

植物学报 ›› 2018, Vol. 53 ›› Issue (4): 456-467.DOI: 10.11983/CBB17226 cstr: 32102.14.CBB17226
收稿日期:2017-11-28
接受日期:2018-02-09
出版日期:2018-07-01
发布日期:2018-09-11
通讯作者:
王雷
作者简介:† 共同第一作者。
基金资助:
Wei Hua1,2, Wang Yan1,2, Liu Baohui3, Wang Lei1,2,*( )
)
Received:2017-11-28
Accepted:2018-02-09
Online:2018-07-01
Published:2018-09-11
Contact:
Wang Lei
About author:† These authors contributed equally to this paper
摘要: 作为植物细胞内部的授时机制, 生物钟系统主要包括信号输入、核心振荡器和信号输出3个主要部分。该系统通过感受外界光照和温度等环境因子的昼夜周期性变化动态, 协调植物生长发育、代谢与生理反应, 赋予植物对生存环境的适应性。植物生物钟系统的核心振荡器通过多层级调控复杂的下游信号转导网络来参与调节植物生长发育及对生物与非生物胁迫的适应性。该文概述了近年来生物钟核心振荡器及其调控植物生长发育过程诸方面的研究进展, 并初步提出了植物时间生物学研究领域一些亟待解决的科学问题, 以期为生物钟领域的研究成果在作物分子育种方面的利用提供理论借鉴。
魏华, 王岩, 刘宝辉, 王雷. 植物生物钟及其调控生长发育的研究进展. 植物学报, 2018, 53(4): 456-467.
Wei Hua, Wang Yan, Liu Baohui, Wang Lei. Deciphering the Underlying Mechanism of the Plant Circadian System and Its Regulation on Plant Growth and Development. Chinese Bulletin of Botany, 2018, 53(4): 456-467.
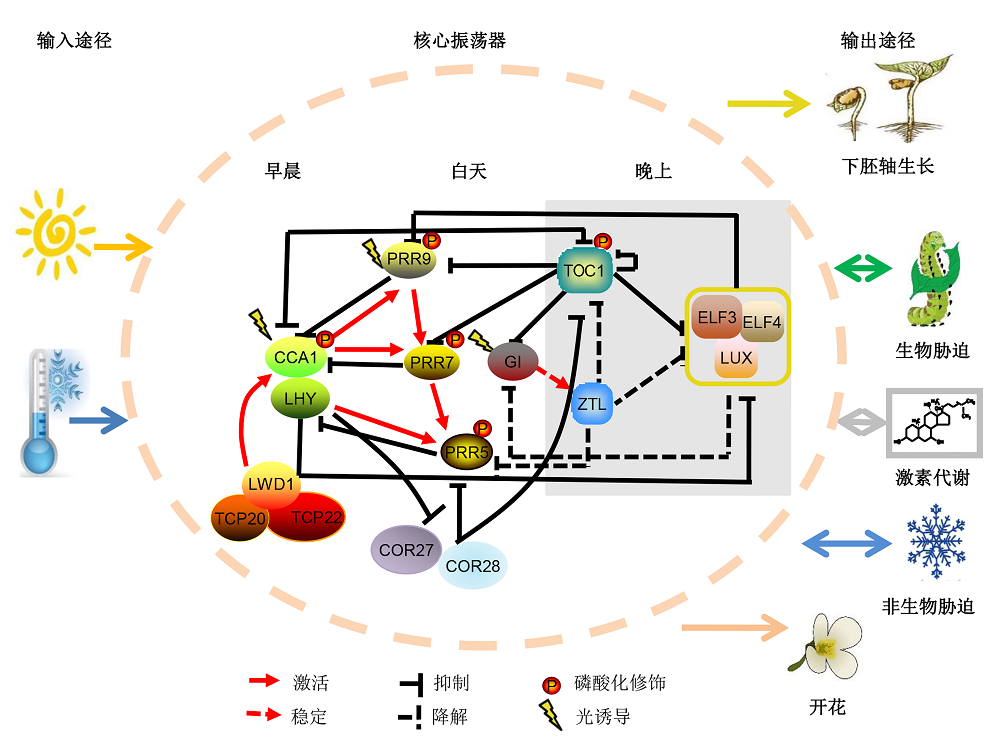
图1 生物钟调控拟南芥生长发育模型拟南芥生物钟系统主要包括输入途径、核心振荡器和输出途径三大部分。其中核心振荡器由多个相互联锁的转录翻译反馈环组成, 是生物钟系统的重要组成部分, 包括早循环、中心循环和晚循环, 它能够整合外界环境信号协调多种生理进程(虚线椭圆内部分)。核心振荡器接收传入的环境时间信号, 在细胞中产生内源性的昼夜节律, 并将时间信息传达到输出途径, 从而控制众多生命活动, 下胚轴伸长、病原体防御、生物和非生物胁迫响应、激素代谢和光周期调控的开花等。
