

Chinese Bulletin of Botany ›› 2021, Vol. 56 ›› Issue (2): 183-190.DOI: 10.11983/CBB20177 cstr: 32102.14.CBB20177
• EXPERIMENTAL COMMUNICATIONS • Previous Articles Next Articles
Fei Zhao1, Liuyi Dang2, Minhui Wei1, Chunying Liu1, Wei Leng1,*( ), Chenjing Shang1,3,*(
), Chenjing Shang1,3,*( )
)
Received:2020-11-01
Accepted:2021-03-01
Online:2021-03-01
Published:2021-03-17
Contact:
Wei Leng,Chenjing Shang
Fei Zhao, Liuyi Dang, Minhui Wei, Chunying Liu, Wei Leng, Chenjing Shang. Expression of Amaranthin-like Lectins Gene and Responses to Abiotic Stresses in Cucumber[J]. Chinese Bulletin of Botany, 2021, 56(2): 183-190.
| Primer name | Forward primer (5'-3') | Reverse primer (5'-3') |
|---|---|---|
| CACS | TGGGAAGATTCTTA- TGAAGTGC | CTCGTCAAATTT- ACACATTGGT |
| PP2A | CAACAGGTGATATT- GGATTATGAT | GCCAGCTCATCC- TCATATAAG |
| AAT4 | TTCGTACACGCAAC- GAGA | TGAAGAGGGTAA- GGCTTG |
| NAAT1 | GAGGAGCTGTGAAA- GGAGCA | CCCTCCACGACA- GTTCCAAT |
| AAT9 | CAGAAACAGCGAAC- CAGAGC | AACTTCATCCCCA- CCGAGTT |
| AAT14 | GGGAATAGAGACGA- TCCGAACT | GCGCAGAAGGCA- GTGTTT |
Table 1 Primer list for qRT-PCR
| Primer name | Forward primer (5'-3') | Reverse primer (5'-3') |
|---|---|---|
| CACS | TGGGAAGATTCTTA- TGAAGTGC | CTCGTCAAATTT- ACACATTGGT |
| PP2A | CAACAGGTGATATT- GGATTATGAT | GCCAGCTCATCC- TCATATAAG |
| AAT4 | TTCGTACACGCAAC- GAGA | TGAAGAGGGTAA- GGCTTG |
| NAAT1 | GAGGAGCTGTGAAA- GGAGCA | CCCTCCACGACA- GTTCCAAT |
| AAT9 | CAGAAACAGCGAAC- CAGAGC | AACTTCATCCCCA- CCGAGTT |
| AAT14 | GGGAATAGAGACGA- TCCGAACT | GCGCAGAAGGCA- GTGTTT |
| Gene | Start position | End position | Orientation | Gene | Start position | End position | Orientation |
|---|---|---|---|---|---|---|---|
| CucsaAAT1 | 5496646 | 5495195 | Reverse | CucsaAAT9 | 5654159 | 5656355 | Forward |
| CucsaAAT2 | 5583941 | 5582553 | Reverse | CucsaAAT10 | 5681664 | 5683484 | Forward |
| CucsaAAT3 | 5590531 | 5589140 | Reverse | CucsaAAT11 | 5691672 | 5690257 | Reverse |
| CucsaAAT4 | 5596447 | 5595167 | Reverse | CucsaAAT12 | 7077986 | 7075008 | Reverse |
| CucsaAAT5 | 5605793 | 5607184 | Forward | CucsaAAT13 | 7083801 | 7085207 | Forward |
| CucsaAAT6 | 5619299 | 5621364 | Forward | CucsaAAT14 | 7090825 | 7094963 | Forward |
| CucsaAAT7 | 5630016 | 5631894 | Forward | CucsaNAAT1 | 5648115 | 5646235 | Reverse |
| CucsaAAT8 | 5636899 | 5638943 | Forward | CucsaNAAT2 | 5670107 | 5668263 | Reverse |
Table 2 Amaranthin-like genes in cucumber and their locations on chromosome 6
| Gene | Start position | End position | Orientation | Gene | Start position | End position | Orientation |
|---|---|---|---|---|---|---|---|
