao-Ying, ZHANG Xiao-Kang, WU Yun-Fei. Remote sensing of solar-induced chlorophyll fluorescence and its applications in terrestrial ecosystem monitoring [J]. Chin J Plant Ecol, 2022, 46(10): 1167-1199. |
| [9] | Lingling Xie, Jinlong Wang, Guoqiang Wu. Regulatory Mechanisms of the Plant CBL-CIPK Signaling System in Response to Abiotic Stress [J]. Chinese Bulletin of Botany, 2021, 56(5): 614-626. |
| [10] | Fei Zhao, Liuyi Dang, Minhui Wei, Chunying Liu, Wei Leng, Chenjing Shang. Expression of Amaranthin-like Lectins Gene and Responses to Abiotic Stresses in Cucumber [J]. Chinese Bulletin of Botany, 2021, 56(2): 183-190. |
| [11] | Dongrui Zhang, Zhigang Bu, Lingling Chen, Ying Chang. Establishment of a Tissue Culture and Rapid Propagation System of Dryopteris fragrans [J]. Chinese Bulletin of Botany, 2020, 55(6): 760-767. |
| [12] | Yingyan Xiao, Weina Yuan, Jing Liu, Jian Meng, Qiming Sheng, Yehuan Tan, Chunxiang Xu. Xyloglucan and the Advances in Its Roles in Plant Tolerance to Stresses [J]. Chinese Bulletin of Botany, 2020, 55(6): 777-787. |
| [13] | Lin Hong,Lei Yang,Haijian Yang,Wu Wang. Research Advances in AP2/ERF Transcription Factors in Regulating Plant Responses to Abiotic Stress [J]. Chinese Bulletin of Botany, 2020, 55(4): 481-496. |
| [14] | Menglong Wang,Xiaoqun Peng,Zhufeng Chen,Xiaoyan Tang. Research Advances on Lectin Receptor-like Kinases in Plants [J]. Chinese Bulletin of Botany, 2020, 55(1): 96-105. |
| [15] | Xun Zhang,Juanjuan Yu,Sizhu Wang,Ying Li,Shaojun Dai. Research Advances in DREPP Gene Family in Plants [J]. Chinese Bulletin of Botany, 2019, 54(5): 582-595. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||