Figure 1 A proposed model of circadian clock regulated growth and development in ArabidopsisIn higher plants, circadian clock is composed of three major parts, including input pathways, core oscillator and output pathways. The core oscillator is formed by interlocked feedback loops, such as morning loop, central loop and evening loop, indicated by the dotted oval in the figure. Circadian core oscillator regulates multiple outputs, such as hypocotyl elongation, pathogen defense, biotic and abiotic stress adaption, plant hormone signaling pathways and flowering time.
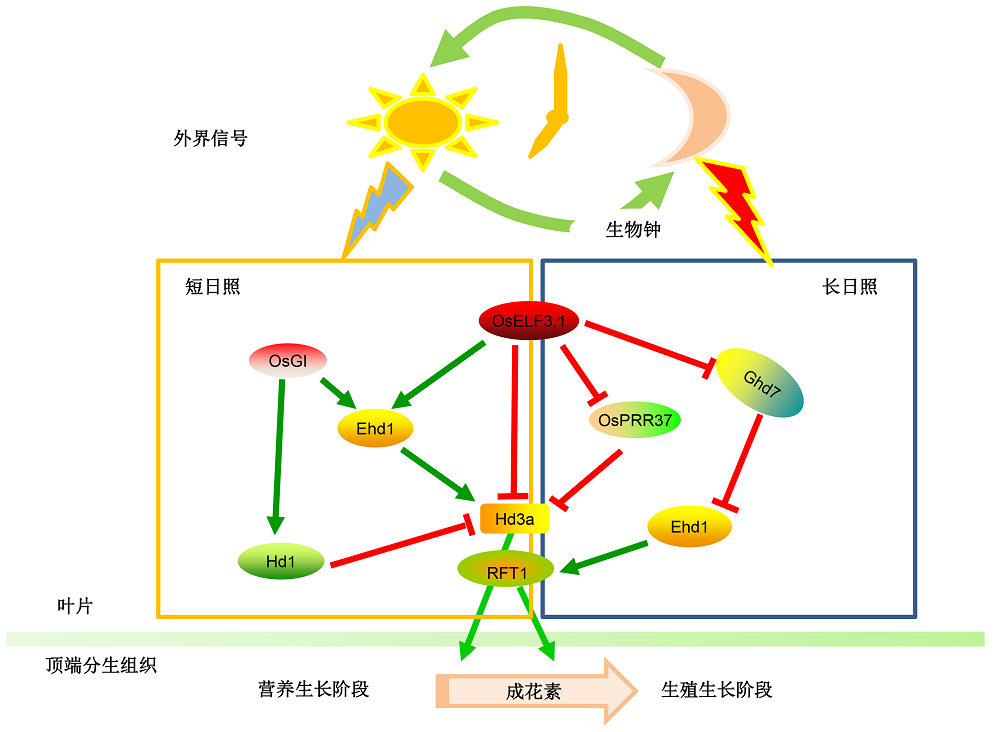
图2 生物钟调控水稻抽穗期模式水稻中OsELF3.1维持生物钟周期并参与调控水稻抽穗期。在长日照条件下, OsELF3.1是开花负调节因子, 作用于OsPRR73和Ghd7的上游, 通过抑制Hd3a和Ehd1调控开花; 在短日照条件下, OsELF3.1是开花正调节因子, 通过感受蓝光信号激活Ehd1的表达进而促进开花。OsPRR37和OsGI也参与维持生物钟周期, 具体调控对光周期的敏感性, 控制水稻抽穗期。
Figure 2 The proposed model for circadian clock regulating heading date in riceIn rice, OsELF3.1 is a key factor to maintain circadian rhythm and regulate heading date. In the long day condition, OsELF3.1 acts as a negative factor by repressing Hd3a and Ehd1 to regulate heading date, genetically works at the upstream of OsPRR73 and Ghd7, while in short day condition, OsELF3.1 can work as a positive regulator by sensing the blue light signaling to promote Ehd1 expression. OsPRR37 and OsGI are other essential components of rice circadian clock to regulate heading date in rice through photoperiod dependent pathway.