| CucsaAAT1 | 5496646 | 5495195 | Reverse | CucsaAAT9 | 5654159 | 5656355 | Forward |
| CucsaAAT2 | 5583941 | 5582553 | Reverse | CucsaAAT10 | 5681664 | 5683484 | Forward |
| CucsaAAT3 | 5590531 | 5589140 | Reverse | CucsaAAT11 | 5691672 | 5690257 | Reverse |
| CucsaAAT4 | 5596447 | 5595167 | Reverse | CucsaAAT12 | 7077986 | 7075008 | Reverse |
| CucsaAAT5 | 5605793 | 5607184 | Forward | CucsaAAT13 | 7083801 | 7085207 | Forward |
| CucsaAAT6 | 5619299 | 5621364 | Forward | CucsaAAT14 | 7090825 | 7094963 | Forward |
| CucsaAAT7 | 5630016 | 5631894 | Forward | CucsaNAAT1 | 5648115 | 5646235 | Reverse |
| CucsaAAT8 | 5636899 | 5638943 | Forward | CucsaNAAT2 | 5670107 | 5668263 | Reverse |
| Protein | Domain architecture | No. of amino acids | Protein size (kDa) | Theoretical pI |
|---|---|---|---|---|
| CucsaAAT1 | AAT | 484 | 54.9 | 8.3 |
| CucsaAAT2 | AAT | 463 | 53.6 | 7.9 |
| CucsaAAT3 | AAT | 463 | 53.5 | 8.0 |
| CucsaAAT4 | AAT | 466 | 53.7 | 8.2 |
| CucsaAAT5 | AAT | 463 | 53.3 | 7.5 |
| CucsaAAT6 | AAT | 464 | 53.4 | 7.5 |
| CucsaAAT7 | AAT | 471 | 53.5 | 5.1 |
| CucsaAAT8 | AAT | 475 | 54.7 | 8.3 |
| CucsaAAT9 | AAT | 474 | 55.0 | 6.7 |
| CucsaAAT10 | AAT | 476 | 54.9 | 5.7 |
| CucsaAAT11 | AAT | 482 | 55.4 | 7.0 |
| CucsaAAT12 | AAT | 502 | 57.6 | 9.0 |
| CucsaAAT13 | AAT | 468 | 53.9 | 7.7 |
| CucsaAAT14 | AAT | 505 | 58.2 | 6.8 |
| CucsaNAAT1 | NAAT | 626 | 70.4 | 5.1 |
| CucsaNAAT2 | NAAT | 614 | 69.4 | 4.9 |
Table 3 Physiochemical properties of amaranthin-like proteins in cucumber
| Protein | Domain architecture | No. of amino acids | Protein size (kDa) | Theoretical pI |
|---|---|---|---|---|
| CucsaAAT1 | AAT | 484 | 54.9 | 8.3 |
| CucsaAAT2 | AAT | 463 | 53.6 | 7.9 |
| CucsaAAT3 | AAT | 463 | 53.5 | 8.0 |
| CucsaAAT4 | AAT | 466 | 53.7 | 8.2 |
| CucsaAAT5 | AAT | 463 | 53.3 | 7.5 |
| CucsaAAT6 | AAT | 464 | 53.4 | 7.5 |
| CucsaAAT7 | AAT | 471 | 53.5 | 5.1 |
| CucsaAAT8 | AAT | 475 | 54.7 | 8.3 |
| CucsaAAT9 | AAT | 474 | 55.0 | 6.7 |
| CucsaAAT10 | AAT | 476 | 54.9 | 5.7 |
| CucsaAAT11 | AAT | 482 | 55.4 | 7.0 |
| CucsaAAT12 | AAT | 502 | 57.6 | 9.0 |
| CucsaAAT13 | AAT | 468 | 53.9 | 7.7 |
| CucsaAAT14 | AAT | 505 | 58.2 | 6.8 |
| CucsaNAAT1 | NAAT | 626 | 70.4 | 5.1 |
| CucsaNAAT2 | NAAT | 614 | 69.4 | 4.9 |
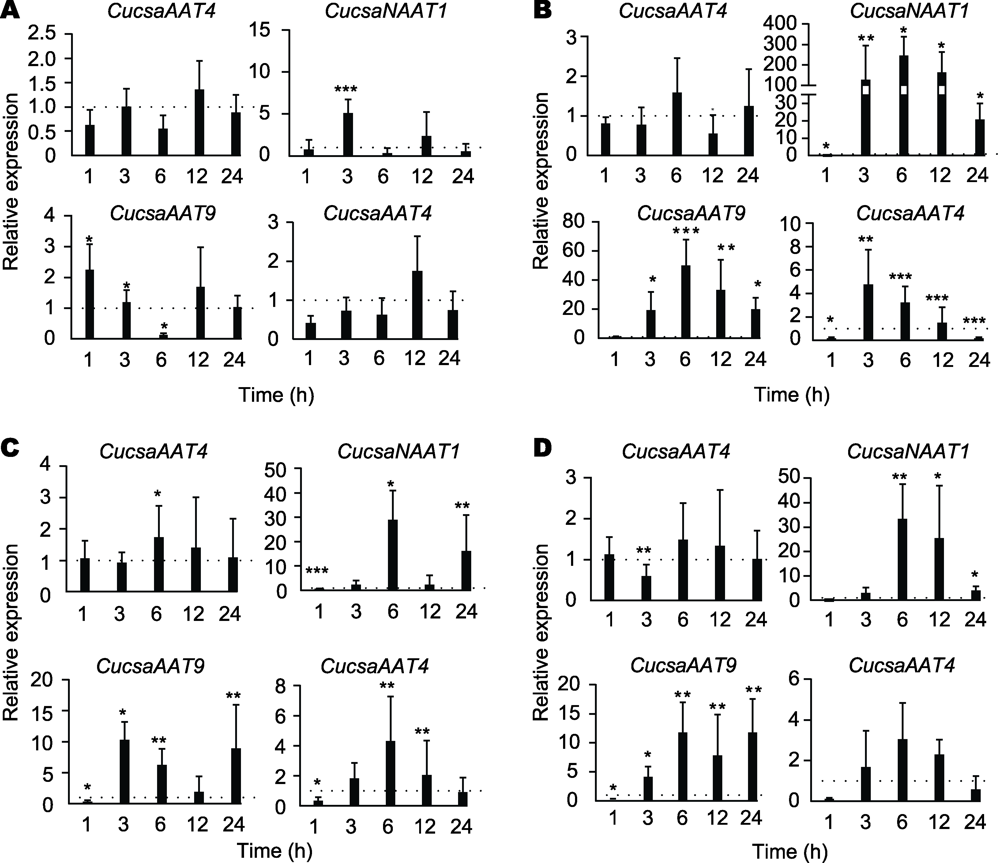
Figure 4 Relative expression levels for amaranthin-like genes in cucumber plants subjected to different abiotic stress conditions (A)Cold treatment; (B) Salt treatment; (C) Drought treatment;(D) ABA treatment. Bars represent means and standard errors from two independent biological replicates, each replicate containing a pool of 4 plants for stress-treated groups as well as control groups. Asterisks indicate statistically signi?cant differences compared to the plants with mock treatments (* P<0.05,** P<0.01, ***P<0.001).