| 1 | 徐江民, 姜洪真, 林晗, 黄苗苗, 符巧丽, 曾大力, 饶玉春 (2016). 水稻ES1参与生物钟基因表达调控以及逆境胁迫响应. 植物学报 51, 743-756. |
| 2 | Adams S, Manfield I, Stockley P, Carré IA (2015). Revised morning loops of the Arabidopsis circadian clock based on analyses of direct regulatory interactions.PLoS One 10, e0143943. |
| 3 | Alabadı? D, Oyama T, Yanovsky MJ, Harmon FG, Más P, Kay SA (2001). Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 293, 880-883. |
| 4 | Atamian HS, Harmer SL (2016). Circadian regulation of hormone signaling and plant physiology.Plant Mol Biol 91, 691-702. |
| 5 | Bhardwaj V, Meier S, Petersen LN, Ingle RA, Roden LC (2011). Defence responses of Arabidopsis thaliana to infection by Pseudomonas syringae are regulated by the circadian clock. PLoS One 6, e26968. |
| 6 | Boden SA, Weiss D, Ross JJ, Davies NW, Trevaskis B, Chandler PM, Swain SM (2014). EARLY FLOWERING 3 regulates flowering in spring barley by mediating gibberellin production and FLOWERING LOCUS T expression. Plant Cell 26, 1557-1569. |
| 7 | Cha JY, Kim J, Kim TS, Zeng QN, Wang L, Lee SY, Kim WY, Somers DE (2017). GIGANTEA is a co-chaperone which facilitates maturation of ZEITLUPE in the Arabidopsis circadian clock.Nat Commun 8, 3. |
| 8 | Chew YH, Halliday KJ (2011). A stress-free walk from Arabidopsis to crops.Curr Opin Biotechnol 22, 281-286. |
| 9 | Chow BY, Sanchez SE, Breton G, Pruneda-Paz JL, Krogan NT, Kay SA (2014). Transcriptional regulation of LUX by CBF1 mediates cold input to the circadian clock in Ara- bidopsis. Curr Biol 24, 1518-1524. |
| 10 | Covington MF, Maloof JN, Straume M, Kay SA, Harmer SL (2008). Global transcriptome analysis reveals circadian regulation of key pathways in plant growth and development.Genome Biol 9, R130. |
| 11 | Ford B, Deng WW, Clausen J, Oliver S, Boden S, Hemming M, Trevaskis B (2016). Barley (Hordeum vulgare) circadian clock genes can respond rapidly to temperature in an EARLY FLOWERING 3-dependent manner. J Exp Bot 67, 5517-5528. |
| 12 | Fowler SG, Cook D, Thomashow MF (2005). Low temperature induction of Arabidopsis CBF1, 2, and 3 is gated by the circadian clock. Plant Physiol 137, 961-968. |
| 13 | Gao H, Jin MN, Zheng XM, Chen J, Yuan DY, Xin YY, Wang MQ, Huang DY, Zhang Z, Zhou KN, Sheng PK, Ma J, Ma WW, Deng HF, Jiang L, Liu SJ, Wang HY, Wu CY, Yuan LP, Wan JM (2014). Days to heading 7, a major quantitative locus determining photoperiod sensitivity and regional adaptation in rice. Proc Natl Acad Sci USA 111, 16337-16342. |
| 14 | Gendron JM, Pruneda-Paz JL, Doherty CJ, Gross AM, Kang SE, Kay SA (2012). Arabidopsis circadian clock protein, TOC1, is a DNA-binding transcription factor.Proc Natl Acad Sci USA 109, 3167-3172. |
| 15 | Greenham K, McClung CR (2015). Integrating circadian dynamics with physiological processes in plants.Nat Rev Genet 16, 598-610. |
| 16 | Grundy J, Stoker C, Carré IA (2015). Circadian regulation of abiotic stress tolerance in plants.Front Plant Sci 6, 648. |
| 17 | Harmer SL (2009). The circadian system in higher plants.Annu Rev Plant Biol 60, 357-377. |
| 18 | Hayama R, Sarid-Krebs L, Richter R, Fernández V, Jang S, Coupland G (2017). PSEUDO RESPONSE REGULATORs stabilize CONSTANS protein to promote flowering in response to day length.EMBO J 36, 904-918. |
| 19 | Huang W, Peréz-García P, Pokhilko A, Millar AJ, Antoshechkin I, Riechmann JL, Mas P (2012). Mapping the core of the Arabidopsis circadian clock defines the network structure of the oscillator.Science 336, 75-79. |
| 20 | Izawa T, Mihara M, Suzuki Y, Gupta M, Itoh H, Nagano AJ, Motoyama R, Sawada Y, Yano M, Hirai MY, Makino A, Nagamura Y (2011). Os-GIGANTEA confers robust diurnal rhythms on the global transcriptome of rice in the field. Plant Cell 23, 1741-1755. |
| 21 | Kamioka M, Takao S, Suzuki T, Taki K, Higashiyama T, Kinoshita T, Nakamichi N (2016). Direct repression of evening genes by CIRCADIAN CLOCK-ASSOCIATED1 in the Arabidopsis circadian clock. Plant Cell 28, 696-711. |
| 22 | Keily J, MacGregor DR, Smith RW, Millar AJ, Halliday KJ, Penfield S (2013). Model selection reveals control of cold signaling by evening-phased components of the plant circadian clock.Plant J 76, 247-257. |
| 23 | Kim WY, Ali Z, Park HJ, Park SJ, Cha JY, Perez- Hormaeche J, Quintero FJ, Shin G, Kim MR, Qiang Z, Ning L, Park HC, Lee SY, Bressan RA, Pardo JM, Bohnert HJ, Yun DJ (2013). Release of SOS2 kinase from sequestration with GIGANTEA determines salt tole- rance in Arabidopsis.Nat Commun 4, 1352. |
| 24 | Ko DK, Rohozinski D, Song QX, Taylor SH, Juenger TE, Harmon FG, Chen ZJ (2016). Temporal shift of circa- dian-mediated gene expression and carbon fixation contributes to biomass heterosis in maize hybrids.PLoS Genet 12, e1006197. |
| 25 | Kobayashi Y, Kaya H, Goto K, Iwabuchi M, Araki T (1999). A pair of related genes with antagonistic roles in mediating flowering signals.Science 286, 1960-1962. |
| 26 | Kolmos E, Chow BY, Pruneda-Paz JL, Kay SA (2014). HsfB2b-mediated repression of PRR7 directs abiotic stress responses of the circadian clock. Proc Natl Acad Sci USA 111, 16172-16177. |
| 27 | Koo BH, Yoo SC, Park JW, Kwon CT, Lee BD, An G, Zhang ZY, Li JJ, Li ZC, Paek NC (2013). Natural variation in OsPRR37 regulates heading date and contributes to rice cultivation at a wide range of latitudes. Mol Plant 6, 1877-1888. |
| 28 | Korneli C, Danisman S, Staiger D (2014). Differential control of pre-invasive and post-invasive antibacterial defense by the Arabidopsis circadian clock.Plant Cell Physiol 55, 1613-1622. |