| [1] | 王梦龙, 彭小群, 陈竹锋, 唐晓艳 (2020). 植物凝集素类受体蛋白激酶研究进展. 植物学报 55,96-105. |
| [2] | 王志斌, 张秀梅, 郭三堆 (2000). 在转基因植物中利用植物凝集素防治害虫的研究. 植物学通报 17,108-113. |
| [3] | 曾日中, 黎瑜 (1998). 橡胶蛋白—一种与胶乳凝固有关的具有抗真菌活性的植物凝集素. 植物学通报 15(增刊),24-28. |
| [4] | Al Atalah B, Fouquaert E, Van Damme EJM (2013). Promoter analysis for three types of EUL-related rice lectins in transgenic Arabidopsis. Plant Mol Biol Rep 31, 1315- 1324. |
| [5] | Chen SC, Jin WJ, Liu AR, Zhang SJ, Liu DL, Wang FH, Lin XM, He CX (2013). Arbuscular mycorrhizal fungi (AMF) increase growth and secondary metabolism in cucumber subjected to low temperature stress. Sci Hortic 160,222-229. |
| [6] | Dang LY, Rougé P, Van Damme EJM (2017). Amaranthin-like proteins with aerolysin domains in plants. Front Plant Sci 8,1368. |
| [7] | Dang LY, Van Damme EJM (2016). Genome-wide identification and domain organization of lectin domains in cucumber. Plant Physiol Biochem 108,165-176. |
| [8] | Faruque K, Begam R, Deyholos MK (2015). The amaranthin-like lectin (LuALL) genes of flax: a unique gene family with members inducible by defence hormones. Plant Mol Biol Rep 33,731-741. |
| [9] | Ghazarian H, Idoni B, Oppenheimer SB (2011). A glycobiology review: carbohydrates, lectins and implications in cancer therapeutics. Acta Histochem 113,236-247. |
| [10] | Higo K, Ugawa Y, Iwamoto M, Korenaga T (1999). Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res 27,297-300. |
| [11] | Huang SW, Li RQ, Zhang ZH, Li L, Gu XF, Fan W, Lucas WJ, Wang XW, Xie BY, Ni PX, Ren YY, Zhu HM, Li J, Lin K, Jin WW, Fei ZJ, Li GC, Staub J, Kilian A, van der Vossen EAG, Wu Y, Guo J, He J, Jia ZQ, Ren Y, Tian G, Lu Y, Ruan J, Qian WB, Wang MW, Huang QF, Li B, Xuan ZL, Cao JJ, Asan, Wu ZG, Zhang JB, Cai QL, Bai YQ, Zhao BW, Han YH, Li Y, Li XF, Wang SH, Shi QX, Liu SQ, Cho WK, Kim JY, Xu Y, Heller-Uszynska K, Miao H, Cheng ZC, Zhang SP, Wu J, Yang YH, Kang HX, Li M, Liang HQ, Ren XL, Shi ZB, Wen M, Jian M, Yang HL, Zhang GJ, Yang ZT, Chen R, Liu SF, Li JW, Ma LJ, Liu H, Zhou Y, Zhao J, Fang XD, Li GQ, Fang L, Li YR, Liu DY, Zheng HK, Zhang Y, Qin N, Li Z, Yang GH, Yang S, Bolund L, Kristiansen K, Zheng HC, Li SC, Zhang XQ, Yang HM, Wang J, Sun RF, Zhang BX, Jiang SZ, Wang J, Du YC, Li SG (2009). The genome of the cucumber, Cucumis sativus L. Nat Genet 41, 1275- 1281. |