| 29 | Kreps JA, Wu YJ, Chang HS, Zhu T, Wang X, Harper JF (2002). Transcriptome changes for Arabidopsis in response to salt, osmotic, and cold stress.Plant Physiol 130, 2129-2141. |
| 30 | Lee YS, Yi J, An G (2016). OsPhyA modulates rice flowering time mainly through OsGI under short days and Ghd7 under long days in the absence of phytochrome B. Plant Mol Biol 91, 413-427. |
| 31 | Li F, Zhang XM, Hu RB, Wu FQ, Ma JH, Meng Y, Fu YF (2013). Identification and molecular characterization of FKF1 and GI homologous genes in soybean. PLoS One 8, e79036. |
| 32 | Li S, Yue WH, Wang M, Qiu WM, Zhou L, Shou HX (2016a). Mutation of OsGIGANTEA leads to enhanced tolerance to polyethylene glycol-generated osmotic stress in rice. Front Plant Sci 7, 465. |
| 33 | Li X, Ma DB, Lu SX, Hu XY, Huang RF, Liang T, Xu TD, Tobin EM, Liu HT (2016b). Blue light- and low temperature-regulated COR27 and COR28 play roles in the Ara- bidopsis circadian clock.Plant Cell 28, 2755-2769. |
| 34 | Liu C, Song GY, Zhou YH, Qu XF, Guo ZB, Liu ZW, Jiang DM, Yang DC (2015). OsPRR37 and Ghd7 are the major genes for general combining ability of DTH, PH and SPP in rice. Sci Rep 5, 12803. |
| 35 | Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi-Shinozaki K, Shinozaki K (1998). Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis.Plant Cell 10, 1391-1406. |
| 36 | Liu T, Carlsson J, Takeuchi T, Newton L, Farré EM (2013). Direct regulation of abiotic responses by the Arabidopsis circadian clock component PRR7.Plant J 76, 101-114. |
| 37 | Lu SJ, Zhao XH, Hu YL, Liu SL, Nan HY, Li XM, Fang C, Cao D, Shi XY, Kong LP, Su T, Zhang FG, Li SC, Wang Z, Yuan XH, Cober ER, Weller JL, Liu BH, Hou XL, Tian ZX, Kong FJ (2017). Natural variation at the soybean J locus improves adaptation to the tropics and enhances yield. Nat Genet 49, 773-779. |
| 38 | Matsubara K, Ogiso-Tanaka E, Hori K, Ebana K, Ando T, Yano M (2012). Natural variation in Hd17, a homolog of Arabidopsis ELF3 that is involved in rice photoperiodic flowering. Plant Cell Physiol 53, 709-716. |
| 39 | Millar AJ, Carre IA, Strayer CA, Chua NH, Kay SA (1995). Circadian clock mutants in Arabidopsis identified by luci- ferase imaging.Science 267, 1161-1163. |
| 40 | Nagel DH, Pruneda-Paz JL, Kay SA (2014). FBH1 affects warm temperature responses in the Arabidopsis circadian clock.Proc Natl Acad Sci USA 111, 14595-14600. |
| 41 | Nakamichi N, Kiba T, Henriques R, Mizuno T, Chua NH, Sakakibara H (2010). PSEUDO-RESPONSE REGULATORS 9, 7, and 5 are transcriptional repressors in the Arabidopsis circadian clock.Plant Cell 22, 594-605. |
| 42 | Nakamichi N, Kiba T, Kamioka M, Suzuki T, Yamashino T, Higashiyama T, Sakakibara H, Mizuno T (2012). Transcriptional repressor PRR5 directly regulates clock-output pathways.Proc Natl Acad Sci USA 109, 17123-17128. |
| 43 | Nakamichi N, Kusano M, Fukushima A, Kita M, Ito S, Yamashino T, Saito K, Sakakibara H, Mizuno T (2009). Transcript profiling of an Arabidopsis PSEUDO RES- PONSE REGULATOR arrhythmic triple mutant reveals a role for the circadian clock in cold stress response. Plant Cell Physiol 50, 447-462. |
| 44 | Nieto C, López-Salmerón V, Davière JM, Prat S (2015). ELF3-PIF4 interaction regulates plant growth independently of the evening complex.Curr Biol 25, 187-193. |
| 45 | Ning YS, Shi XT, Wan RY, Fan JB, Park CH, Zhang CY, Zhang T, Ouyang XH, Li SG, Wang GL (2015). OsELF3- 2, an ortholog of Arabidopsis ELF3, interacts with the E3 ligase APIP6 and negatively regulates immunity against Magnaporthe oryzae in rice. Mol Plant 8, 1679-1682. |
| 46 | Nohales MA, Kay SA (2016). Molecular mechanisms at the core of the plant circadian oscillator.Nat Struct Mol Biol 23, 1061-1069. |
| 47 | Nolte C, Staiger D (2015). RNA around the clock-regulation at the RNA level in biological timing.Front Plant Sci 6, 311. |
| 48 | Nomoto Y, Kubozono S, Yamashino T, Nakamichi N, Mizuno T (2012). Circadian clock- and PIF4-controlled plant growth: a coincidence mechanism directly integrates a hormone signaling network into the photoperiodic control of plant architectures inArabidopsis thaliana. Plant Cell Physiol 53, 1950-1964. |
| 49 | Nozue K, Covington MF, Duek PD, Lorrain S, Fankhauser C, Harmer SL, Maloof JN (2007). Rhythmic growth explained by coincidence between internal and external cues.Nature 448, 358-361. |
| 50 | Nusinow DA, Helfer A, Hamilton EE, King JJ, Imaizumi T, Schultz TF, Farré EM, Kay SA (2011). The ELF4-ELF3- LUX complex links the circadian clock to diurnal control of hypocotyl growth.Nature 475, 398-402. |
| 51 | Romanowski A, Yanovsky MJ (2015). Circadian rhythms and post-transcriptional regulation in higher plants.Front Plant Sci 6, 437. |
| 52 | Sakuraba Y, Han SH, Yang HJ, Piao WL, Paek NC (2016). Mutation of Rice Early Flowering 3.1 (OsELF3.1) delays leaf senescence in rice. Plant Mol Biol 92, 223-234. |
| 53 | Sakuraba Y, Jeong J, Kang MY, Kim J, Paek NC, Choi G (2014). Phytochrome-interacting transcription factors PIF4 and PIF5 induce leaf senescence in Arabidopsis.Nat Commun 5, 4636. |
| 54 | Seo PJ, Mas P (2014). Multiple layers of posttranslational regulation refine circadian clock activity in Arabidopsis.Plant Cell 26, 79-87. |
| 55 | Shin J, Heidrich K, Sanchez-Villarreal A, Parker JE, Davis SJ (2012). TIME FOR COFFEE represses accumulation of the MYC2 transcription factor to provide time-of-day regulation of jasmonate signaling in Arabidopsis.Plant Cell 24, 2470-2482. |