| [12] | Li YM, Li SH, He XR, Jiang WL, Zhang DL, Liu BB, Li QM (2020). CO2 enrichment enhanced drought resistance by regulating growth, hydraulic conductivity and phytohormone contents in the root of cucumber seedlings. Plant Physiol Biochem 152,62-71. |
| [13] | Migocka M, Papierniak A (2011). Identification of suitable reference genes for studying gene expression in cucumber plants subjected to abiotic stress and growth regulators. Mol Breed 28,343-357. |
| [14] | Pfaffl MW, Horgan GW, Dempfle L (2002). Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res 30,e36. |
| [15] | Shang CJ, Dang LY, Van Damme EJM (2017). Plant AB toxins with lectin domains. In: Gopalakrishnakone P, Carlini RC, Ligabue-Braun R, eds. Plant Toxins. Dordrecht: Springer. pp.1-14. |
| [16] | Szczesny P, Iacovache I, Muszewska A, Ginalski K, van der Goot FG, Grynberg M (2011). Extending the aerolysin family: from bacteria to vertebrates. PLoS One 6,e20349. |
| [17] | Tsaneva M, Van Damme EJM (2020). 130 years of plant lectin research. Glycoconj J 37,533-551. |
| [18] | Tuteja N (2007). Abscisic acid and abiotic stress signaling. Plant Signal Behav 2,135-138. |
| [19] | Van Holle S, Van Damme EJM (2019). Messages from the past: new insights in plant lectin evolution. Front Plant Sci 10,36. |
| [20] | Wang J, Pan CT, Wang Y, Ye L, Wu J, Chen LF, Zou T, Lu G (2015). Genome-wide identification of MAPK, MAPKK, and MAPKKK gene families and transcriptional profiling analysis during development and stress response in cucumber. BMC Genomics 16,386. |
| [21] | Yan SS, Che G, Ding L, Chen ZJ, Liu XF, Wang HY, Zhao WS, Ning K, Zhao JY, Tesfamichael K, Wang Q, Zhang XL (2016). Different cucumber CsYUC genes regulate response to abiotic stresses and flower development. Sci Rep 6,20760. |
| [22] | Zhu YX, Jia JH, Yang L, Xia YC, Zhang HL, Jia JB, Zhou R, Nie PY, Yin JL, Ma DF, Liu LC (2019). Identification of cucumber circular RNAs responsive to salt stress. BMC Plant Biol 19,164. |
| [1] | Xiong Lianglin, Liang Guolu, Guo Qigao, Jing Danlong. Advances in the Regulation of Alternative Splicing of Genes in Plants in Response to Abiotic Stress [J]. Chinese Bulletin of Botany, 2025, 60(3): 435-448. |
| [2] | Qingguo Du, Wenxue Li. Research Progress in the Regulation of Development and Stress Responses by Long Non-coding RNAs in Maize [J]. Chinese Bulletin of Botany, 2024, 59(6): 950-962. |
| [3] | Xiang Song, Luyao Wang, Boxiao Fu, Shuangda Li, Yuanyuan Wei, Yan Hong, Silan Dai. Advances in Identification and Synthesis of Promoter Elements in Higher Plants [J]. Chinese Bulletin of Botany, 2024, 59(5): 691-708. |