| 56 | Song SS, Huang H, Gao H, Wang JJ, Wu DW, Liu XL, Yang SH, Zhai QZ, Li CY, Qi TC, Xie DX (2014). Interaction between MYC2 and ETHYLENE INSENSITIVE3 mo- dulates antagonism between jasmonate and ethylene signaling in Arabidopsis.Plant Cell 26, 263-279. |
| 57 | Song YH, Shim JS, Kinmonth-Schultz HA, Imaizumi T (2015). Photoperiodic flowering: time measurement mech- anisms in leaves.Annu Rev Plant Biol 66, 441-464. |
| 58 | Soy J, Leivar P, González-Schain N, Martín G, Diaz C, Sentandreu M, Al-Sady B, Quail PH, Monte E (2016). Molecular convergence of clock and photosensory pathways through PIF3-TOC1 interaction and co-occupancy of target promoters.Proc Natl Acad Sci USA 113, 4870-4875. |
| 59 | Tripathi P, Carvallo M, Hamilton EE, Preuss S, Kay SA (2017). Arabidopsis B-BOX32 interacts with CONSTANS- LIKE3 to regulate flowering.Proc Natl Acad Sci USA 114, 172-177. |
| 60 | Wang GY, Zhang C, Battle S, Lu H (2014). The phosphate transporter PHT4;1 is a salicylic acid regulator likely controlled by the circadian clock protein CCA1.Front Plant Sci 5, 701. |
| 61 | Wang L, Fujiwara S, Somers DE (2010). PRR5 regulates phosphorylation, nuclear import and subnuclear localization of TOC1 in the Arabidopsis circadian clock.EMBO J 29, 1903-1915. |
| 62 | Wang L, Kim J, Somers DE (2013). Transcriptional corepressor TOPLESS complexes with pseudoresponse regulator proteins and histone deacetylases to regulate circadian transcription.Proc Natl Acad Sci USA 110, 761-766. |
| 63 | Wang P, Cui X, Zhao CS, Shi LY, Zhang GW, Sun FL, Cao XF, Yuan L, Xie QG, Xu XD (2017). COR27 and COR28 encode nighttime repressors integrating Arabidopsis circadian clock and cold response. J Integr Plant Biol 59, 78-85. |
| 64 | Wang W, Barnaby JY, Tada Y, Li Hr, Tör M, Caldelari D, Lee DU, Fu XD, Dong XN (2011a). Timing of plant immune responses by a central circadian regulator.Nature 470, 110-114. |
| 65 | Wang XT, Wu LJ, Zhang SF, Wu LC, Ku LX, Wei XM, Xie LL, Chen YH (2011b). Robust expression and association of ZmCCA1 with circadian rhythms in maize. Plant Cell Rep 30, 1261-1272. |
| 66 | Wang XX, Wu FM, Xie QG, Wang HM, Wang Y, Yue YL, Gahura O, Ma SS, Liu L, Cao Y, Jiao YL, Puta F, McClung CR, Xu XD, Ma LG (2012). SKIP is a component of the spliceosome linking alternative splicing and the circadian clock in Arabidopsis.Plant Cell 24, 3278-3295. |
| 67 | Wang ZY, Tobin EM (1998). Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell 93, 1207-1217. |
| 68 | Wu JF, Tsai HL, Joanito I, Wu YC, Chang CW, Li YH, Wang Y, Hong JC, Chu JW, Hsu CP, Wu SH (2016). LWD-TCP complex activates the morning gene CCA1 in Arabidopsis. Nat Commun 7, 13181. |
| 69 | Yang Y, Peng Q, Chen GX, Li XH, Wu CY (2013). OsELF3 is involved in circadian clock regulation for promoting flow- ering under long-day conditions in rice.Mol Plant 6, 202-215. |