| [4] | Wenjie Zhou, Wenhan Zhang, Wei Jia, Zicheng Xu, Wuxing Huang. Advances in Plant miRNAs Responses to Abiotic Stresses [J]. Chinese Bulletin of Botany, 2024, 59(5): 810-833. |
| [5] | Yuejing Zhang, Hetian Sang, Hanqi Wang, Zhenzhen Shi, Li Li, Xin Wang, Kun Sun, Ji Zhang, Hanqing Feng. Research Progress of Plant Signaling in Systemic Responses to Abiotic Stresses [J]. Chinese Bulletin of Botany, 2024, 59(1): 122-133. |
| [6] | Zhaoxuan Zhong, Dongrui Zhang, Lu Li, Ying Su, Daining Wang, Zeran Wang, Yang Liu, Ying Chang. Bioinformatic and Expression Pattern Analysis of dfr-miR160a and Target Gene DfARF10 in Dryopteris fragrans [J]. Chinese Bulletin of Botany, 2024, 59(1): 22-33. |
| [7] | Yanan Xu, Jiarong Yan, Xin Sun, Xiaomei Wang, Yufeng Liu, Zhouping Sun, Mingfang Qi, Tianlai Li, Feng Wang. Red and Far-red Light Regulation of Plant Growth, Development, and Abiotic Stress Responses [J]. Chinese Bulletin of Botany, 2023, 58(4): 622-637. |
| [8] | Jia Zhang, Qidong Li, Cui Li, Qinghai Wang, Xincun Hou, Chunqiao Zhao, Shuhe Li, Qiang Guo. Research Progress on MATE Transporters in Plants [J]. Chinese Bulletin of Botany, 2023, 58(3): 461-474. |
| [9] | Xiaotong Ren, Ranran Zhang, Shaowei Wei, Xiaofeng Luo, Jiahui Xu, Kai Shu. Research Progress of Spermosphere Microorganisms [J]. Chinese Bulletin of Botany, 2023, 58(3): 499-509. |
| [10] | WU Lin-Sheng, ZHANG Yong-Guang, ZHANG Zhao-Ying, ZHANG Xiao-Kang, WU Yun-Fei. Remote sensing of solar-induced chlorophyll fluorescence and its applications in terrestrial ecosystem monitoring [J]. Chin J Plant Ecol, 2022, 46(10): 1167-1199. |
| [11] | Lingling Xie, Jinlong Wang, Guoqiang Wu. Regulatory Mechanisms of the Plant CBL-CIPK Signaling System in Response to Abiotic Stress [J]. Chinese Bulletin of Botany, 2021, 56(5): 614-626. |
| [12] | Yingyan Xiao, Weina Yuan, Jing Liu, Jian Meng, Qiming Sheng, Yehuan Tan, Chunxiang Xu. Xyloglucan and the Advances in Its Roles in Plant Tolerance to Stresses [J]. Chinese Bulletin of Botany, 2020, 55(6): 777-787. |
| [13] | Lin Hong,Lei Yang,Haijian Yang,Wu Wang. Research Advances in AP2/ERF Transcription Factors in Regulating Plant Responses to Abiotic Stress [J]. Chinese Bulletin of Botany, 2020, 55(4): 481-496. |
| [14] | Huijin Fan, Kangming Jin, Renying Zhuo, Guirong Qiao. Cloning and Expression Analysis of Different Truncated U3 Promoters in Phyllostachys edulis [J]. Chinese Bulletin of Botany, 2020, 55(3): 299-307. |
| [15] | Menglong Wang,Xiaoqun Peng,Zhufeng Chen,Xiaoyan Tang. Research Advances on Lectin Receptor-like Kinases in Plants [J]. Chinese Bulletin of Botany, 2020, 55(1): 96-105. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||