| 70 | Yue YL, Liu NX, Jiang BJ, Li M, Wang HJ, Jiang Z, Pan HT, Xia QJ, Ma QB, Han TF, Nian H (2017). A single nucleotide deletion in J encoding GmELF3 confers long juvenility and is associated with adaption of tropic soybean. Mol Plant 10, 656-658. |
| 71 | Zhang C, Xie QG, Anderson RG, Ng G, Seitz NC, Peterson T, McClung CR, McDowell JM, Kong DD, Kwak JM, Lu H (2013). Crosstalk between the circadian clock and innate immunity in Arabidopsis.PLoS Pathog 9, e1003370. |
| 72 | Zhang YY, Wang Y, Wei H, Li N, Tian WW, Chong K, Wang L (2018). Circadian evening complex represses jasmo- nate-induced leaf senescence in Arabidopsis.Mol Plant 11, 326-337. |
| 73 | Zhao JM, Huang X, Ouyang XH, Chen WL, Du AP, Zhu L, Wang SG, Deng XW, Li SG (2012). OsELF3-1, an ortholog of Arabidopsis EARLY FLOWERING 3, regulates rice circadian rhythm and photoperiodic flowering. PLoS One 7, e43705. |
| 74 | Zheng XY, Zhou M, Yoo H, Pruneda-Paz JL, Spivey NW, Kay SA, Dong XN (2015). Spatial and temporal regulation of biosynthesis of the plant immune signal salicylic acid.Proc Natl Acad Sci USA 112, 9166-9173. |
| 75 | Zhu JY, Oh E, Wang TN, Wang ZY (2016). TOC1-PIF4 interaction mediates the circadian gating of thermores- ponsive growth in Arabidopsis.Nat Commun 7, 13692. |
| [1] | 苏晨, 牛钰凡, 徐航, 王希岭, 于英俊, 何雨晴, 王雷. 生物钟与光温环境信号互作网络研究进展[J]. 植物学报, 2025, 60(3): 315-341. |
| [2] | 王亚萍, 包文泉, 白玉娥. 单细胞转录组学在植物生长发育及胁迫响应中的应用进展[J]. 植物学报, 2025, 60(1): 101-113. |
| [3] | 李青洋, 刘翠, 何李, 彭姗, 马嘉吟, 胡子祎, 刘宏波. 甘蓝型油菜BnaA02.CPSF6基因的克隆及功能分析(长英文摘要)[J]. 植物学报, 2025, 60(1): 62-73. |
| [4] | 王涛, 冯敬磊, 张翠. 高温胁迫影响玉米生长发育的分子机制研究进展[J]. 植物学报, 2024, 59(6): 963-977. |
| [5] | 杜庆国, 李文学. lncRNA调控玉米生长发育和非生物胁迫研究进展[J]. 植物学报, 2024, 59(6): 950-962. |
| [6] | 闫恒宇, 李朝霞, 李玉斌. 高温对玉米生长的影响及中国耐高温玉米筛选研究进展[J]. 植物学报, 2024, 59(6): 1007-1023. |
| [7] | 路笃贤, 张严妍, 刘艳, 李岩竣, 左新秀, 林金星, 崔亚宁. 非编码RNA在植物生长发育及逆境响应中的研究进展[J]. 植物学报, 2024, 59(5): 709-725. |
| [8] | 谢启光, 徐小冬. 植物生物钟在农业生产中应对全球变暖的应用[J]. 植物学报, 2024, 59(4): 635-650. |
| [9] | 蒋海港, 曾云鸿, 唐华欣, 刘伟, 李杰林, 何国华, 秦海燕, 王丽超, 姚银安. 三种藓类植物固碳耗水节律调节作用[J]. 植物生态学报, 2023, 47(7): 988-997. |
| [10] | 曾鑫海, 陈锐, 师宇, 盖超越, 范凯, 李兆伟. 植物SPL转录因子的生物功能研究进展[J]. 植物学报, 2023, 58(6): 982-997. |
| [11] | 许亚楠, 闫家榕, 孙鑫, 王晓梅, 刘玉凤, 孙周平, 齐明芳, 李天来, 王峰. 红光和远红光在调控植物生长发育及应答非生物胁迫中的作用[J]. 植物学报, 2023, 58(4): 622-637. |
| [12] | 张嘉, 李启东, 李翠, 王庆海, 侯新村, 赵春桥, 李树和, 郭强. 植物MATE转运蛋白研究进展[J]. 植物学报, 2023, 58(3): 461-474. |
| [13] | 王琪, 吴允哲, 刘学英, 孙丽莉, 廖红, 傅向东. 类受体激酶调控水稻生长发育和环境适应研究进展[J]. 植物学报, 2023, 58(2): 199-213. |
| [14] | 和璐璐, 张萱, 章毓文, 王晓霞, 刘亚栋, 刘岩, 范子莹, 何远洋, 席本野, 段劼. 辽东山区不同坡向长白落叶松人工林树冠特征与林木生长关系[J]. 植物生态学报, 2023, 47(11): 1523-1539. |
| [15] | 张宏祥, 闻志彬, 王茜. 新疆野苹果种群遗传结构及其环境适应性[J]. 植物生态学报, 2022, 46(9): 1098-1108. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||